Machine Learning Approaches to Predict In-Hospital Mortality among Neonates with Clinically Suspected Sepsis in the Neonatal Intensive Care Unit
Abstract
:1. Introduction
2. Methods
2.1. Patients, Setting, and Study Design
2.2. Study Variables and Data Pre-Processing
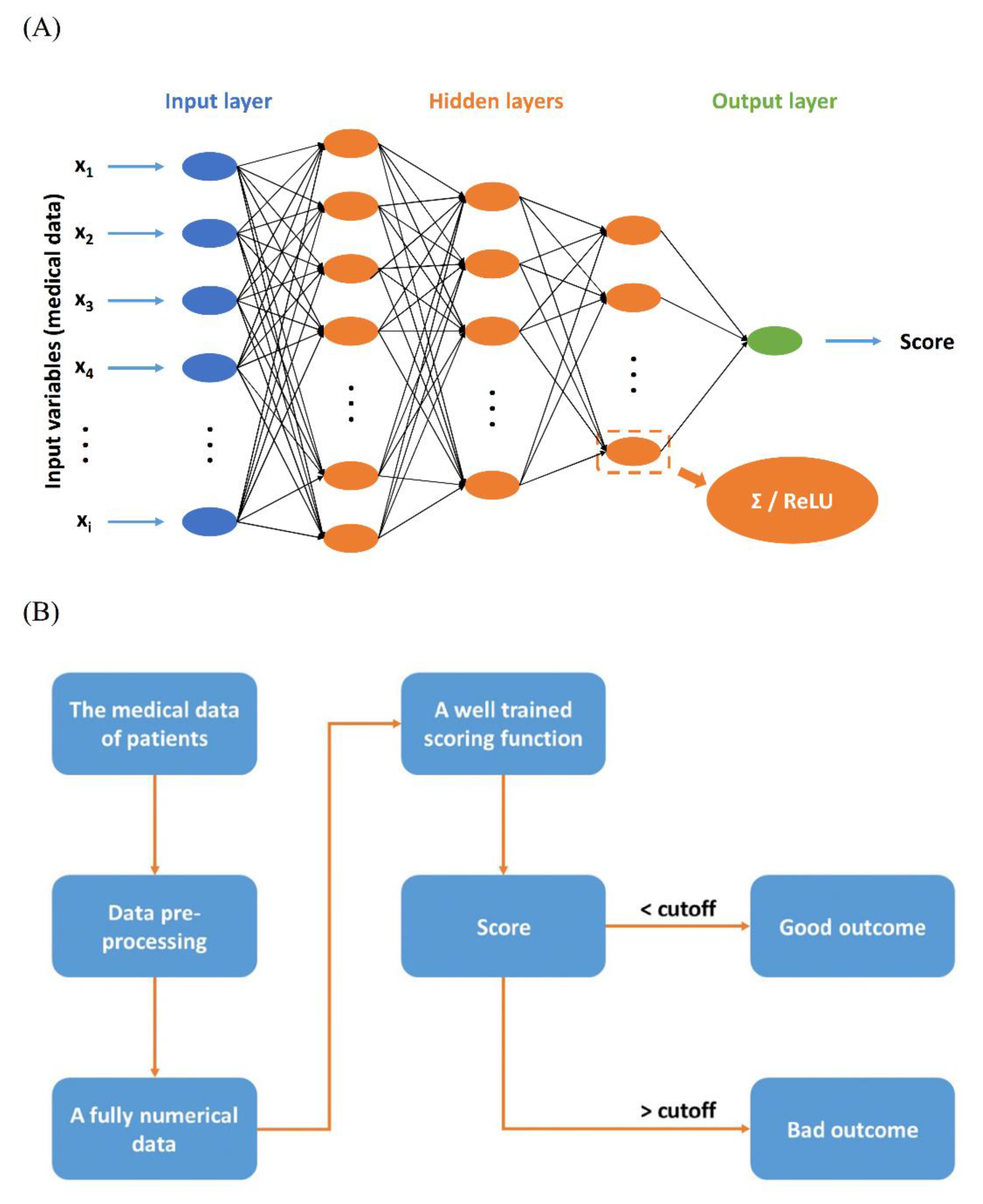
2.3. Scoring Function
2.4. Feature Selection of Scoring Function
2.5. Assay Procedure
2.6. Statistical Analysis
Machine Learning Procedures
3. Results
3.1. Feature Selection of DNN Model
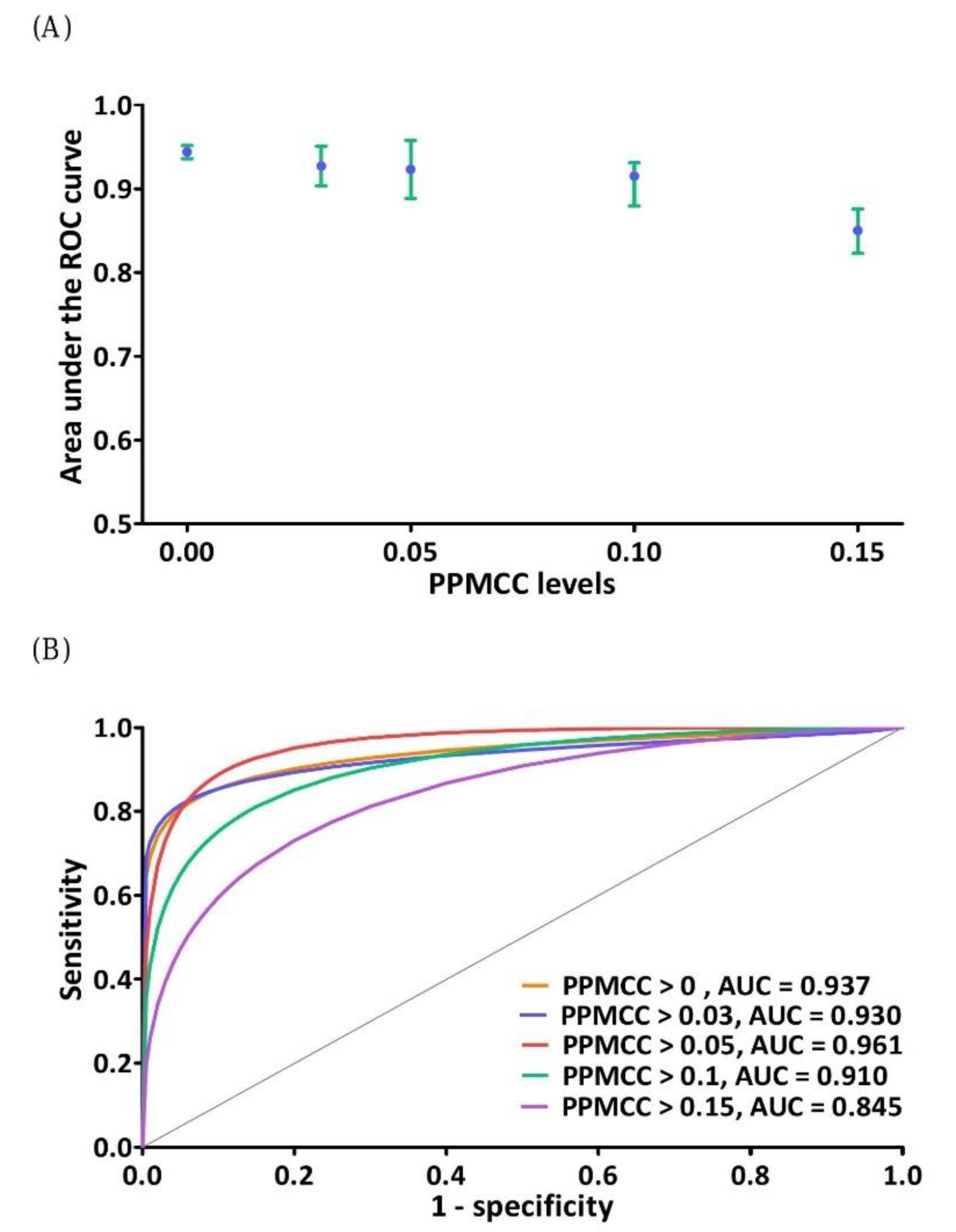
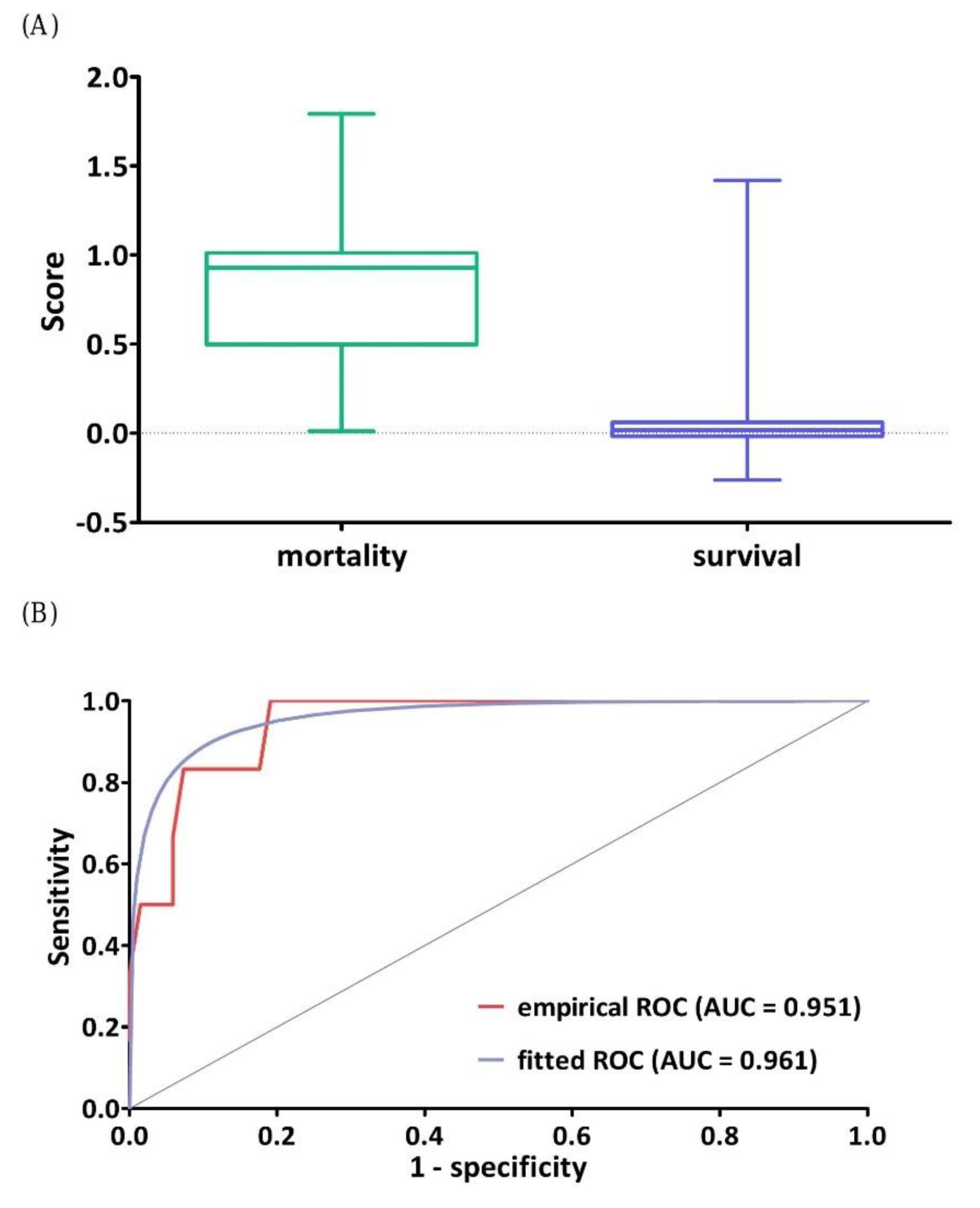
3.2. Effectiveness Evaluation of the DNN Model
3.3. Rank of Predictors in the Prediction Scoring Function
3.4. Classification Performance of DNN Model
3.5. Comparisons of the Performance of Various Machine Learning Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Horbar, J.D.; Edwards, E.M.; Greenberg, L.T.; Morrow, K.A.; Soll, R.F.; Buus-Frank, M.E.; Buzas, J.S. Variation in performance of neonatal intensive care units in the United States. JAMA Pediatr. 2017, 171, e164396. [Google Scholar] [CrossRef]
- Abdel-Latif, M.E.; Nowak, G.; Bajuk, B.; Glass, K.; Harley, D. Variation in hospital mortality in an Australian neonatal intensive care unit network. Arch. Dis. Child. Fetal. Neonatal Ed. 2017, 103, F331–F336. [Google Scholar] [CrossRef]
- Tsai, M.H.; Hsu, J.F.; Chu, S.M.; Lien, R.; Huang, H.R.; Chiang, M.C.; Fu, R.-H.; Lee, C.-W.; Huang, Y.-C. Incidence, clinical characteristics, and risk factors of adverse outcome in neonates with late-onset sepsis. Pediatr. Infect. Dis. J. 2014, 33, e7–e13. [Google Scholar] [CrossRef]
- Tsai, M.H.; Chu, S.M.; Lee, C.W.; Hsu, J.F.; Huang, H.R.; Chiang, M.C.; Fu, R.-H.; Lien, R.; Huang, Y.-C. Recurrent late-onset sepsis in the neonatal intensive care unit: Incidence, clinical characteristics and risk factors. Clin. Microbiol. Infect. 2014, 20, O928–O935. [Google Scholar] [CrossRef] [Green Version]
- Muhe, L.M.; McClure, E.M.; Nigussie, A.K.; Mekasha, A.; Worku, B.; Worku, A.; Demtse, A.; Eshetu, B.; Tigabu, Z.; Gizaw, M.A.; et al. Major causes of death in preterm infants in selected hospitals in Ethiopia (SIP): A prospective, cross-sectional, observational study. Lancet Glob. Health 2019, 7, e1130–e1138. [Google Scholar] [CrossRef] [Green Version]
- Shane, A.L.; Sánchez, P.J.; Stoll, B.J. Neonatal sepsis. Lancet 2017, 390, 1770–1780. [Google Scholar] [CrossRef]
