Multiple Single Nucleotide Polymorphism Testing Improves the Prediction of Diabetic Retinopathy Risk with Type 2 Diabetes Mellitus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. SNP Genotyping
2.3. Construction of the Combined Nongenetic and Genetic Risk Model
2.4. Statistical Analysis
3. Results
3.1. Subject Characteristics
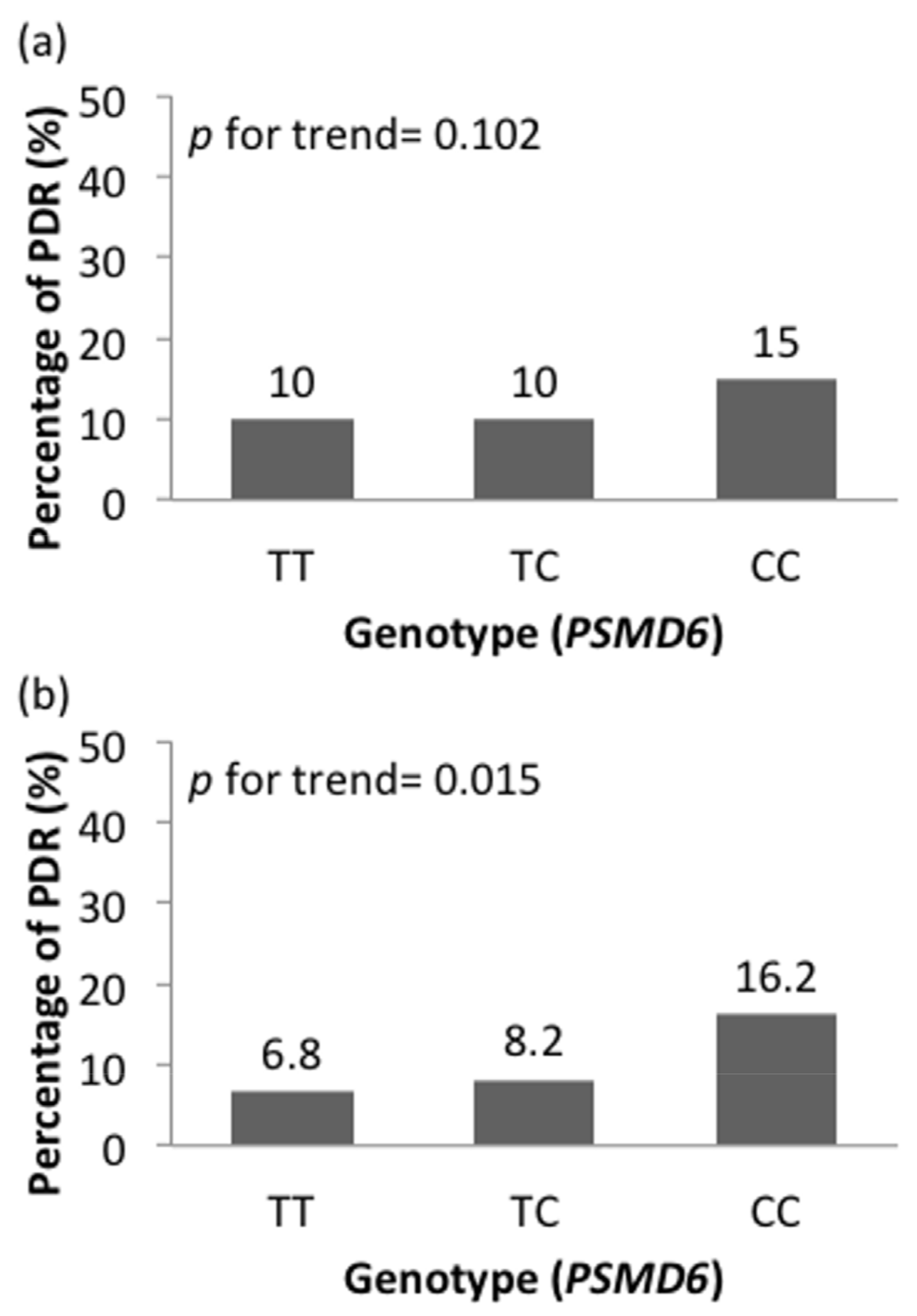
3.2. Association between SNPs and DR Risk
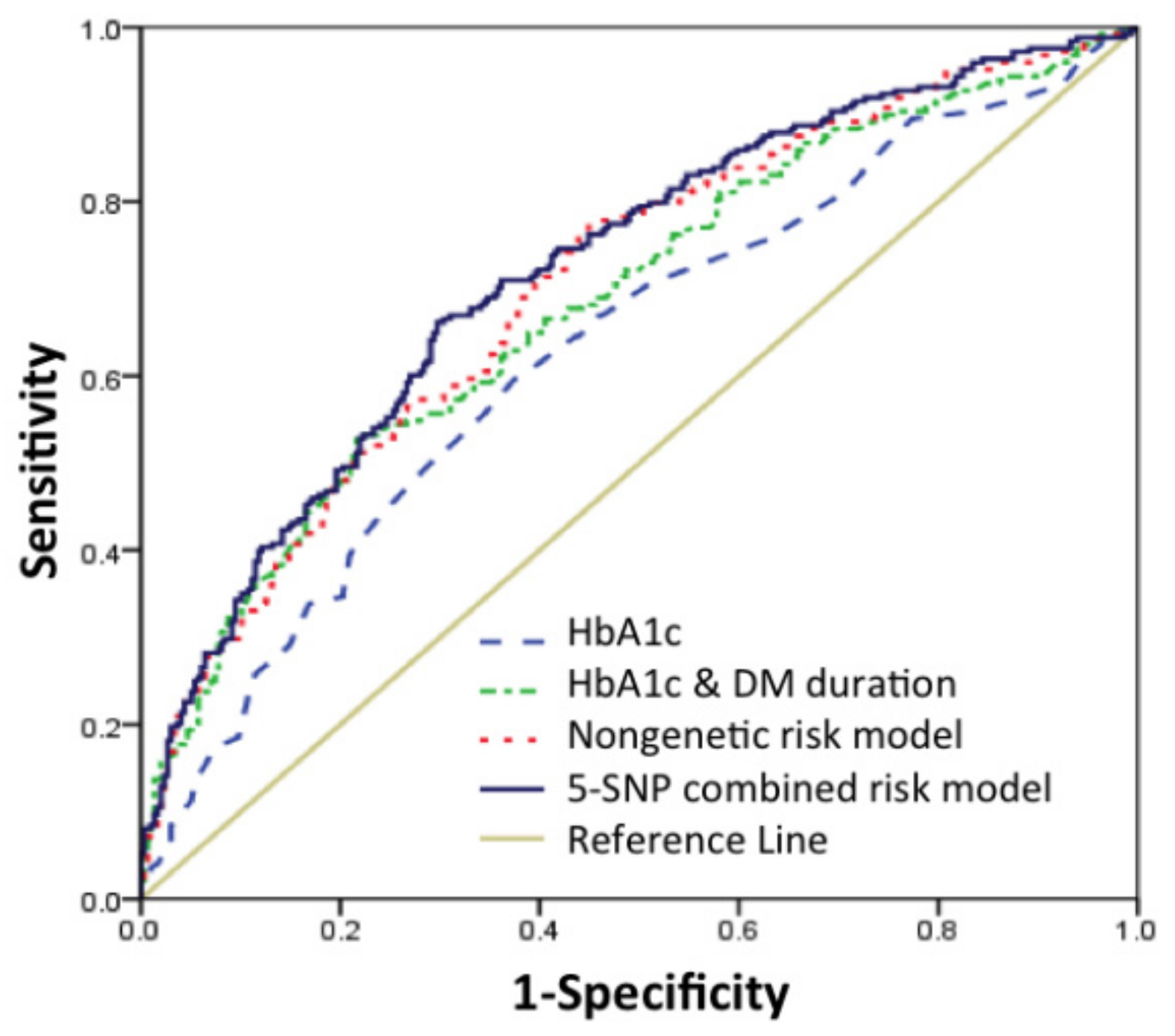
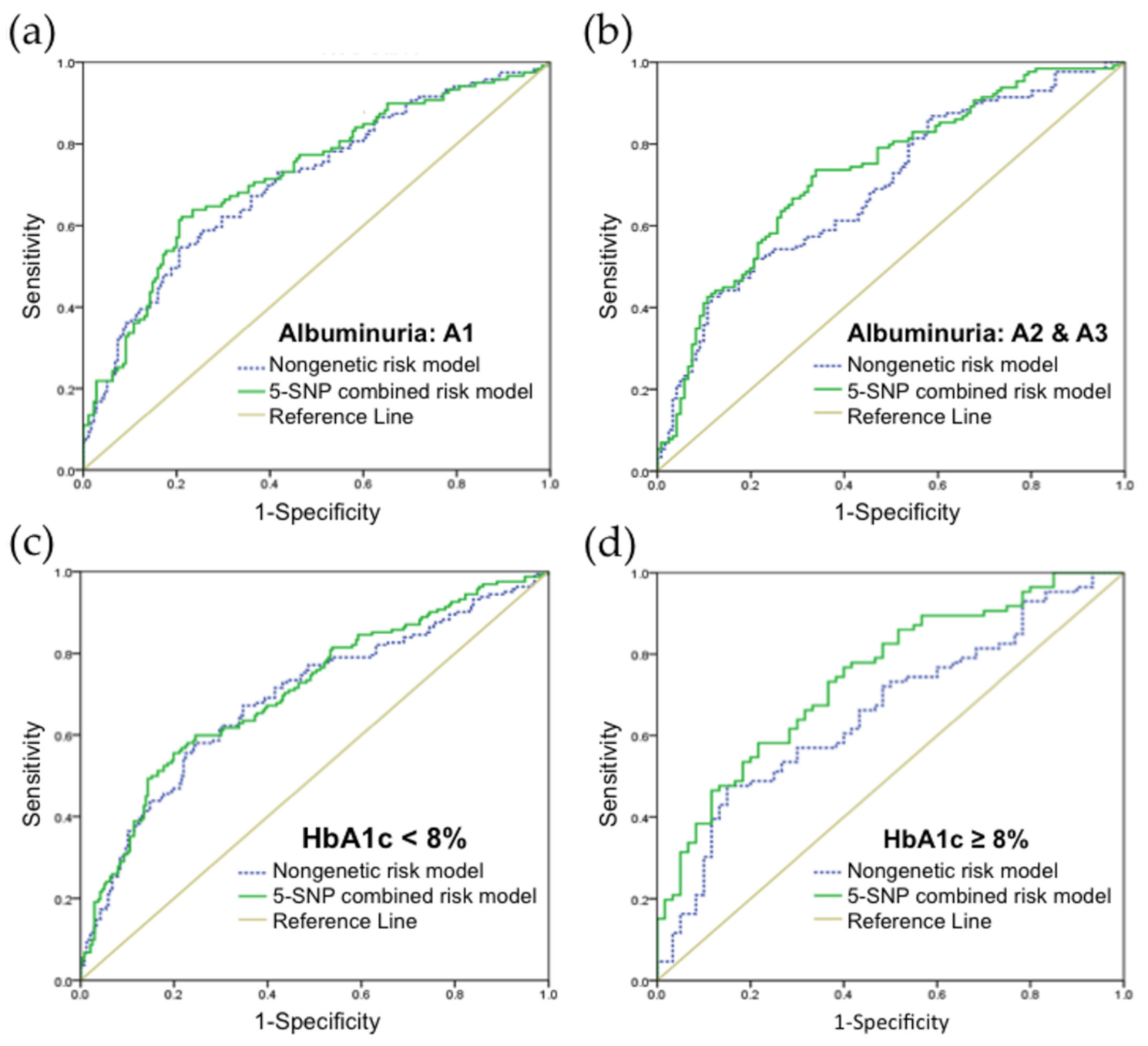
3.3. Validation of Risk Prediction Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruta, L.M.; Magliano, D.J.; LeMesurier, R.; Taylor, H.R.; Zimmet, P.Z.; Shaw, J.E. Prevalence of diabetic retinopathy in Type 2 diabetes in developing and developed countries. Diabet. Med. 2013, 30, 387–398. [Google Scholar] [CrossRef]
