Abstract
Purpose: This study aimed to examine OATP1B1 (SLCO1B1) and OATP1B3 (SLCO1B3) on the pharmacokinetics of valsartan. Twenty-five subjects were genotyped for 16 single-nucleotide polymorphisms of the SLCO1B1 and SLCO1B3 genes. Methods: After a single dose of 160 mg of valsartan was orally administered to healthy male volunteers, drug concentrations were assayed up to 48 h. The 25 subjects were genotyped for 16 single-nucleotide polymorphisms (SNPs) of the SLCO1B1 and SLCO1B3 genes. Subjects were classified into groups according to their SLCO1B1*1B haplotype; 23 subjects were carriers of SLCO1B1*1B and two subjects were included in the reference group with SLCO1B1*1A/*1A. Alternations of the splicing factor-binding site pattern caused by the given mutation were evaluated with the Human Splicing Finder (HSF) 3.1. Results: The subjects who carried SLCO1B1*1B showed a 2.3-fold higher clearance than those without the *1B haplotype. Mean Cmax and AUCinf were reduced by 45% and 54%, respectively, in the SLCO1B1*1B genotype group compared to the reference group with the *1A/*1A genotype (p < 0.01). The carriers of the rs4149153 T allele of SLCO1B3 had a 27% lower mean Cmax and a 1.5-fold higher Vd compared to homozygotic CC carriers (p < 0.05). In a combined analysis of SLCO1B1 and SLCO1B3, subjects not carrying SLCO1B1 *1B and carrying SLCO1B3 rs4149153 T allele showed a 1.6-fold higher clearance than those with the other genotypes, whereas mean Cmax and AUClast were reduced by 35% and 42%, respectively (p < 0.05), in the subjects. HSF 3.1 analysis showed that rs4149153 could cause alterations of the acceptor splice site (TAAATACTAAAGAC to TAAATATTAAAGAC) with scoring change (from 72.57 to 71.92, difference = −0.9). Conclusion: It was found that plasma exposure to valsartan is significantly decreased in SLCO1B1*1B carriers and carriers of the rs4149153 T allele of SLCO1B3, possibly as a result of increased hepatic uptake.
1. Introduction
Valsartan is an orally active angiotensin receptor blocker often prescribed to treat hypertension. It selectively inhibits the binding of angiotensin II to the angiotensin type 1 receptor in many tissues, including vascular smooth muscle and the adrenal gland, and blocks vasoconstriction and aldosterone-secreting effects of angiotensin II. Valsartan undergoes a minor degree of metabolism involving 4-hydroxylation, whereas 85% of orally administered valsartan is excreted in an unchanged form [1]. At physiologic pH, valsartan exists as a di-anion and is selectively distributed to the liver. Because of this negative charge, the hepatic uptake of valsartan through anion transporters is likely to be a major pathway for drug disposition. It has been reported that approximately 70% of the total clearance of valsartan is accounted for by hepatic clearance [2].
Organic anion transporting polypeptide (OATP) family mediates hepatic uptake of endogenous compounds, drugs, xenobiotics, and anionic peptides [3,4]. Among the OATP family transporters, organic anion-transporting polypeptide 1B1 (OATP1B1) and organic anion-transporting polypeptide 1B3 (OATP1B3) are exclusively expressed in human hepatocytes [5]; it suggests that two transporters play a crucial role in the hepatic uptake and clearance of organic compounds [6]. As valsartan has an anionic carboxyl moiety, it is efficiently taken up into hepatocytes by OATP family transporters [7]. Yamashiro et al. reported that valsartan was transported by OATP1B1- and OATP1B3-expressing HEK293 cells [8], and Hanna et al. showed that OATP1B1 contributed 27% of hepatic uptake of valsartan, whereas OATP1B3 contributed 73% [9]. In in vitro kinetic study with transporter-transfected cells, OATP1B1 and OATP1B3 accounted for 55% and 42% of intrinsic hepatic clearance of valsartan, respectively [10]. Taken together, both OATP1B1 and OATP1B3 are major hepatic uptake transporters for valsartan.