- Hentschel, R.; Guenther, K.; Vach, W.; Bruder, I. Risk-adjusted mortality of VLBW infants in high-volume versus low-volume NICUs. Arch. Dis. Child. Fetal Neonatal Ed. 2018, 104, F390–F395. [Google Scholar] [CrossRef]
- Yu, R.; Zhou, Q.; Jiang, S.; Mei, Y.; Wang, M. Combination of 16S rRNA and procalcitonin in diagnosis of neonatal clinically suspected sepsis. J. Int. Med. Res. 2020, 48, 300060519892418. [Google Scholar] [CrossRef] [PubMed]
- El Gawhary, S.; El-Anany, M.; Hassan, R.; Ali, D.; El Gameel, E.Q. The role of 16S rRNA gene sequencing in confirmation of suspected neonatal sepsis. J. Trop. Pediatr. 2016, 62, 75–80. [Google Scholar] [CrossRef] [Green Version]
- Cuttano, A.; Scaramuzzo, R.T.; Gentile, M.; Ciantelli, M.; Sigali, E.; Boldrini, A. Education in neonatology by simulation: Between reality and declaration of intent. J. Matern. Fetal Neonatal Med. 2011, 24 (Suppl. 1), 2497–2498. [Google Scholar] [CrossRef]
- Ostfeld, B.M.; Schwartz-Soicher, O.; Reichman, N.E.; Teitler, J.O.; Hegyi, T. Prematurity and sudden unexpected infant deaths in the United States. Pediatrics 2017, 140, e20163334. [Google Scholar] [CrossRef] [Green Version]
- Houweling, T.A.J.; van Klaveren, D.; Das, S.; Azad, K.; Tripathy, P.; Manandhar, D.; Neuman, M.; De Jonge, E.; Been, J.V.; Steyerberg, E.; et al. A prediction model for neonatal mortality in low- and middle-income countries: An analysis of data from population surveillance sites in India, Nepal and Bangladesh. Int. J. Epidemiol. 2019, 48, 186–198. [Google Scholar] [CrossRef] [Green Version]
- Harsha, S.S.; Archana, B.R. SNAPPE-II (Score for neonatal acute physiology with perinatal extension-II) in predicting mortality and morbidity in NICU. J. Clin. Diagn. Res. 2015, 9, SC10. [Google Scholar] [CrossRef]
- Lee, S.M.; Lee, M.H.; Chang, Y.S. The Korean Neonatal Network. The clinical risk index for babies II for prediction of time-dependent mortality and short-term morbidities in very low birth weight infants. Neonatology 2019, 116, 244–251. [Google Scholar] [CrossRef]