- Flaxman, S.R.; Bourne, R.R.A.; Resnikoff, S.; Ackland, P.; Braithwaite, T.; Cicinelli, M.V.; Das, A.; Jonas, J.B.; Keeffe, J.; Kempen, J.H.; et al. Global causes of blindness and distance vision impairment 1990-2020: A systematic review and meta-analysis. Lancet Glob. Health 2017, 5, e1221–e1234. [Google Scholar] [CrossRef] [Green Version]
- Ting, D.S.; Cheung, G.C.; Wong, T.Y. Diabetic retinopathy: Global prevalence, major risk factors, screening practices and public health challenges: A review. Clin. Exp. Ophthalmol. 2016, 44, 260–277. [Google Scholar] [CrossRef] [Green Version]
- Cheung, N.; Mitchell, P.; Wong, T.Y. Diabetic retinopathy. Lancet 2010, 376, 124–136. [Google Scholar] [CrossRef]
- Federation, I.D. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2019. [Google Scholar]
- Liu, W.J.; Lee, L.T.; Yen, M.F.; Tung, T.H.; Williams, R.; Duffy, S.W.; Chen, T.H. Assessing progression and efficacy of treatment for diabetic retinopathy following the proliferative pathway to blindness: Implication for diabetic retinopathy screening in Taiwan. Diabet Med. 2003, 20, 727–733. [Google Scholar] [CrossRef]
- World Health Organization. Strengthening Diagnosis and Treatment of Diabetic Retinopathy in the South-East Asia Region; World Health Organization, Regional Office for South-East Asia: New Delhi, India, 2020; Available online: https://apps.who.int/iris/bitstream/handle/10665/334224/9789290227946-eng.pdf (accessed on 18 June 2021).