OATP1B1, which is encoded by SLCO1B1, is a glycoprotein consisting of 691 amino acids [11]. A large number of sequence variants have been found in the SLCO1B1 gene. Two common SNPs of SLCO1B1, A388G (rs2306283) and T521C (rs4149056), which have been associated with altered PK parameters of OATP1B1 substrates, form 4 haplotypes: SLCO1B1*1A (388A–521T), *1B (388G–521T), *5 (388A–521C), and *15 (388G–521C) [12,13,14]. OATP1B3, which is encoded by SLCO1B3, shares 80% of the OATP1B1 amino acid sequence and shows overlapping substrate specificities with OATP1B1 [15]. Considering the genetic and functional similarities between OATP1B1 and OATP1B3, both may affect PK [16,17]. However, there is no study on the effects of OATP1B1 and OATP1B3 polymorphisms on valsartan PK. Therefore, in this study, we aimed to evaluate the contribution of the SLCO1B1 and SLCO1B3 polymorphisms to valsartan PK parameters in healthy Korean individuals.
2. Methods
2.1. Study Population
The study population was enrolled from among 50 healthy male volunteers who had participated in a bioequivalence study of a composite product comprised of 160 mg valsartan and 10 mg amlodipine [18]. Among these volunteers, 25 healthy men participated in this study after providing additional written consent for genotyping. Eligible subjects were men between the ages of 20 and 50 years who were within 20% of their ideal body weight, with no congenital abnormalities or chronic disease. Exclusion criteria were: (1) use of prescription drugs or herbal medications within 2 weeks or use of nonprescription drugs within the week before the study that potentially interact with valsartan, and (2) use of drugs that induce or inhibit drug-metabolizing enzymes within the month before the study that potentially interact with the study medications. Vital signs monitoring, physical examination, and routine laboratory tests were performed before the start of the study. The study protocol was approved by the Ethics Committee of the Institutional Review Board (IRB No. 2012-4-0283). Informed consent was obtained from all study subjects before their participation in the study.
2.2. Clinical Study
The PK data of the study population were obtained from the previous single-dose study of valsartan [18]. Subjects took a 160 mg tablet of valsartan orally with 240 mL of water at 8 a.m. after an overnight fast for 10 h. Venous blood samples were collected into ethylenediaminetetraacetic acid-containing tubes by an indwelling catheter inserted into the forearm at 0 (pre-dose) and 0.5, 1, 1.5, 2, 3, 4, 6, 8, 10, 12, 16, 24, and 48 h after dosing. Blood samples for genotyping were also collected, and genotyping was performed after the end of the study.
2.3. Analysis of Valsartan Concentrations and Genotyping
Plasma valsartan concentrations were analyzed with a validated UPLC-MS/MS method as reported in a previous study [18]. Genomic DNA was prepared from blood samples using the QIAamp DNA Blood Mini Kit (QIAGEN GmbH, Hilden, Germany), according to the manufacturer’s standard recommended procedures.
SNPs of SLCO1B1 and SLCO1B3 were selected based on other studies and genetic information from the UCSC Genome Browser. Linkage disequilibrium data and minor allele frequency (MAF) data in Japanese and Han Chinese populations from Haploreg ver. 2 and the tagger function within the Haploview v4.2 program were incorporated to assort SLCO1B1 and SLCO1B3 gene SNPs. Tag SNPs of the SLCO1B1 and SLCO1B3 genes were assigned with a condition of MAF ≥ 30% and an r2 threshold of 0.8 in Japanese and Han Chinese populations. In addition, based on the functional effects and previous reports [13,19,20], 11 SNPs (rs12317268, rs4149071, rs4149042, rs2417954, rs4149022, rs4149081, rs2306283, rs4149085, rs4149032, rs4149031, and rs4149056) were selected for SLCO1B1 and 5 SNPs (rs4149118, rs7311358, rs10841661, rs4149153, and rs11045585) were selected for SLCO1B3. Genotyping of SLCO1B1 and SLCO1B3 polymorphisms was conducted with a single-base primer extension assay using ABI PRISM SNaPShot Multiplex kits (ABI, Foster City, CA, USA) according to the manufacturer’s recommendations.