- Dorling, J.S.; Field, D.J.; Manktelow, B. Neonatal disease severity scoring systems. Arch. Dis. Child. Fetal Neonatal Ed. 2005, 90, F11–F16. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A.; Pany, M.J.; Parikh, R.B.; Obermeyer, Z. Development and application of a machine learning approach to assess short-term mortality risk among patients with cancer starting chemotherapy. JAMA Netw Open 2018, 1, e180926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shillan, D.; Sterne, J.A.C.; Champneys, A.; Gibbison, B. Use of machine learning to analyze routinely collected intensive care unit data: A systemic review. Crit. Care 2019, 23, 284. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.W.; Kim, J.; Kim, D.K.; Oh, K.M.; Joo, K.W.; Kim, Y.S.; Han, S.S. Machine learning algorithm to predict mortality in patients undergoing continuous renal replacement therapy. Crit. Care 2020, 24, 42. [Google Scholar] [CrossRef] [Green Version]
- Meyer, A.; Zverinski, D.; Pfahringer, B.; Kempfert, J.; Kuehne, T.; Sündermann, S.H.; Stamm, C.; Hofmann, T.; Falk, V.; Eickhoff, C. Machine learning for real-time prediction of complications in critical care: A retrospective study. Lancet Respir. Med. 2018, 6, 905–914. [Google Scholar] [CrossRef]
- Wang, P.; Ge, R.; Xiao, X.; Cai, Y.; Wang, G.; Zhou, F. Rectified-Linear-Unit-based deep learning for biomedical multi-label data. Interdiscip. Sci. 2017, 9, 419–422. [Google Scholar] [CrossRef]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation coefficients: Appropriate use and interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef]
- Hsu, J.F.; Chu, S.M.; Huang, Y.C.; Lien, R.; Huang, H.R.; Lee, C.W.; Chiang, M.-C.; Fu, R.-H.; Tsai, M.-H. Predictors of clinical and microbiological treatment failure in neonatal bloodstream infections. Clin. Microbiol. Infect. 2015, 21, 482.e9–482.e17. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.H.; Chu, S.M.; Hsu, J.F.; Lien, R.; Huang, H.R.; Chiang, M.C.; Fu, R.-H.; Lee, C.-W.; Huang, Y.-C. Risk factors and outcomes for multidrug-resistant Gram-negative bacteremia in the NICU. Pediatrics 2014, 133, e322–e329. [Google Scholar] [CrossRef] [Green Version]
- Chu, S.M.; Hsu, J.F.; Lai, M.Y.; Huang, H.R.; Chiang, M.C.; Fu, R.-H.; Tsai, M.H. Risk factors of initial inappropriate antibiotic therapy and the impacts on outcomes of neonates with gram-negative bacteremia. Antibiotics 2020, 9, 203. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.C.; Liao, C.C.; Chu, S.M.; Lai, M.Y.; Huang, H.R.; Chiang, M.C.; Fu, R.-H.; Hsu, J.F.; Tsai, M.H. Impacts of multidrug-resistant pathogens and inappropriate initial antibiotic therapy on the outcomes of neonates with ventilator-associated pneumonia. Antibiotics 2020, 9, 760. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.; Kamitsuka, M.; Clark, R.H.; Kelleher, A.S.; Spitzer, A.R. Etiologies of NICU deaths. Pediatrics 2015, 135, e59–e65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podda, M.; Bacciu, D.; Micheli, A.; Bellù, R.; Placidi, G.; Gagliardi, L. A machine learning approach to estimating preterm infant survival: Development of the preterm infants survival assessment (PISA) predictor. Sci. Rep. 2018, 8, 13743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turova, V.; Sidorenko, I.; Eckardt, L.; Rieger-Fackeldey, E.; Felderhoff-Müser, U.; Alves-Pinto, A.; Lampe, R. Machine learning models for identifying preterm infants at risk of cerebral hemorrhage. PLoS ONE 2020, 15, e0227419. [Google Scholar] [CrossRef] [Green Version]
- Ball, G.; Aljabar, P.; Arichi, T.; Tusor, N.; Cox, D.; Merchant, N.; Nongena, P.; Hajnal, J.V.; Edwards, A.D.; Counsell, S.J. Machine-learning to characterize neonatal functional connectivity in the preterm brain. Neuroimage 2016, 124 Pt A, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, M.L.; Wang, Z.; Aljabar, P.; Ball, G.; Mirza, G.; Saxena, A.; Counsell, S.; Hajnal, J.; Montana, G.; Edwards, A.D. Machine learning shows association between genetic variability in PPARG and cerebral connectivity in preterm infants. Proc. Natl. Acad. Sci. USA 2017, 114, 13744–13749. [Google Scholar] [CrossRef] [Green Version]
- Cantey, J.B.; Baird, S.D. Ending the culture of culture-negative sepsis in the neonatal ICU. Pediatrics 2017, 140, e20170044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ting, J.Y.; Synnes, A.; Roberts, A.; Deshpandey, A.; Dow, K.; Yoon, E.W.; Lee, K.-S.; Dobson, S.; Lee, S.K.; Shah, P.S.; et al. Association between antibiotic use and neonatal mortality and morbidities in very low-birth-weight infants without culture-proven sepsis or necrotizing enterocolitis. JAMA Pediatr. 2016, 170, 1181–1187. [Google Scholar] [CrossRef]
- van den Brand, M.; van den Dungen, F.A.M.; Bos, M.P.; van Weissenbruch, M.M.; van Furth, A.M.; de Lange, A.; Rubenjan, A.; Peters, R.P.H.; Savelkoul, P.H.M. Evaluation of a real-time PCR assay for detection and quantification of bacterial DNA directly in blood of preterm neonates with suspected late-onset sepsis. Crit. Care 2018, 22, 105. [Google Scholar] [CrossRef] [PubMed]