- Das, T.; Takkar, B.; Sivaprasad, S.; Thanksphon, T.; Taylor, H.; Wiedemann, P.; Nemeth, J.; Nayar, P.D.; Rani, P.K.; Khandekar, R. Recently updated global diabetic retinopathy screening guidelines: Commonalities, differences, and future possibilities. Eye 2021, 1–14. [Google Scholar] [CrossRef]
- Storey, P.P.; Murchison, A.P.; Pizzi, L.T.; Hark, L.A.; Dai, Y.; Leiby, B.E.; Haller, J.A. Impact of physician communication on diabetic eye examination adherence: Results From a Retrospective Cohort Analysis. Retina 2016, 36, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Ogunyemi, O.; Kermah, D. Machine Learning Approaches for Detecting Diabetic Retinopathy from Clinical and Public Health Records. AMIA Annu. Symp. Proc. 2015, 2015, 983–990. [Google Scholar] [PubMed]
- Ogunyemi, O.I.; Gandhi, M.; Tayek, C. Predictive Models for Diabetic Retinopathy from Non-Image Teleretinal Screening Data. AMIA Summits Transl. Sci. Proc. 2019, 2019, 472–477. [Google Scholar] [PubMed]
- Yau, J.W.; Rogers, S.L.; Kawasaki, R.; Lamoureux, E.L.; Kowalski, J.W.; Bek, T.; Chen, S.J.; Dekker, J.M.; Fletcher, A.; Grauslund, J.; et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care 2012, 35, 556–564. [Google Scholar] [CrossRef] [Green Version]
- Vujosevic, S.; Aldington, S.J.; Silva, P.; Hernández, C.; Scanlon, P.; Peto, T.; Simó, R. Screening for diabetic retinopathy: New perspectives and challenges. Lancet Diabetes Endocrinol. 2020, 8, 337–347. [Google Scholar] [CrossRef]
- Lachin, J.M.; Genuth, S.; Nathan, D.M.; Zinman, B.; Rutledge, B.N. Effect of glycemic exposure on the risk of microvascular complications in the diabetes control and complications trial—Revisited. Diabetes 2008, 57, 995–1001. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Lando, L.; Skowronska-Krawczyk, D.; Chao, D.L. Genetics of Diabetic Retinopathy. Curr. Diabetes Rep. 2019, 19, 67. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Valle, M.L.; Beveridge, C.; Liu, Y.; Sharma, S. Unraveling the role of genetics in the pathogenesis of diabetic retinopathy. Eye 2019, 33, 534–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, W.Y.; Gu, H.; Yang, X.F.; She, C.Y.; Liu, X.P.; Liu, N.P. Association of candidate gene polymorphisms with diabetic retinopathy in Chinese patients with type 2 diabetes. Int. J. Ophthalmol. 2020, 13, 301–308. [Google Scholar] [CrossRef]
- Irace, C.; Scarinci, F.; Scorcia, V.; Bruzzichessi, D.; Fiorentino, R.; Randazzo, G.; Scorcia, G.; Gnasso, A. Association among low whole blood viscosity, haematocrit, haemoglobin and diabetic retinopathy in subjects with type 2 diabetes. Br. J. Ophthalmol. 2011, 95, 94–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Miao, Z.; Liu, T.; Liu, M.; Grdinovac, K.; Song, X.; Liang, Y.; Delen, D.; Paiva, W. Derivation and Validation of Essential Predictors and Risk Index for Early Detection of Diabetic Retinopathy Using Electronic Health Records. J. Clin. Med. 2021, 10, 1473. