Abstract
UDP-glucuronosyltransferases (UGTs) are phase II drug-metabolizing enzymes that metabolize endogenous fatty acids such as arachidonic acid metabolites, as well as many prescription drugs, such as opioids, antiepileptics, and antiviral drugs. The UGT1A and 2B genes are highly polymorphic, and their genetic variants may affect the pharmacokinetics and hence the responses of many drugs and fatty acids. This study collected data and updated the current view of the molecular functionality of genetic variants on UGT genes that impact drug responses and the susceptibility to human diseases. The functional information of UGT genetic variants with clinical associations are essential to understand the inter-individual variation in drug responses and susceptibility to toxicity.
1. Introduction
The UDP-glucuronosyltransferase (UGT) enzymes are phase II drug-metabolizing enzymes that catalyze the glucuronidation reaction. This chemical reaction involves the formation of a covalent bond between the endogenous polar glucuronic acid with drugs and endogenous lipophilic compounds [1]. The glucuronidated compounds have chemical functional groups that accept glucuronic acid. These functional groups include hydroxyl, carboxylic acid, amine, and thiol [2]. The UGTs glucuronidate endogenous compounds, such as bilirubin, bile acids, and steroid hormones. Additionally, the UGTs glucuronidate exogenous compounds such as opioid analgesics, non-steroidal anti-inflammatory agents (NSAIDs), anticonvulsants, and antiviral drugs [3].
Glucuronidation mainly terminates and enhances the elimination of chemical compounds by enhancing their solubility in urine. Additionally, glucuronidated compounds are large, which favors their elimination through biliary excretion [4]. Therefore, the glucuronidation reaction can increase the efficacy and toxicity of some drugs, and glucuronide morphine is reportedly 100 times more potent than the morphine substrate itself [5].
Glucuronidation occurs in mammalian species, although significant inter-species differences exist in the rate of glucuronidation, expression, and selectivity [6]. For example, codeine is glucuronidated at higher rates among humans than rats [7,8]. Additionally, cat livers cannot glucuronidate the analgesic paracetamol drug [9]. Therefore, any information obtained about glucuronidation in animals is not directly applicable to humans.
2. UGT Isoforms and Genes
The UGT superfamily includes many isoforms with different substrate selectivity and expression [10]. Twenty-two UGTs have been identified in humans [10,11]. Almost all UGT isoforms consist of 29 conserved amino acids involved in the binding to the UDP-glucuronic acid [8]. The UGT isoforms are classified into four major families depending on the DNA sequence similarity: UGT1, 2, 3, and 8 [1]. The UGT1 and 2 families are involved mainly in xenobiotic metabolism, while the UGT3 and 8 families only metabolize endogenous compounds [12].
The UGT1 isoform genes consist of five exons. The gene sequence of the first exon is distinct, while they share the remaining four exons. Alternative splicing of the distinct first exons with the common four exons results in the synthesis of nine different isoforms of the UGT1 family; A1 and A3–10 [11]. The isoforms of the UGT2 family contain an entirely different polypeptide sequence; their isoform genes do not share common exons, as in the UGT1 isoforms. The UGT2 family is subdivided into the UGT2A and B subfamilies [13].
3. Expression of UGT Isoforms
The liver has the greatest abundance of UGT expression [14,15]. UGTs 1A1, 1A3, 1A4, 1A6, 1A9, 2B7, and 2B15 play major roles in the glucuronidation of drugs in the liver. Additionally, the UGT1A and 2B subfamilies are also expressed in the kidneys, small intestine, colon, stomach, lungs, epithelium, ovaries, testes, mammary glands, prostate, and heart [16,17]. The UGT3 family is not expressed in the liver; it is mainly expressed in the thymus, testes, and kidneys [12]. Therefore, the UGT3 family members are considered extrahepatic UGT enzymes. The UGT2B subfamily isoforms are expressed at higher rates than the UGT1A subfamily isoforms [14,15,17]. UGTs are transmembrane proteins located in the smooth endoplasmic reticulum of cells [18].