- Muktan, D.; Singh, R.R.; Bhatta, N.K.; Shah, D. Neonatal mortality risk assessment using SNAPPE-II score in a neonatal intensive care unit. BMC Pediatr. 2019, 19, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dammann, O.; Shah, B.; Naples, M.; Bednarek, F.; Zupancic, J.; Allred, E.N.; Leviton, A. Interinstitutional variation in prediction of death by SNAP-II and SNAPPE-II among extremely preterm infants. Pediatrics 2009, 124, e1001–e1006. [Google Scholar] [CrossRef] [Green Version]
- Aluvaala, J.; Collins, G.S.; Maina, M.; Berkley, J.A.; English, M. A systemic review of neonatal treatment intensity scores and their potential application in low-resource setting hospitals for predicting mortality, morbidity and estimating resource use. Syst. Rev. 2017, 6, 248. [Google Scholar] [CrossRef] [Green Version]
- Mansoor, K.P.; Ravikiran, S.R.; Kulkarni, V.; Baliga, K.; Rao, S.; Kamalakshi, G.B.; Shantharam Baliga, B.; Kamath, N. Modified sick neonatal score (MSNS): A novel neonatal disease severity scoring system for resource-limited settings. Crit. Care Res. Pract. 2019, 2019, 9059073. [Google Scholar] [CrossRef]
- Mesquita Ramirez, M.N.; Godoy, L.E.; Alvarez Barrrientos, E. SNAP II and SNAPPE II as predictors of neonatal mortality in a pediatric intensive care unit: Does postnatal age play a role? Int. J. Pediatr. 2014, 2014, 298198. [Google Scholar] [CrossRef] [Green Version]
- Cicchetti, D.V.; Klin, A.; Volkmar, F.R. Assessing binary diagnoses of bio-behavior disorders: The clinical relevance of Cohen’s Kappa. J. Nerv. Ment. Dis. 2017, 205, 58–65. [Google Scholar] [CrossRef]
- Russo, D.P.; Zorn, K.M.; Clark, A.M.; Zhu, H.; Ekin, S. Comparing multiple machine learning algorithms and metrics for estrogen receptor binding prediction. Mol. Pharm. 2018, 15, 4361–4370. [Google Scholar] [CrossRef]
- Farrahi, V.; Niemela, M.; Tjurin, P.; Kangas, M.; Korpelainen, R.; Jamsa, T. Evaluating and enhancing the generalization performance of machine learning models for physical activity intensity prediction from raw acceleration data. IEEE J. Biomed. Health Inform. 2019, 24, 27–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Characteristics | All Study Subjects (Total n = 1095) | The Training Set (Total n = 765) | The Test Set (Total n = 330) | p Values |
|---|---|---|---|---|
| Cases demographics | ||||