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.H.; Fan, Q.; Tham, Y.C.; Gan, A.; Tan, S.P.; Tan, G.; Wang, J.J.; Mitchell, P.; Wong, T.Y.; Cheng, C.Y. Type 2 Diabetes Genetic Variants and Risk of Diabetic Retinopathy. Ophthalmology 2017, 124, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.L.; Lin, J.M.; Chen, W.L.; Hsieh, M.C.; Wu, C.M.; Chang, Y.W.; Huang, Y.C.; Tsai, F.J. Multilocus genetic risk score for diabetic retinopathy in the Han Chinese population of Taiwan. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Marathe, P.H.; Gao, H.X.; Close, K.L. American Diabetes Association Standards of Medical Care in Diabetes 2017. J. Diabetes 2017, 9, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E., Jr.; Collins, K.J.; Dennison Himmelfarb, C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2018, 71, e127–e248. [Google Scholar] [CrossRef] [PubMed]
- Pettitt, D.J.; Talton, J.W.; Liese, A.D.; Liu, L.L.; Crimmins, N.; West, N.A.; D′Agostino, R.B., Jr.; Kahn, H.S.; SEARCH for Diabetes in Youth Study Group. Comparison of two waist circumference measurement protocols: The SEARCH for diabetes in youth study. Pediatric Obes. 2012, 7, e81–e85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, P.E.; Levin, A. Evaluation and Management of Chronic Kidney Disease: Synopsis of the Kidney Disease: Improving Global Outcomes 2012 Clinical Practice Guideline. Ann. Intern. Med. 2013, 158, 825–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, C.P.; Ferris, F.L., 3rd; Klein, R.E.; Lee, P.P.; Agardh, C.D.; Davis, M.; Dills, D.; Kampik, A.; Pararajasegaram, R.; Verdaguer, J.T. Proposed international clinical diabetic retinopathy and diabetic macular edema disease severity scales. Ophthalmology 2003, 110, 1677–1682. [Google Scholar] [CrossRef]
- Wong, T.Y.; Cheung, N.; Tay, W.T.; Wang, J.J.; Aung, T.; Saw, S.M.; Lim, S.C.; Tai, E.S.; Mitchell, P. Prevalence and risk factors for diabetic retinopathy: The Singapore Malay Eye Study. Ophthalmology 2008, 115, 1869–1875. [Google Scholar] [CrossRef]
- Ahmad, A.; Sundquist, K.; Palmér, K.; Svensson, P.J.; Sundquist, J.; Memon, A.A. Risk prediction of recurrent venous thromboembolism: A multiple genetic risk model. J. Thromb. Thrombolysis 2019, 47, 216–226. [Google Scholar] [CrossRef] [Green Version]
- de Haan, H.G.; Bezemer, I.D.; Doggen, C.J.; Le Cessie, S.; Reitsma, P.H.; Arellano, A.R.; Tong, C.H.; Devlin, J.J.; Bare, L.A.; Rosendaal, F.R.; et al. Multiple SNP testing improves risk prediction of first venous thrombosis. Blood 2012, 120, 656–663. [Google Scholar] [CrossRef] [Green Version]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferris, F.L., III; Nathan, D.M. Preventing Diabetic Retinopathy Progression. Ophthalmology 2016, 123, 1840–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, M.L.; Pérez, S.; Mena-Mollá, S.; Desco, M.C.; Ortega, Á.L. Oxidative Stress and Microvascular Alterations in Diabetic Retinopathy: Future Therapies. Oxid. Med. Cell. Longev. 2019, 2019, 4940825. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yu, X.; Zheng, B.; Ding, S.; Mu, Z.; Guo, L. Early neurovascular changes in the retina in preclinical diabetic retinopathy and its relation with blood glucose. BMC Ophthalmol. 2021, 21, 1–9. [Google Scholar] [CrossRef]
- Stehouwer, C.D.A. Microvascular Dysfunction and Hyperglycemia: A Vicious Cycle with Widespread Consequences. Diabetes 2018, 67, 1729–1741. [Google Scholar] [CrossRef] [Green Version]