Many transcriptional factors can regulate the expression of UGT genes. Hepatocyte nuclear factors (HNFs) 1 and 4, the aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), pregnane X receptor (PXR), farnesoid X receptor (FXR), liver X receptor (LXR), and peroxisome proliferator-activated receptors (PPARs) regulate the expression of UGTs in the liver and other tissues [3,19,20]. CAR induces UGT1A1 and PXR regulates the expression of the UGT1A1, 1A3, 1A4, and 1A6 genes [21,22]. Activation of FXR upregulates UGT2B4 and downregulates UGT2B7 [23,24] and LXR induces the expression of the UGT1A3 gene [25]. PPARα regulates the expression of the UGT1A1, 1A3, 1A4, 1A6, 1A9, and 2B4 genes in a tissue-specific manner [26]. Furthermore, the UGT1A1, 1A3, 1A4, 1A6, and 1A9 genes are upregulated after the activation of AhR nuclear receptor ligands, such as polycyclic aromatic hydrocarbons [27]. Steroid hormones are regulators of UGT expression in the breast and prostate, and 19β-estradiol and dihydrotestosterone increase the expression of UGT genes responsible for glucuronidation of androgens [28]. Furthermore, Jarrar et al. (2019) showed that NSAIDs downregulated the mRNA expression of the mouse ugt2b1 gene in the liver and kidneys and upregulated the expression of ugt2b1 in the heart. However, the underlying mechanisms of how NSAIDs regulate the expression of ugt2b1 in an organ-specific manner remain to be investigated [6].
4. The Role of UGTs in Xenobiotic Metabolism
UGT1A1, 1A3, 1A4, 1A6, 1A9, and 2B7 play major roles in drug metabolism in humans [3]. UGT1A1 glucuronidates R-carvedilol [29], etoposide [30], B-estradiol [31], ezetimibe [32], and the active metabolite of irinotecan, SN-38 [33]. UGT1A3 glucuronidates ezetimibe [34] and telmisartan [35]. UGT1A4 glucuronidates amitriptyline [36], lamotrigine [37], midazolam [38], olanzapine [39], and trifluoperazine [40]. UGT1A6 metabolizes deferiprone [41] and paracetamol [42] and UGT1A9 glucuronidates propofol [43], entacapone [44], indomethacin [45], mycophenolic acid [46], and oxazepam [47]. UGT2B7 metabolizes carvedilol [29], codeine [48], diclofenac [45], epirubicin [49], flurbiprofen [45], morphine [50], naloxone [51], and zidovudine [52], while UGT2B15 glucuronidates lorazepam [53] and oxazepam [47].
Glucuronidation of certain drugs, such as cyclooxygenase (COX)-2 selective NSAIDs rofecoxib and celecoxib, requires a hydroxyl group on the drug, which is obtained through a cytochrome P450 (CYP450) oxidative reaction [54,55]. However, glucuronidation of many drugs, such as morphine, can be done without the need for the CYP450 oxidation reaction [50].
UGTs also play a role in the metabolism of phytochemical compounds. For example, glycyrrhetinic acid, which is found in licorice, is glucuronidated through UGT1A1, 1A3, 2B4, and 2B7 [56]. The hepatotoxic alkaloid senecionine is glucuronidated by UGT1A4 [57]. This herbal metabolism by UGTs forms part of the drug–herb interaction and influences the metabolism and hence the efficacy of the drugs.
5. Factors Affecting UGT Activity
Table 1 summarizes the factors that affect glucuronidation capacity, such as age, gender, diseases, and genetic variants. Owens et al. [58] found that paracetamol glucuronidation was affected by age and renal function. In another study, activity and expression of UGT1A4, the main UGT in paracetamol glucuronidation, differed widely according to age, and the maximum UGT1A4 protein levels peaked at around 20 months old [59]. The expression of UGTs in prenatal children and infants is low, possibly contributing to the susceptibility of neonates to certain drug toxicities [60].

Table 1.
Factors affecting glucuronidation capacity.
In terms of the effect of human diseases on drug glucuronidation, microsomes isolated from cirrhotic human livers showed reduced glucuronidation capacities for zidovudine and lidocaine [61]. Additionally, glucuronidation was decreased by two-fold in hepatic cancer tissues treated with the anti-hepatic cancer drug sorafenib compared to normal liver tissues, and this was associated with the decreased hepatic protein expression of UGTs [62]. Mouse ugt2b1 and ugt1a1 genes were downregulated in the liver of uncontrolled diabetic mice, but this downregulation was normalized after insulin treatment [63]. This may explain, at least in part, the reduced capacity of drug glucuronidation in diabetic patients [64].