2.4. Pharmacokinetic Analysis
PK parameters were calculated using actual sampling times. Maximum blood concentration (Cmax) and time to maximum concentration (Tmax) were obtained from the observed data. The area under the plasma concentration-time curve from time zero to the time of the last concentration (AUClast) was calculated using the linear trapezoidal rule. The AUC from time zero to infinity (AUCinf) was the sum of AUClast and Clast/ke, where Clast is the last quantifiable concentration and ke is the terminal elimination rate constant; the half-life was 0.693/ke. Plasma concentrations during the terminal phase were fitted to a log linear line by the least squares method to obtain the ke. PK parameters were analyzed by a non-compartmental method using WinNonlin5.3 (Pharsight Corporation, Mountain View, CA, USA).
2.5. In Silico Analysis
To predict the possible effects of given variants on splicing, different computational tools were used. Netgene2 and Splice Site Prediction by Neural Network (NNSPLICE) were used for splice site predictions [21,22]. Alternations of the splicing factor-binding site pattern caused by the given mutation were evaluated with the Human Splicing Finder (HSF) 3.1 [23]. We used the default threshold values, and a score for a given sequence was considered to be potentially significant if it was above the threshold value.
2.6. Statistical Analysis
All PK data were expressed as mean ± SD. The median difference with 95% confidence interval was calculated with the Hodges-Lehmann approach and the method of Moses [24]. Differences in PK parameters among the genotype groups were evaluated using the Kruskal-Wallis test for three-group comparison and the Mann Whitney rank sum test for two-group comparison following normality and equal variance tests; p values < 0.05 were considered statistically significant. Statistical analyses were performed using SPSS 20.0 (International Business Machines Corp., New York City, NY, USA).
3. Results
All subjects who completed blood sampling as scheduled were enrolled, and only those subjects who gave their consent for the genotyping study were included. Mean age, weight, and height of the 25 subjects were 26.8 ± 5.9 years, 67.9 ± 8.22 kg, and 174.2 ± 4.79 cm, respectively. There were no statistically significant differences in PK parameters according to clinical characteristics (Table 1). The mean PK parameter values were: AUCinf 33.23 ± 14.53 h·μg/mL, CL/F 5.68 ± 2.39 L/h, Cmax 4.39 ± 1.45 μg/mL, t1/2 6.86 ± 3.02 h, ke 0.11 ± 0.04 h−1, Tmax 3.20 ± 1.25 h, and Vd/F 51.29 ± 18.40 L. According to the SLCO1B1 haplotype, the numbers of carriers of SLCO1B1*1A/*1A, *1A/*1B, *1B/*15 and *1B/*1B were 2, 9, 6, and 8, respectively. None of the study subjects had the SLCO1B1*5 haplotype.

Table 1.
Effects of baseline characteristics on valsartan pharmacokinetics.
As shown in Table 2, subjects who carried SLCO1B1*1B showed more than 2.3-fold higher clearance (CL/F) compared to those with *1A/*1A (p < 0.01), resulting in a significantly lower AUC, including AUClast (p < 0.05) and AUCinf (p < 0.01). Vd/F increased by 87%, whereas Cmax decreased by 45% in subjects with the SLCO1B1*1B haplotype, compared to those with *1A/*1A (p < 0.01). There were no significant differences in the Tmax, t1/2, and ke.

Table 2.
Effects of SLCO1B1 polymorphisms on valsartan pharmacokinetics.
In the case of SLCO1B3, Vd/F increased by 50% (p < 0.05), whereas Cmax decreased by 27% (p < 0.05) in subjects who were T allele carriers of rs4149153 compared to those with CC genotype. When subjects were classified according to SLCO1B3 haplotype on the basis of nucleotide base differences in IVS4+76/699/IVS12-5676 (rs4149118/rs7311358/ rs11045585), there was no statistically significant difference between those haplotypes (Table 3).

Table 3.
Effects of SLCO1B3 polymorphisms on valsartan pharmacokinetics.