| Gestational age (weeks), median (IQR) | 30.0 (27.0–35.0) | 30.0 (27.0–35.0) | 31.0 (27.8–36.0) | 0.338 |
| Birth weight (g), median (IQR) | 1360 (1020–2150.0) | 1355.0 (1040.0–2020.0) | 1365 (897.5–2500.0) | 0.758 |
| Gender (male/female), n (%) | 580 (53.0)/515 (47.0) | 406 (53.1)/359 (46.9) | 174 (52.7)/156 (47.3) | 0.947 |
| 5 min Apgar score ≤ 7, n (%) | 219 (20.0) | 151 (19.7) | 68 (20.6) | 0.742 |
| Inborn/outborn, n (%) | 873 (79.9)/222 (20.1) | 606 (79.2)/159 (20.8) | 267 (80.9)/63 (19.1) | 0.567 |
| Birth by NSD/Cesarean section, n (%) | 358 (32.7)/736 (67.3) | 256 (33.5)/508 (66.5) | 102 (30.9)/228 (69.1) | 0.440 |
| Respiratory distress syndrome (≥Gr II), n (%) | 467 (42.6) | 325 (42.5) | 142 (43.0) | 0.894 |
| Perinatal asphyxia, n (%) | 84 (7.7) | 55 (7.2) | 29 (8.9) | 0.387 |
| Underlying Chronic Comorbidities, n (%) | ||||
| Neurological sequelae | 202 (18.4) | 146 (19.1) | 56 (17.0) | 0.445 |
| Bronchopulmonary dysplasia | 189 (17.3) | 139 (18.2) | 50 (15.2) | 0.257 |
| Complicated cardiovascular diseases | 27 (2.5) | 23 (3.0) | 4 (1.2) | 0.091 |
| Symptomatic patent ductus arteriosus | 356 (32.5) | 258 (33.7) | 98 (29.7) | 0.206 |
| Gastrointestinal sequelae | 55 (5.0) | 34 (4.4) | 21 (6.4) | 0.227 |
| Renal disorders | 8 (0.7) | 6 (0.8) | 2 (0.6) | 0.753 |
| Congenital anomalies | 36 (6.5) | 29 (8.4) | 7 (5.6) | 0.196 |
| Presences of any chronic comorbidities | 365 (33.3) | 266 (34.8) | 99 (30.0) | 0.142 |
| Day of life at onset of suspected sepsis (day) # | 19.0 (13.0–39.0) | 19.0 (14.0–37.0) | 20.0 (10.0–41.3) | 0.232 |
| Previous antibiotic exposure, n (%) | 662 (60.5) | 472 (61.7) | 190 (57.6) | 0.202 |
| Use of TPN and/or intrafat, n (%) | 527 (48.1) | 367 (47.8) | 160 (48.5) | 0.895 |
| Use of central venous catheter, n (%) | 741 (67.7) | 521 (68.1) | 220 (66.7) | 0.673 |
| Feeding situation *, n (%) | 0.186 | |||
| Nothing by mouth (NPO) | 700 (63.9) | 479 (62.6) | 211 (63.9) | |
| Half amount | 353 (32.2) | 259 (33.9) | 94 (28.5) | |
| Full amount | 42 (3.8) | 27 (3.5) | 15 (4.5) | |
| Ventilator requirement *, n (%) | 0.123 | |||
| O2 hood | 260 (23.7) | 197 (25.7) | 63 (19.1) | |
| Nasal cannula | 45 (4.1) | 34 (4.4) | 11 (3.3) | |
| Nasal continuous positive airway pressure | 151 (13.8) | 90 (11.8) | 61 (18.5) | |
| Nasal intermittent mandatory ventilation | 174 (15.9) | 124 (11.3) | 50 (15.2) | |