Gender affected the drug glucuronidation of (S)-oxazepam [60,65], which was higher in males because males had higher levels of UGT2B15 activity than females.
Genetic variants in UGT genes play major roles in drugs glucuronidation. Multiple UGT genetic polymorphisms (UGT1A8*3, 1A9*3, and 2B7*2) influenced immunosuppressant mycophenolic acid glucuronidation [66]. In addition, UGT2B7*2 reportedly affected tamoxifen plasma levels [67] and was associated with diclofenac-induced hepatic toxicity [68]. Many studies showed that UGT1A1*28 significantly affected the pharmacokinetics and activities of the anticancer drug irinotecan [69,70]. Additionally, using in vitro methods, we showed that the UGT2B7*2 genetic polymorphism reduced 20-hydroxyeicosatetraenoic acid (20-HETE) glucuronidation, and this reduction was increased after incubating liver microsomes with diclofenac, which is a potent NSAID inhibitor of 20-HETE glucuronidation [71]. These results may explain one of the mechanisms underlying NSAID-induced cardiotoxicity. Figure 1 shows the roles of UGTs in the metabolism of the arachidonic acid metabolite 20-HETE.
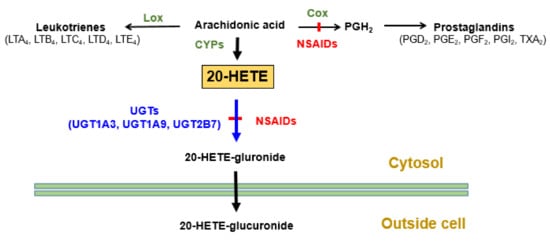
Figure 1.
Summary of enzymes affecting 20-HETE synthesis and removal. LTA4, Leukotriene A4; LOX, Lipoxygenase; COX, Cyclooxygenase; NSAIDs, Non-steroidal anti-inflammatory drugs; PGH2, Prostaglandin H2; TXA2, Thromboxane A2; HETE, Hydroxyeicosatetraenoic acid; CYP, Cytochrome P450; UGT, Uridine 5′-diphospho-glucuronosyltransferase.
Enzyme inducers and smoking can alter the glucuronidation of drugs. Rifampin, the PXR nuclear receptor agonist, decreased the plasma levels of the human immune-deficiency virus (HIV) antiviral zidovudine and accelerated its inactivation by glucuronidation [72]. The plasma levels of SN-38, the active metabolite of the anticancer drug irinotecan, were decreased by approximately 40% in smokers [73].
Collectively, many factors affect the glucuronidation of xenobiotic compounds and hence their influences on the human body. Identification of these factors can decrease xenobiotic toxicity and help to optimize drug therapies.
6. The Clinical Impact of UGT1A Genotype on Drug Response and Toxicity
UGT1A1 contains UGT genetic variants with high clinical impacts on drug responses, as illustrated by the Pharmacogenomics Knowledge Base (PharmGKB) website [74]. Patients homozygous for the UGT1A1*28/*28 genotype (rs8175347) and infected with HIV had a higher risk of hyperbilirubinemia after treatment with the protease inhibitor atazanavir [75]. These patients had a TA nucleotide inserted in the promoter region of the UGT1A1 gene that affects gene expression. The UGT1A1*28 and intronic UGT1A1*6 (rs4148323) alleles increased the likelihood of neutropenia among Asian patients treated with the anticancer drug irinotecan, compared to the wild-type UGT1A1*1 allele [76]. The intronic UGT1A1 rs34650714 T-allele reduced the metabolism of allopurinol and was associated with a decreased dose of allopurinol in patients with gout [77]. Among cardiovascular patients with angina or heart failure, the UGT1A1*6/*6 genotype had a lower capacity to glucuronidate the beta-blocker carvedilol compared with patients with the wild-type UGT1A1*1/*1 genotype [78].
The UGT1A3 rs3806596 CC genotype, with the promotor genetic T > C variant, was associated with hyperbilirubinemia in HIV patients treated with atazanavir and ritonavir [79]. In addition, the intronic UGT1A3 rs7604115 T-allele was associated with decreased concentrations of plasma montelukast levels in healthy individuals compared to those with the C-allele [80]. Further, the UGT1A3 *2 allele (rs1983023, −751T > C) increased the response to atorvastatin in healthy subjects compared to the wild-type UGT1A3 *1 allele [81]. Additionally, beta-thalassemia patients with the UGT1A3*2 TT genotype had a higher response to deferasirox, as measured by lower liver stiffness values than those with the UGT1A3*2 CC genotype [82].