To predict a splicing effect, analysis by HSF 3.1 showed that SLCO1B3 rs4149153, which had significant associations with Cmax and Vd/F of valsartan, could cause alterations of the acceptor splice site (TAAATACTAAAGAC to TAAATATTAAAGAC) with scoring change (from 72.57 to 71.92, difference = −0.9). These results are shown in Table 4. The results generated by Netgene2 and NNSPLICE did not show the presence of altered splicing donor or acceptor by rs4149153. By HSF 3.1, rs4149153 was predicted to alter potential branch points, enhancer motifs, and silencer motifs. This mutation decreased the branch points calculation score from 73.16 to 49.32, breaking a potential branch point (threshold: 67). Two types of enhancer motifs (Predicted Exonic Splicing Enhancers octamers and Exon-Identity Elements) were disrupted by rs4149153 (CTAAAGAC and AATAC, respectively). This mutation was also predicted to break one Sironi’s Motif 1 (CTAAAGAC) and three Predicted Exonic Splicing Silencers octamers (TTAAATAT, TAAATATT, and ATATTAAA) [25]. The mean plasma concentration-time profiles of valsartan after administration to subjects with SLCO1B1*1B haplotypes and rs4149153 of SLCO1B3 are shown in Figure 1. The profiles for rs2306283 and rs4149056, which consist of SLCO1B1*1B haplotype, are shown in Figure S1.

Table 4.
Potential splicing regulatory sequences in wild-type and mutant type of rs4149153 by Human Splicing Finder (HSF) 3.1.
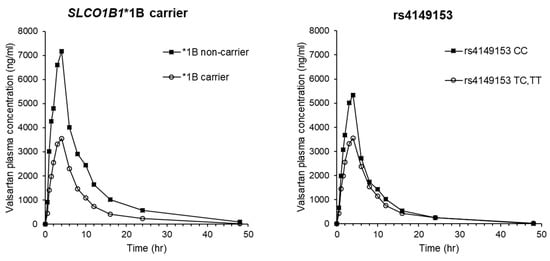
Figure 1.
Mean plasma concentration-time profiles of valsartan after a single oral dose of valsartan in 25 healthy Korean male participants with SLCO1B1*1B haplotypes and rs4149153 in SLCO1B3.
4. Discussion
The main findings of this study are that SLCO1B1*1B gene polymorphism and rs4149153 of the SLCO1B3 gene can influence valsartan PK. Subjects who carried SLCO1B1*1B showed higher CL/F and lower AUC values (AUClast and AUCinf) compared to those without the *1B haplotype. The carriers of the rs4149153 T allele in SLCO1B3 had a 27% lower mean Cmax and 1.5-fold higher Vd/F compared to homozygotic CC carriers.
A388G (rs2306283, Asn130Asp) and T521C (rs4149056, Val174Ala) are the most frequently studied SNPs related to drug disposition. These polymorphisms have been mainly studied with respect to statins, which are the substrates of OATP1B1. The functional effect of the SLCO1B1*1B haplotype, which is an A388G allele, has been variously reported in different in vitro studies showing increased activity, decreased activity, or no change in the activity [26]. This may be because of substrate-specific effects or differences in the experimental conditions. The inconsistent results of the *1B haplotype also have been reported in clinical studies. SLCO1B1*1B alleles showed accelerated transporter activity and were associated with decreased plasma concentrations of pravastatin. A study showed that the AUC of pravastatin was 35% lower in participants with the *1B/*1B genotype than in participants with the *1A/*1A genotype [19]. In contrast, the SLCO1B1*1B haplotype was associated with increased plasma concentrations of pitavastatin and unchanged AUC of rosuvastatin in Asian populations [27,28,29].
It has been reported that the 521C allele is associated with decreased OATP1B1 activity, and the *5 and *15 haplotypes that contain this allele are low-activity haplotypes [30]. The 521C allele has been reported to be associated with decreased hepatic uptake and increased plasma concentrations of statins including atorvastatin, pitavastatin, pravastatin, rosuvastatin, and simvastatin [14,27,31,32,33]. Through various in vitro and in vivo studies of the SLCO1B1 haplotype, the effects of the SLCO1B1 *5 and *15 haplotypes are well documented. Since SLCO1B1*5 is very rarely found in the East Asian population [34], clinical studies in Asian populations have focused on evaluating the increased exposure to substrate and increased risk of toxicity in SLCO1B1*15 carriers [20,34,35]. However, our study did not show significant differences in any valsartan PK parameters between subjects with and without *15.
It has also been reported that the relative contributions of OATP1B1 and OATP1B3 to the hepatic uptake of valsartan are similar [7]. In our study, subjects who carried the T allele of rs4149153 showed lower AUC values (AUClast and AUCinf) and higher CL/F and Vd/F as compared to subjects with the CC genotype (p < 0.05). The SNP of rs4149153 is located in an intron region, which is not thought to be involved in protein production. However, intronic regions have the potential to affect mRNA splicing and alter protein expression or action [36,37]. In our in silico analysis, although no significant splicing motif alteration was detected, a few site broken variations and new sites were detected. Therefore, rs4149513 may be a candidate SNP affecting valsartan PKs.