- Zhao, N.N.; Dong, G.P.; Wu, W.; Wang, J.L.; Ullah, R.; Fu, J.F. FTO gene polymorphisms and obesity risk in Chinese population: A meta-analysis. World J. Pediatr. 2019, 15, 382–389. [Google Scholar] [CrossRef]
- Wu, J.; Xu, J.; Zhang, Z.; Ren, J.; Li, Y.; Wang, J.; Cao, Y.; Rong, F.; Zhao, R.; Huang, X.; et al. Association of FTO polymorphisms with obesity and metabolic parameters in Han Chinese adolescents. PLoS ONE 2014, 9, e98984. [Google Scholar] [CrossRef]
- Xiao, S.; Zeng, X.; Quan, L.; Zhu, J. Correlation between polymorphism of FTO gene and type 2 diabetes mellitus in Uygur people from northwest China. Int. J. Clin. Exp. Med. 2015, 8, 9744–9750. [Google Scholar] [PubMed]
- Bravard, A.; Lefai, E.; Meugnier, E.; Pesenti, S.; Disse, E.; Vouillarmet, J.; Peretti, N.; Rabasa-Lhoret, R.; Laville, M.; Vidal, H.; et al. FTO is increased in muscle during type 2 diabetes, and its overexpression in myotubes alters insulin signaling, enhances lipogenesis and ROS production, and induces mitochondrial dysfunction. Diabetes 2011, 60, 258–268. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, N.; Shara, N.M.; Wang, H.; Voruganti, V.S.; Laston, S.; Haack, K.; Lee, E.T.; Best, L.G.; MacCluer, J.W.; Cochran, B.J.; et al. The association of genetic variants of type 2 diabetes with kidney function. Kidney Int. 2012, 82, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Mottl, A.K.; Kwon, K.S.; Garg, S.; Mayer-Davis, E.J.; Klein, R.; Kshirsagar, A.V. The association of retinopathy and low GFR in type 2 diabetes. Diabetes Res. Clin. Pract. 2012, 98, 487–493. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, C.-F.; Baker, P.R.; Chen, P.-l.; Kaiser, P.; Huang, L. Mass Spectrometric Characterization of the Affinity-Purified Human 26S Proteasome Complex. Biochemistry 2007, 46, 3553–3565. [Google Scholar] [CrossRef]
- Zhang, S.; Jamaspishvili, E.; Tong, H.; Chen, Y.; Zhou, Z.; Sun, L.; Kazakova, E.; Hong, Q. East Asian Genome-wide association study derived loci in relation to type 2 diabetes in the Han Chinese population. Acta Biochim. Pol. 2019, 66, 159–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, Y.S.; Chen, C.H.; Hu, C.; Long, J.; Ong, R.T.; Sim, X.; Takeuchi, F.; Wu, Y.; Go, M.J.; Yamauchi, T.; et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat. Genet. 2011, 44, 67–72. [Google Scholar] [CrossRef]
- Aghdam, S.Y.; Sheibani, N. The ubiquitin-proteasome system and microvascular complications of diabetes. J. Ophthalmic Vis. Res. 2013, 8, 244–256. [Google Scholar]
- Chen, M.; Hu, C.; Zhang, R.; Jiang, F.; Wang, J.; Peng, D.; Tang, S.; Sun, X.; Yan, J.; Wang, S.; et al. A variant of PSMD6 is associated with the therapeutic efficacy of oral antidiabetic drugs in Chinese type 2 diabetes patients. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, Y.; Shi, K.; Wang, C. Body mass index and risk of diabetic retinopathy: A meta-analysis and systematic review. Medicine 2017, 96, e6754. [Google Scholar] [CrossRef]