Colorectal cancer patients with the UGT1A6 rs2070959 AA genotype may have an increased risk for severe neutropenia when treated with irinotecan compared to patients with the wild-type UGT1A6 rs2070959 GG genotype [83]. In addition, pediatric patients with the UGT1A6 rs6759892 GG genotype, with a substitution of serine to alanine at amino acid 7 of the UGT1A6 protein sequence, may have an increased likelihood of cardiotoxicity when treated with anticancer anthracyclines compared to patients with the wild-type UGT1A6 genotype [84]. Additionally, the UGT 1A6 rs6759892 GG genotype was associated with adverse drug reactions to deferiprone in patients with beta-thalassemia, as patients with this genotype showed a decreased metabolism of deferiprone [85].
Colorectal cancer patients who carried the non-synonymous UGT1A7*3 T > C rs11692021 allele had an increased risk of vomiting when treated with a combination of anticancer drugs S-1, irinotecan, and oxaliplatin [86].
Kidney transplant patients homozygous for the UGT1A8*2 rs1042597 CC genotype had increased diarrhea occurrences when administrated the immune suppressants mycophenolate mofetil and cyclosporine compared to patients with the heterozygous and wild-type genotypes [87]. Patients with epilepsy who had the intronic UGT1A8 rs2741049 TT genotype had a lower response to oxcarbazepine than patients with the CC genotype [88].
Non-small cell lung cancer patients with the UGT1A9 rs3832043 T9/T9 genotype, which resulted in the deletion of the thymine nucleotide in the −118 promotor sequence of the UGT1A9 gene, had decreased gene expression and hence reduced glucuronidation capacity of UGT1A9. As a result, these patients showed a reduced elimination rate of the active SN-38 metabolite of irinotecan than those with the wild-type genotype [89]. In addition, hepatotoxicity of a paracetamol overdose was increased in patients with the UGT1A9 rs8330 CC genotype [90]. This variant increases the glucuronidation of paracetamol by altering the splicing of exon 5b of UGT1A9 [90]. Table 2 summarizes the major UGT1A genetic variants that have clinical impacts on drug responses.

Table 2.
UGT1A genetic variants with reported clinical impacts on drugs responses.
7. The Clinical Impact of the UGT2B7 Genotype on Drug Responses and Toxicity
PharmGKB categorizes the UGT2B7 genetic variants within levels 3 and 4, indicating that further clinical evidence is needed before UGT2B genetic variants can be used as biomarkers for drug responses. Patients with the UGT2B7*2 (rs7439366) TT genotype had a reduced response to oxycodone and reduced requirement for codeine compared to the wild-type UGT2B7 genotype [91]. However, the UGT2B7*2 rs7439366 allele was not associated with increased morphine doses in patients with neoplasms and pain [92]. However, sickle-cell anemia patients with the promotor variant UGT2B7 rs7668282 TT genotype required a lower morphine dose because they had a higher capacity of morphine glucuronidation compared to patients with the UGT2B7 rs7668282 CC genotype [93]. Opioid-related disordered patients carrying the UGT2B7 rs7438135 GG, UGT2B7 rs6600880 TT, and UGT2B7 rs11940316 TT genotypes had reduced severity of opiate withdrawal symptoms than patients with the UGT2B7 rs7438135 AA, UGT2B7 rs6600880 AA, and UGT2B7 rs11940316 CC genotypes [94]. Epilepsy patients with the promotor UGT2B7 rs28365063 −161C > T genetic variant showed an increased clearance of the antiepileptic drug lamotrigine due to higher gene expression of the UGT2B7 enzyme [95]. However, epileptic patients with the loss-of-function UGT2B7*2 TT genotype showed an improved oxcarbazepine response due to a reduced oxcarbazepine metabolism rate [96]. The loss-of-function UGT 2B7*3 (rs12233719) G-allele was associated with increased concentrations of valproic acid in the plasma of epilepsy patients compared to patients with the UGT 2B7*3 T-allele [97]. In terms of the influence of the UGT2B7 genotype on drug-induced toxicity, the intronic UGT2B7 rs7438135 G-allele was associated with mycophenolate mofetil-induced anemia in kidney transplant patients, whereas the A-allele was not associated with drug-induced toxicity [98]. Table 3 summarizes the reported UGT2B genetic variants and the associated clinical impacts on drug responses.