To the best of our knowledge, this is the first evaluation of the association between SLCO1B1 and SLCO1B3 gene polymorphisms and valsartan PKs. However, this study had some limitations. The sample size was too small to yield statistically significant results. Our study population consisting of only male subjects precluded analysis of gender differences. Since this study is not a hypothesis-generating study but is intended to find candidate genes affecting valsartan PKs, multiple test correction was not performed. Therefore, it should be implemented with a caution with the risk of false-positive results and needs to be verified by further replication studies.
In conclusion, we investigated that plasma exposure to valsartan is significantly decreased in SLCO1B1*1B carriers and carriers of the rs4149153 T allele of SLCO1B3, possibly as a result of increased hepatic uptake. As these associations have not been proven in cell lines and preclinical models, further in vitro and in vivo studies are needed to strengthen the findings of our clinical researches.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jpm11090862/s1, Figure S1: Mean plasma concentration-time profiles of valsartan after a single oral dose of valsartan in 25 healthy Korean male participants with rs2306283 and rs4149056 in SLCO1B1.
Author Contributions
G.S., J.-E.C., K.P. and H.-S.G. participated in research design. G.S., J.-E.C. and J.Y. conducted experiments. G.S., J.-E.C., K.-E.L., K.P. and H.-S.G. performed data analysis. G.S. and J.-E.C. contributed to the writing of the manuscript, and K.P. and H.-S.G. finalized it. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the Institutional Review Board (IRB No. 2012-4-0283).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Waldmeier, F.; Flesch, G.; Ller, P.M.; Winkler, T.; Kriemler, H.-P.; Buhlmayer, P.; De Gasparo, M. Pharmacokinetics, disposition and biotransformation of [14C]-radiolabelled valsartan in healthy male volunteers after a single oral dose. Xenobiotica 1997, 27, 59–71. [Google Scholar] [CrossRef]
- Flesch, G.; Müller, P.; Lloyd, P. Absolute bioavailability and pharmacokinetics of valsartan, an angiotensin II receptor antagonist, in man. Eur. J. Clin. Pharmacol. 1997, 52, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Kullak-Ublick, G.A.; Hagenbuch, B.; Stieger, B.; Wolkoff, A.W.; Meier, P.J. Functional characterization of the basolateral rat liver organic anion transporting polypeptide. Hepatology 1994, 20, 411–416. [Google Scholar] [PubMed]
- Hagenbuch, B.; Meier, P. The superfamily of organic anion transporting polypeptides. Biochim. Biophys. Acta (BBA) Biomembr. 2003, 1609, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Hagenbuch, B.; Meier, P.J. Organic anion transporting polypeptides of the OATP/SLC21 family: Phylogenetic classification as OATP/SLCO superfamily, new nomenclature and molecular/functional properties. Pflügers Archiv 2004, 447, 653–665. [Google Scholar] [CrossRef] [Green Version]
- Mahagita, C.; Grassl, S.M.; Piyachaturawat, P.; Ballatori, N. Human organic anion transporter 1B1 and 1B3 function as bidirectional carriers and do not mediate GSH-bile acid cotransport. Am. J. Physiol. Liver Physiol. 2007, 293, G271–G278. [Google Scholar] [CrossRef] [Green Version]
- Maeda, K. Organic Anion Transporting Polypeptide (OATP)1B1 and OATP1B3 as Important Regulators of the Pharmacokinetics of Substrate Drugs. Biol. Pharm. Bull. 2015, 38, 155–168. [Google Scholar] [CrossRef] [Green Version]