- Panagiotakos, D.B.; Pitsavos, C.; Yannakoulia, M.; Chrysohoou, C.; Stefanadis, C. The implication of obesity and central fat on markers of chronic inflammation: The ATTICA study. Atherosclerosis 2005, 183, 308–315. [Google Scholar] [CrossRef]
- Fujimoto, W.Y.; Abbate, S.L.; Kahn, S.E.; Hokanson, J.E.; Brunzell, J.D. The visceral adiposity syndrome in Japanese-American men. Obes. Res. 1994, 2, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Haghvirdizadeh, P.; Mohamed, Z.; Abdullah, N.A.; Haghvirdizadeh, P.; Haerian, M.S.; Haerian, B.S. KCNJ11: Genetic Polymorphisms and Risk of Diabetes Mellitus. J. Diabetes Res. 2015, 2015, 908152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosengren, A.H.; Braun, M.; Mahdi, T.; Andersson, S.A.; Travers, M.E.; Shigeto, M.; Zhang, E.; Almgren, P.; Ladenvall, C.; Axelsson, A.S.; et al. Reduced Insulin Exocytosis in Human Pancreatic β-Cells With Gene Variants Linked to Type 2 Diabetes. Diabetes 2012, 61, 1726–1733. [Google Scholar] [CrossRef] [Green Version]
- Unoki, H.; Takahashi, A.; Kawaguchi, T.; Hara, K.; Horikoshi, M.; Andersen, G.; Ng, D.P.K.; Holmkvist, J.; Borch-Johnsen, K.; Jørgensen, T.; et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet. 2008, 40, 1098–1102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, H.; Guan, X.; Niu, Q.; Li, W. Variant rs2237892 of KCNQ1 Is Potentially Associated with Hypertension and Macrovascular Complications in Type 2 Diabetes Mellitus in A Chinese Han Population. Genom. Proteom. Bioinform. 2015, 13, 364–370. [Google Scholar] [CrossRef] [Green Version]
- Rattanatham, R.; Settasatian, N.; Komanasin, N.; Kukongviriyapan, U.; Sawanyawisuth, K.; Intharaphet, P.; Senthong, V.; Settasatian, C. Association of Combined TCF7L2 and KCNQ1 Gene Polymorphisms with Diabetic Micro- and Macrovascular Complications in Type 2 Diabetes Mellitus. Diabetes Metab. J. 2021. [Google Scholar] [CrossRef]
- van Vliet-Ostaptchouk, J.V.; van Haeften, T.W.; Landman, G.W.; Reiling, E.; Kleefstra, N.; Bilo, H.J.; Klungel, O.H.; de Boer, A.; van Diemen, C.C.; Wijmenga, C.; et al. Common variants in the type 2 diabetes KCNQ1 gene are associated with impairments in insulin secretion during hyperglycaemic glucose clamp. PLoS ONE 2012, 7, e32148. [Google Scholar] [CrossRef]
- Včelák, J.; Vejražková, D.; Vaňková, M.; Lukášová, P.; Bradnová, O.; Hálková, T.; Bešťák, J.; Andělová, K.; Kvasničková, H.; Hoskovcová, P.; et al. T2D risk haplotypes of the TCF7L2 gene in the Czech population sample: The association with free fatty acids composition. Physiol. Res. 2012, 61, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Chew, E.Y.; Klein, M.L.; Ferris, F.L., 3rd; Remaley, N.A.; Murphy, R.P.; Chantry, K.; Hoogwerf, B.J.; Miller, D. Association of elevated serum lipid levels with retinal hard exudate in diabetic retinopathy. Early Treatment Diabetic Retinopathy Study (ETDRS) Report 22. Arch. Ophthalmol. 1996, 114, 1079–1084. [Google Scholar] [CrossRef]
- Kowluru, R.A. Retinopathy in a Diet-Induced Type 2 Diabetic Rat Model and Role of Epigenetic Modifications. Diabetes 2020, 69, 689–698. [Google Scholar] [CrossRef] [PubMed]




| Non-DR | DR | p-Value | |
|---|---|---|---|
| n | 297 | 254 | |
| Age (year) | 60.78 ± 9.81 | 61.26 ± 9.71 | 0.648 a |