Table 3.
UGT2B genetic variants with reported clinical impacts on drug responses.
8. The Role of UGTs in Endogenous Metabolism and Susceptibility to Human Diseases
UGTs also play a role in the metabolism of endogenous chemicals, including steroids and unsaturated long-chain fatty acids [99,100]. Besides serving as a substrate for UGTs, these endogenous compounds can also inhibit UGTs. Unsaturated long-chain fatty acids are the most potent inhibitors of several UGT enzymes, including UGT1A3, 1A9, and 2B7 [101]. UGT2B7 metabolizes dietary fatty acids and show inter-individual variations in the glucuronidation of these fatty acids in the intestines [102].
Bile acids are common endogenous compounds that undergo glucuronidation. Bile acids are glucuronidated in different human body tissues, but especially in the liver [103]. Biliary glucuronidation is an important pathway in the excretion of bile acids, and impaired biliary secretion leads to hyperbilirubinemia [104]. The UGT2B7*2 genetic variant possibly changes the glucuronidation of chenodeoxycholic acid, affecting the health of individuals [103]. Steroidal hormones are further examples of endogenous compounds that undergo glucuronidation. Sex hormones, thyroxin, and retinoic acid are glucuronidated in different organs [105,106]. Additionally, the UGT1A1*28 genetic variant is associated with plasma estrogen levels in women with breast cancer [107]. Turgeon et al. [108] showed that leukotriene B4 was glucuronidated by UGT1A1, 1A3, 1A4, and 2B7, whereas UGT1A1, 1A3, 1A4, and 1A9 also conjugated most of the HETEs. In addition, the UGT2 family members, especially UGT2B4 and 2B7, conjugated all HETEs. The author suggested that glucuronidation of arachidonic acid metabolites is an irreversible step to inactivate and eliminate endogenous arachidonic acid metabolites from the body. In another in vitro study, arachidonic acid was glucuronidated by UGT1A1, 1A3, 1A4, 1A9, and 1A10, whereas prostaglandin B1 was glucuronidated by UGT1A1, 1A9, and 1A10. All of the arachidonic acid metabolites were glucuronidated by UGT2B7, and arachidonic acid and 20-HETE were the best substrates [109]. UGT1A1, 1A3, 1A9, and 2B7 also glucuronidated 20-HETE [109]. Multiple UGT isoforms are involved in the glucuronidation of arachidonic acid and its metabolites, and these have different enzymatic affinities and maximum capacity rates. As a result, it might be expected that the inhibition or alteration of specific UGT isoforms, such as UGT2B7, have a more significant effect on certain arachidonic acid metabolites, such as 20-HETE and 15-HETE, than other arachidonic acid metabolites. The changes in the arachidonic acid metabolite ratio might affect human homeostasis and lead to a predisposition to certain diseases. Interestingly, one study screened the metabolic and endogenous plasma metabolite changes in human volunteers following administration of diclofenac, a potent UGT2B7 substrate. The results showed that plasma cardiotoxic 20-HETE was significantly increased compared to other endogenous metabolites [110]. In addition, morphine, a strong UGT2B7 inhibitor, altered the arachidonic acid metabolism and activity in a vascular in vitro system [111]. Arachidonic acid metabolites such as 20-HETE were excreted in the glucuronidated form in human urine [112]. Interestingly, the UGT substrate indomethacin reduced urinary 20-HETE levels [113]. It is suggested that inhibition of arachidonic acid-UGT metabolizing enzymes might be one mechanism underlying NSAID-induced hepato- and nephrotoxicity [114]. We showed previously that NSAIDs and the UGT2B7*2 genetic variant inhibited the glucuronidation of 20-HETE [45]. Additionally, NSAIDs inhibited the in vitro glucuronidation of the endogenous hypertensive aldosterone [115] and blood levels of aldosterone increased following NSAID treatment, especially diclofenac and celecoxib [116,117,118]. The increase in aldosterone correlated with decreased aldosterone-glucuronide levels in the urine. Furthermore, in vitro methods showed that diclofenac inhibited testosterone glucuronidation and potentially increased testosterone plasma levels, leading to a hormonal imbalance [119]. The anticonvulsant valproic acid, which causes hormonal imbalance [120], inhibited the endogenous steroidal glucuronidation by inhibiting UGT2B15, the enzyme responsible for steroid metabolism [121]. These data indicate that chemical inhibition of UGTs or loss-of-function genetic variants in the UGT genes can contribute to human disease susceptibility by increasing levels of harmful non-metabolized fatty acids in the plasma, such as 20-HETE. Table 4 summarizes the possible mechanisms of drug-induced toxicity involved in the inhibition of endogenous glucuronidation.