- Yamashiro, W.; Maeda, K.; Hirouchi, M.; Adachi, Y.; Hu, Z.; Sugiyama, Y. Involvement of transporters in the hepatic uptake and biliary excretion of valsartan, a selective antagonist of the angiotensin II AT1-receptor, in humans. Drug Metab Dispos. 2006, 34, 1247–1254. [Google Scholar] [CrossRef] [Green Version]
- Hanna, I.; Alexander, N.; Crouthamel, M.H.; Davis, J.; Natrillo, A.; Tran, P.; Vapurcuyan, A.; Zhu, B. Transport properties of valsartan, sacubitril and its active metabolite (LBQ657) as determinants of disposition. Xenobiotica 2018, 48, 300–313. [Google Scholar] [CrossRef]
- Poirier, A.; Cascais, A.-C.; Funk, C.; Lavé, T. Prediction of pharmacokinetic profile of valsartan in human based on in vitro uptake transport data. J. Pharmacokinet. Pharmacodyn. 2009, 36, 585–611. [Google Scholar] [CrossRef]
- Nakashima, A.; Kawashita, H.; Masuda, N.; Saxer, C.; Niina, M.; Nagae, Y.; Iwasaki, K. Identification of cytochrome P450 forms involved in the 4-hydroxylation of valsartan, a potent and specific angiotensin II receptor antagonist, in human liver microsomes. Xenobiotica 2005, 35, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Kameyama, Y.; Yamashita, K.; Kobayashi, K.; Hosokawa, M.; Chiba, K. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharm. Genom. 2005, 15, 513–522. [Google Scholar] [CrossRef]
- Xiang, X.; Jada, S.R.; Li, H.H.; Fan, L.; Tham, L.S.; Wong, C.I.; Lee, S.C.; Lim, R.; Zhou, Q.Y.; Goh, B.C.; et al. Pharmacogenetics of SLCO1B1 gene and the impact of *1b and *15 haplotypes on irinotecan disposition in Asian cancer patients. Pharm. Genom. 2006, 16, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.W.; Song, I.-S.; Shin, H.; Yeo, C.-W.; Cho, D.-Y.; Shon, J.-H.; Shin, J.-G. The effect of SLCO1B1*15 on the disposition of pravastatin and pitavastatin is substrate dependent: The contribution of transporting activity changes by SLCO1B1*15. Pharm. Genom. 2008, 18, 424–433. [Google Scholar] [CrossRef]
- König, J.; Cui, Y.; Nies, A.T.; Keppler, D. Localization and Genomic Organization of a New Hepatocellular Organic Anion Transporting Polypeptide. J. Biol. Chem. 2000, 275, 23161–23168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boivin, A.-A.; Cardinal, H.; Barama, A.; Pichette, V.; Hébert, M.-J.; Roger, M. Organic Anion Transporting Polypeptide 1B1 (OATP1B1) and OATP1B3: Genetic Variability and Haplotype Analysis in White Canadians. Drug Metab. Pharmacokinet. 2010, 25, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.-J.; Lee, K.-R.; Noh, C.-K.; Chong, S.; Kim, D.-D.; Shim, C.-K.; Chung, S.-J. Functional Consequences of Genetic Variations in the Human Organic Anion Transporting Polypeptide 1B3 (OATP1B3) in the Korean Population. J. Pharm. Sci. 2012, 101, 1302–1313. [Google Scholar] [CrossRef]
- Kim, Y.; Son, M.; Lee, D.; Roh, H.; Son, H.; Chae, D.; Bahng, M.Y.; Park, K. Pharmacokinetic Comparison of 2 Fixed-Dose Combination Tablets of Amlodipine and Valsartan in Healthy Male Korean Volunteers: A Randomized, Open-Label, 2-Period, Single-Dose, Crossover Study. Clin. Ther. 2013, 35, 934–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, K.; Ieiri, I.; Yasuda, K.; Fujino, A.; Fujiwara, H.; Otsubo, K.; Hirano, M.; Watanabe, T.; Kitamura, Y.; Kusuhara, H. Effects of organic anion transporting polypeptide 1B1 haplotype on pharmacokinetics of pravastatin, valsartan, and temocapril. Clin. Pharmacol. Ther. 2006, 79, 427–439. [Google Scholar] [CrossRef]
- Jiang, J.; Tang, Q.; Feng, J.; Dai, R.; Wang, Y.; Yang, Y.; Tang, X.; Deng, C.; Zeng, H.; Zhao, Y.; et al. Association between SLCO1B1 −521T>C and −388A>G polymorphisms and risk of statin-induced adverse drug reactions: A meta-analysis. SpringerPlus 2016, 5, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Brunak, S.; Engelbrecht, J.; Knudsen, S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991, 220, 49–65. [Google Scholar] [CrossRef]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved Splice Site Detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef]