| Male sex, no. (%) | 155 (53.4) | 142 (54.4) | 0.822 b |
| Waist circumference (cm) | 90.30 ± 10.38 | 91.64 ± 11.29 | 0.143 a |
| SBP (mmHg) | 136.56 ± 18.80 | 140.90 ± 19.86 | 0.002 a |
| DBP (mmHg) | 77.53 ± 11.89 | 78.41 ± 12.49 | 0.375 a |
| Duration of DM (year) | 14.70 ± 6.47 | 19.15 ± 8.73 | <0.001 a |
| HbA1c (%) | 7.24 ± 0.88 | 7.72 ± 1.14 | <0.001 a |
| Total cholesterol (mg/dL) | 168.98 ± 1.75 | 167.27 ± 2.07 | 0.305 a |
| HDL (mg/dL) | 47.42 ± 0.70 | 46.89 ± 0.89 | 0.090 a |
| LDL (mg/dL) | 89.94 ± 1.32 | 90.18 ± 1.58 | 0.601 a |
| Triglycerides (mg/dL) | 139.92 ± 5.26 | 134.51 ± 4.81 | 0.678 a |
| Albuminuria categories c | 0.02 b | ||
| A1, no. (%) | 176 (59.3) | 121 (47.6) | |
| A2, no. (%) | 102 (34.3) | 101 (39.8) | |
| A3, no. (%) | 19 (6.4) | 32 (12.6) |
| Genetic Variables | Genotypic Value | 1 vs. 0 | 2 vs. 1 | 2 vs. 0 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | S.E. | OR (95% CI) | p-Value | S.E. | OR (95% CI) | p-Value | S.E. | OR (95% CI) | p-Value | |
| FTO (rs8050136) | CC | CA | AA | 1.120 | 0.789 (0.509, 1.223) | 0.290 | 0.829 | 5.650 (1.111, 28.571) | 0.044 | 3.376 | 4.451 (0.905, 21.888) | 0.095 |
| PSMD6 (rs831571) | TT | TC | CC | 2.125 | 0.666 (0.386, 1.150) | 0.174 | 0.204 | 1.692 (1.135, 2.525) | 0.020 | 0.177 | 1.127 (0.645, 1.972) | 0.674 |
| Genetic Variables | Alleles | S.E. | OR (95% CI) | p-Value |
|---|---|---|---|---|
| FTO (rs8050136) | CC + CA vs. AA | 3.569 | 4.605 (0.944, 22.454) | 0.071 |
| PSMD6 (rs831571) | TT + TC vs. CC | 4.762 | 1.519 (1.044, 2.212) | 0.044 |
| Variables | S.E. | OR (95% CI) | p-Value |
|---|---|---|---|
| DM medications | |||
| Insulin | 0.262 | 2.609 (1.561, 4.360) | 0.003 |
| GLP-1 receptor agonists | 0.499 | 2.838 (1.067, 7.553) | 0.043 |
| genetics | |||
| FTO (rs8050136) | 4.780 | 5.851 (1.201, 28.510) | 0.041 |
| PSMD6 (rs831571) | 3.144 | 1.414 (0.964, 2.074) | 0.076 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsiao, Y.-T.; Shen, F.-C.; Weng, S.-W.; Wang, P.-W.; Chen, Y.-J.; Lee, J.-J. Multiple Single Nucleotide Polymorphism Testing Improves the Prediction of Diabetic Retinopathy Risk with Type 2 Diabetes Mellitus. J. Pers. Med. 2021, 11, 689. https://doi.org/10.3390/jpm11080689
Hsiao Y-T, Shen F-C, Weng S-W, Wang P-W, Chen Y-J, Lee J-J. Multiple Single Nucleotide Polymorphism Testing Improves the Prediction of Diabetic Retinopathy Risk with Type 2 Diabetes Mellitus. Journal of Personalized Medicine. 2021; 11(8):689. https://doi.org/10.3390/jpm11080689
Chicago/Turabian StyleHsiao, Yu-Ting, Feng-Chih Shen, Shao-Wen Weng, Pei-Wen Wang, Yung-Jen Chen, and Jong-Jer Lee. 2021. "Multiple Single Nucleotide Polymorphism Testing Improves the Prediction of Diabetic Retinopathy Risk with Type 2 Diabetes Mellitus" Journal of Personalized Medicine 11, no. 8: 689. https://doi.org/10.3390/jpm11080689
APA StyleHsiao, Y.-T., Shen, F.-C., Weng, S.-W., Wang, P.-W., Chen, Y.-J., & Lee, J.-J. (2021). Multiple Single Nucleotide Polymorphism Testing Improves the Prediction of Diabetic Retinopathy Risk with Type 2 Diabetes Mellitus. Journal of Personalized Medicine, 11(8), 689. https://doi.org/10.3390/jpm11080689