| Intubation with mechanical ventilation | 380 (34.7) | 260 (34.0) | 120 (36.4) | |
| On high frequency oscillatory ventilation | 85 (7.8) | 60 (7.8) | 25 (7.6) | |
| Clinical features *, n (%) | ||||
| Intravascular volume expansion | 185 (16.9) | 137 (17.9) | 48 (14.5) | 0.188 |
| Requirement of cardiac inotropic agents | 91 (8.3) | 62 (8.1) | 29 (8.8) | 0.721 |
| Metabolic acidosis | 201 (18.4) | 141 (18.4) | 60 (18.2) | 0.932 |
| Coagulopathy | 175 (16.0) | 120 (15.7) | 55 (16.7) | 0.187 |
| Requirement of blood transfusion ** | 627 (57.3) | 445 (58.2) | 182 (55.2) | 0.387 |
| Laboratory data | ||||
| Leukocytosis or leucopenia | 269 (24.6) | 193 (25.2) | 76 (23.0) | 0.491 |
| Shift to left in WBC (immature > 20%) | 65 (5.9) | 48 (6.3) | 17 (5.1) | 0.577 |
| Anemia (hemoglobin level < 11.5 g/dL) | 717 (65.5) | 501 (65.5) | 216 (65.4) | 0.991 |
| Thrombocytopenia (platelet < 150,000/uL) | 243 (22.2) | 166 (21.7) | 77 (23.3) | 0.526 |
| C-reactive protein (mg/dL), median (IQR) | 11.3 (8.2–23.3) | 11.4 (7.5–24.0) | 11.3 (8.1–22.5) | 0.456 |
| PPMCC > 0.00 | PPMCC > 0.03 | PPMCC > 0.05 | PPMCC > 0.10 | PPMCC > 0.15 | |
|---|---|---|---|---|---|
| Area under ROC curve (AUC) | 94.40 ± 0.49% | 92.72 ± 2.40% | 92.32 ± 3.45% | 91.52 ± 1.56% | 85.02 ± 2.57% |
| Sensitivity (true positive rate) | 87.14 ± 3.19% | 81.41 ± 2.99% | 82.86 ± 5.30% | 82.86 ± 1.60% | 82.86 ± 4.66% |
| Specificity (true negative rate) | 92.78 ± 3.18% | 95.63 ± 2.61% | 96.82 ± 1.58% | 94.70 ± 2.12% | 85.36 ± 4.14% |
| False-positive rate (FPR) | 7.22 ± 3.18% | 4.37 ± 2.61% | 3.18 ± 1.58% | 5.30 ± 2.12% | 14.64 ± 4.14% |
| False-negative rate (FNR) | 12.86 ± 3.19% | 18.57 ± 2.99% | 17.14 ± 5.30% | 17.14 ± 1.60% | 17.14 ± 4.66% |
| Positive predictive value (PPV or Recall) | 52.27 ± 13.88% | 65.71 ± 12.88% | 72.18 ± 11.51% | 60.29 ± 8.04% | 35.23 ± 5.02% |
| Negative predictive value (NPV) | 98.74 ± 0.27% | 98.23 ± 0.26% | 98.39 ± 0.48% | 98.32 ± 0.12% | 98.19 ± 0.42% |
| F1 score [ = 2×TPR×Recall/(TPR + Recall)] | 0.67 ± 0.08% | 0.72 ± 0.08% | 0.77 ± 0.06% | 0.69 ± 0.05% | 0.49 ± 0.04% |
| Accuracy | 92.30 ± 2.69% | 94.42 ± 2.25% | 95.64 ± 1.28% | 93.70 ± 1.81% | 85.15 ± 3.45% |
| Variables | Weights | Variables | Weights |
|---|---|---|---|
| Requirement of ventilator support | 0.6016 | Birth body weight (g) | 0.0735 |
| Feeding conditions | 0.3938 | Bronchopulmonary dysplasia | 0.0628 |