- Desmet, F.-O.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Béroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef] [Green Version]
- Hollander, M.; Wolfe, D.A. Nonparametric Statistical Methods; John Wiley & Sons: New York, NY, USA, 1973. [Google Scholar]
- Zhang, X.H.-F.; Chasin, L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004, 18, 1241–1250. [Google Scholar] [CrossRef] [Green Version]
- Niemi, M.; Pasanen, M.K.; Neuvonen, P. Organic Anion Transporting Polypeptide 1B1: A Genetically Polymorphic Transporter of Major Importance for Hepatic Drug Uptake. Pharmacol. Rev. 2011, 63, 157–181. [Google Scholar] [CrossRef]
- Choi, J.; Lee, M.; Cho, J.-Y.; Lee, J.-E.; Kim, K.; Park, K. Influence of OATP1B1 Genotype on the Pharmacokinetics of Rosuvastatin in Koreans. Clin. Pharmacol. Ther. 2008, 83, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Xiong, Y. OATP1B1 388A>G polymorphism and pharmacokinetics of pitavastatin in Chinese healthy volunteers. J. Clin. Pharm. Ther. 2010, 35, 99–104. [Google Scholar] [CrossRef]
- Hu, M.; Mak, V.W.L.; Yin, O.Q.P.; Chu, T.T.W.; Tomlinson, B. Effects of Grapefruit Juice and SLCO1B1 388A>G Polymorphism on the Pharmacokinetics of Pitavastatin. Drug Metab. Pharmacokinet. 2013, 28, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, I.Y.; Kim, R.B. Impact of Genetic Variation in OATP Transporters to Drug Disposition and Response. Drug Metab. Pharmacokinet. 2013, 28, 4–18. [Google Scholar] [CrossRef] [Green Version]
- The SEARCH Collaborative Group; Link, E.M.; Parish, S.; Armitage, J.M.; Bowman, L.H.; Heath, S.; Matsuda, F.; Gut, I.; Lathrop, M.; A Collins, R. SLCO1B1 Variants and Statin-Induced Myopathy—A Genomewide Study. N. Engl. J. Med. 2008, 359, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lee, M.; Lim, L.; Jang, S.; Chung, J. Effects of SLCO1B1 and ABCB1 genotypes on the pharmacokinetics of atorvastatin and 2-hydroxyatorvastatin in healthy Korean subjects. Int. J. Clin. Pharmacol. Ther. 2010, 48, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wu, X.-J.; Zhao, G.-L.; Zhang, T.; Xu, S.-S.; Sun, Y.-X.; Qiu, F.; Zhao, L.-M. Effects of Polymorphisms in NR1H4, NR1I2, SLCO1B1, and ABCG2 on the Pharmacokinetics of Rosuvastatin in Healthy Chinese Volunteers. J. Cardiovasc. Pharmacol. 2016, 68, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-Y.; Cho, D.-Y.; Shin, H.; Lee, S.-S.; Shon, J.-H.; Shin, J.-G.; Shin, S.-G. Duplex pyrosequencing assay of the 388A>G and 521T>C SLCO1B1 polymorphisms in three Asian populations. Clin. Chim. Acta 2008, 388, 68–72. [Google Scholar] [CrossRef]
- Oh, E.S.; Kim, C.O.; Cho, S.K.; Park, M.S.; Chung, J.-Y. Impact of ABCC2, ABCG2 and SLCO1B1 Polymorphisms on the Pharmacokinetics of Pitavastatin in Humans. Drug Metab. Pharmacokinet. 2013, 28, 196–202. [Google Scholar] [CrossRef] [Green Version]
- Kiyotani, K.; Mushiroda, T.; Kubo, M.; Zembutsu, H.; Sugiyama, Y.; Nakamura, Y. Association of genetic polymorphisms in SLCO1B3 and ABCC2 with docetaxel-induced leukopenia. Cancer Sci. 2008, 99, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Chew, S.C.; Sandanaraj, E.; Singh, O.; Chen, X.; Tan, E.H.; Lim, D.W.-T.; Lee, E.J.D.; Chowbay, B. Influence of SLCO1B3 haplotype-tag SNPs on docetaxel disposition in Chinese nasopharyngeal cancer patients. Br. J. Clin. Pharmacol. 2012, 73, 606–618. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).