Table 4.
Mechanisms of drug-induced toxicity involved in the inhibition of endogenous glucuronidation.
9. Conclusions
Chemical inhibition and genetic variants of the UGT genes play important roles in the drug response, toxicity, and susceptibility to human diseases. However, clinical evidence has shown that the UGT1A1 isoform genetic variants can be considered biomarkers for drug responses and susceptibility to diseases. Additionally, inhibition of endogenous glucuronidation can lead to an imbalance in the levels of endogenous fatty acids and steroidal hormones and cause human diseases. Further clinical studies are needed to validate the clinical impacts of the UGT1A and UGT2B genes for personalized medicine and human diseases.
Author Contributions
Supervision, S.-J.L.; Writing—original draft, Y.J. and S.-J.L. Both authors have read and agreed to the published version of the manuscript.
Funding
The article was supported by grants from the National Research Foundation of Korea funded by the Korean government (NRF-2020R1I1A3073778) and the National Research Foundation of Korea funded by Korea (MIST) (number 2018R1A5A2021242).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tukey, R.H.; Strassburg, C.P. Human UDP-glucuronosyltransferases: Metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 581–616. [Google Scholar] [CrossRef]
- Miners, J.O.; Mackenzie, P.I.; Knights, K.M. The prediction of drug-glucuronidation parameters in humans: UDP-glucuronosyltransferase enzyme-selective substrate and inhibitor probes for reaction phenotyping and in vitro-in vivo extrapolation of drug clearance and drug-drug interaction potential. Drug Metab. Rev. 2010, 42, 196–208. [Google Scholar] [CrossRef]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef]
- Perreault, M.; Wunsch, E.; Bialek, A.; Trottier, J.; Verreault, M.; Caron, P.; Poirier, G.G.; Milkiewicz, P.; Barbier, O. Urinary Elimination of Bile Acid Glucuronides under Severe Cholestatic Situations: Contribution of Hepatic and Renal Glucuronidation Reactions. Can. J. Gastroenterol. Hepatol. 2018, 2018, 8096314. [Google Scholar] [CrossRef]
- Klimas, R.; Mikus, G. Morphine-6-glucuronide is responsible for the analgesic effect after morphine administration: A quantitative review of morphine, morphine-6-glucuronide, and morphine-3-glucuronide. Br. J. Anaesth. 2014, 113, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Jarrar, Y.; Jarrar, Q.; Abu-Shalhoob, M.; Abed, A.; Sha’ban, E. Relative Expression of Mouse Udp-glucuronosyl Transferase 2b1 Gene in the Livers, Kidneys, and Hearts: The Influence of Nonsteroidal Anti-inflammatory Drug Treatment. Curr. Drug Metab. 2019, 20, 918–923. [Google Scholar] [CrossRef] [PubMed]
- Mays, D.C.; Dixon, K.F.; Balboa, A.; Pawluk, L.J.; Bauer, M.R.; Nawoot, S.; Gerber, N. A nonprimate animal model applicable to zidovudine pharmacokinetics in humans: Inhibition of glucuronidation and renal excretion of zidovudine by probenecid in rats. J. Pharmacol. Exp. Ther. 1991, 259, 1261–1270. [Google Scholar] [PubMed]
- Oguri, K.; Hanioka, N.; Yoshimura, H. Species differences in metabolism of codeine: Urinary excretion of codeine glucuronide, morphine-3-glucuronide and morphine-6-glucuronide in mice, rats, guinea pigs and rabbits. Xenobiotica 1990, 20, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Court, M.H. Feline drug metabolism and disposition: Pharmacokinetic evidence for species differences and molecular mechanisms. Vet. Clin. N. Am. Small Anim. Pract. 2013, 43, 1039–1054. [Google Scholar] [CrossRef]