| Intravascular volume expansion | 0.3623 | PaO2 (mmHg) | 0.0605 |
| Shift to left in white blood cell (immature > 20%) | 0.2397 | Blood transfusion with fresh frozen plasma | 0.0493 |
| Requirement of cardiac inotropic agents | 0.2258 | PH value | 0.0385 |
| Peak end expiratory pressure | 0.2201 | Use of diuretics/bronchodilator | 0.0333 |
| Metabolic acidosis | 0.2111 | C-reactive protein | 0.0331 |
| Central venous catheter | 0.1785 | Previous antibiotic exposure * | 0.0248 |
| Apnea | 0.1411 | Perinatal asphyxia | 0.0204 |
| Apgar score at 5 min | 0.1402 | Thrombocytopenia | 0.0068 |
| PaCO2 (mmHg) | 0.1372 | Gender | 0.0057 |
| Presences of any chronic comorbidities | 0.1358 | On corticosteroid | 0.0029 |
| Previous positive sputum cultures * | 0.1262 | Fever | 0.0022 |
| Red blood cell count | 0.1008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, J.-F.; Chang, Y.-F.; Cheng, H.-J.; Yang, C.; Lin, C.-Y.; Chu, S.-M.; Huang, H.-R.; Chiang, M.-C.; Wang, H.-C.; Tsai, M.-H. Machine Learning Approaches to Predict In-Hospital Mortality among Neonates with Clinically Suspected Sepsis in the Neonatal Intensive Care Unit. J. Pers. Med. 2021, 11, 695. https://doi.org/10.3390/jpm11080695
Hsu J-F, Chang Y-F, Cheng H-J, Yang C, Lin C-Y, Chu S-M, Huang H-R, Chiang M-C, Wang H-C, Tsai M-H. Machine Learning Approaches to Predict In-Hospital Mortality among Neonates with Clinically Suspected Sepsis in the Neonatal Intensive Care Unit. Journal of Personalized Medicine. 2021; 11(8):695. https://doi.org/10.3390/jpm11080695
Chicago/Turabian StyleHsu, Jen-Fu, Ying-Feng Chang, Hui-Jun Cheng, Chi Yang, Chun-Yuan Lin, Shih-Ming Chu, Hsuan-Rong Huang, Ming-Chou Chiang, Hsiao-Chin Wang, and Ming-Horng Tsai. 2021. "Machine Learning Approaches to Predict In-Hospital Mortality among Neonates with Clinically Suspected Sepsis in the Neonatal Intensive Care Unit" Journal of Personalized Medicine 11, no. 8: 695. https://doi.org/10.3390/jpm11080695
APA StyleHsu, J.-F., Chang, Y.-F., Cheng, H.-J., Yang, C., Lin, C.-Y., Chu, S.-M., Huang, H.-R., Chiang, M.-C., Wang, H.-C., & Tsai, M.-H. (2021). Machine Learning Approaches to Predict In-Hospital Mortality among Neonates with Clinically Suspected Sepsis in the Neonatal Intensive Care Unit. Journal of Personalized Medicine, 11(8), 695. https://doi.org/10.3390/jpm11080695