- Miners, J.O.; Smith, P.A.; Sorich, M.J.; McKinnon, R.A.; Mackenzie, P.I. Predicting human drug glucuronidation parameters: Application of in vitro and in silico modeling approaches. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 1–25. [Google Scholar] [CrossRef]
- Mackenzie, P.I.; Bock, K.W.; Burchell, B.; Guillemette, C.; Ikushiro, S.; Iyanagi, T.; Miners, J.O.; Owens, I.S.; Nebert, D.W. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 2005, 15, 677–685. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, P.I.; Rogers, A.; Elliot, D.J.; Chau, N.; Hulin, J.A.; Miners, J.O.; Meech, R. The novel UDP glycosyltransferase 3A2: Cloning, catalytic properties, and tissue distribution. Mol. Pharmacol. 2011, 79, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Kiang, T.K.; Ensom, M.H.; Chang, T.K. UDP-glucuronosyltransferases and clinical drug-drug interactions. Pharmacol. Ther. 2005, 106, 97–132. [Google Scholar] [CrossRef] [PubMed]
- Court, M.H.; Zhang, X.; Ding, X.; Yee, K.K.; Hesse, L.M.; Finel, M. Quantitative distribution of mRNAs encoding the 19 human UDP-glucuronosyltransferase enzymes in 26 adult and 3 fetal tissues. Xenobiotica 2012, 42, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Izukawa, T.; Nakajima, M.; Fujiwara, R.; Yamanaka, H.; Fukami, T.; Takamiya, M.; Aoki, Y.; Ikushiro, S.; Sakaki, T.; Yokoi, T. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab. Dispos. 2009, 37, 1759–1768. [Google Scholar] [CrossRef]
- Gaganis, P.; Miners, J.O.; Knights, K.M. Glucuronidation of fenamates: Kinetic studies using human kidney cortical microsomes and recombinant UDP-glucuronosyltransferase (UGT) 1A9 and 2B7. Biochem. Pharmacol. 2007, 73, 1683–1691. [Google Scholar] [CrossRef]
- Ohno, S.; Nakajin, S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab. Dispos. 2009, 37, 32–40. [Google Scholar] [CrossRef]
- Radominska-Pandya, A.; Czernik, P.J.; Little, J.M.; Battaglia, E.; Mackenzie, P.I. Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab. Rev. 1999, 31, 817–899. [Google Scholar] [CrossRef]
- Tien, E.S.; Negishi, M. Nuclear receptors CAR and PXR in the regulation of hepatic metabolism. Xenobiotica 2006, 36, 1152–1163. [Google Scholar] [CrossRef]
- Mackenzie, P.I.; Hu, D.G.; Gardner-Stephen, D.A. The regulation of UDP-glucuronosyltransferase genes by tissue-specific and ligand-activated transcription factors. Drug Metab. Rev. 2010, 42, 99–109. [Google Scholar] [CrossRef]
- Sugatani, J.; Nishitani, S.; Yamakawa, K.; Yoshinari, K.; Sueyoshi, T.; Negishi, M.; Miwa, M. Transcriptional regulation of human UGT1A1 gene expression: Activated glucocorticoid receptor enhances constitutive androstane receptor/pregnane X receptor-mediated UDP-glucuronosyltransferase 1A1 regulation with glucocorticoid receptor-interacting protein 1. Mol. Pharmacol. 2005, 67, 845–855. [Google Scholar] [PubMed]
- Gardner-Stephen, D.; Heydel, J.M.; Goyal, A.; Lu, Y.; Xie, W.; Lindblom, T.; Mackenzie, P.; Radominska-Pandya, A. Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab. Dispos. 2004, 32, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Barbier, O.; Duran-Sandoval, D.; Pineda-Torra, I.; Kosykh, V.; Fruchart, J.C.; Staels, B. Peroxisome proliferator-activated receptor alpha induces hepatic expression of the human bile acid glucuronidating UDP-glucuronosyltransferase 2B4 enzyme. J. Biol. Chem. 2003, 278, 32852–32860. [Google Scholar] [CrossRef]
- Lu, Y.; Heydel, J.M.; Li, X.; Bratton, S.; Lindblom, T.; Radominska-Pandya, A. Lithocholic acid decreases expression of UGT2B7 in Caco-2 cells: A potential role for a negative farnesoid X receptor response element. Drug Metab. Dispos. 2005, 33, 937–946. [Google Scholar] [CrossRef]
- Verreault, M.; Senekeo-Effenberger, K.; Trottier, J.; Bonzo, J.A.; Belanger, J.; Kaeding, J.; Staels, B.; Caron, P.; Tukey, R.H.; Barbier, O. The liver X-receptor alpha controls hepatic expression of the human bile acid-glucuronidating UGT1A3 enzyme in human cells and transgenic mice. Hepatology 2006, 44, 368–378. [Google Scholar] [CrossRef]
- Senekeo-Effenberger, K.; Chen, S.; Brace-Sinnokrak, E.; Bonzo, J.A.; Yueh, M.F.; Argikar, U.; Kaeding, J.; Trottier, J.; Remmel, R.P.; Ritter, J.K.; et al. Expression of the human UGT1 locus in transgenic mice by 4-chloro-6-(2,3-xylidino)-2-pyrimidinylthioacetic acid (WY-14643) and implications on drug metabolism through peroxisome proliferator-activated receptor alpha activation. Drug Metab. Dispos. 2007, 35, 419–427. [Google Scholar] [CrossRef]
- Lankisch, T.O.; Gillman, T.C.; Erichsen, T.J.; Ehmer, U.; Kalthoff, S.; Freiberg, N.; Munzel, P.A.; Manns, M.P.; Strassburg, C.P. Aryl hydrocarbon receptor-mediated regulation of the human estrogen and bile acid UDP-glucuronosyltransferase 1A3 gene. Arch. Toxicol. 2008, 82, 573–582. [Google Scholar] [CrossRef]
- Hu, D.G.; Mackenzie, P.I. Estrogen receptor alpha, fos-related antigen-2, and c-Jun coordinately regulate human UDP glucuronosyltransferase 2B15 and 2B17 expression in response to 17beta-estradiol in MCF-7 cells. Mol. Pharmacol. 2009, 76, 425–439. [Google Scholar] [CrossRef]
- Ohno, A.; Saito, Y.; Hanioka, N.; Jinno, H.; Saeki, M.; Ando, M.; Ozawa, S.; Sawada, J. Involvement of human hepatic UGT1A1, UGT2B4, and UGT2B7 in the glucuronidation of carvedilol. Drug Metab. Dispos. 2004, 32, 235–239. [Google Scholar] [CrossRef]
- Watanabe, Y.; Nakajima, M.; Ohashi, N.; Kume, T.; Yokoi, T. Glucuronidation of etoposide in human liver microsomes is specifically catalyzed by UDP-glucuronosyltransferase 1A1. Drug Metab. Dispos. 2003, 31, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tracy, T.S.; Remmel, R.P. Correlation between bilirubin glucuronidation and estradiol-3-gluronidation in the presence of model UDP-glucuronosyltransferase 1A1 substrates/inhibitors. Drug Metab. Dispos. 2011, 39, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Nguyen, N.; Peterkin, V.; Yang, Y.S.; Hotz, K.; La Placa, D.B.; Chen, S.; Tukey, R.H.; Stevens, J.C. A humanized UGT1 mouse model expressing the UGT1A1*28 allele for assessing drug clearance by UGT1A1-dependent glucuronidation. Drug Metab. Dispos. 2010, 38, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Ando, Y.; Saka, H.; Asai, G.; Sugiura, S.; Shimokata, K.; Kamataki, T. UGT1A1 genotypes and glucuronidation of SN-38, the active metabolite of irinotecan. Ann. Oncol. 1998, 9, 845–847. [Google Scholar] [CrossRef]
- Ghosal, A.; Hapangama, N.; Yuan, Y.; Achanfuo-Yeboah, J.; Iannucci, R.; Chowdhury, S.; Alton, K.; Patrick, J.E.; Zbaida, S. Identification of human UDP-glucuronosyltransferase enzyme(s) responsible for the glucuronidation of ezetimibe (Zetia). Drug Metab. Dispos. 2004, 32, 314–320. [Google Scholar] [CrossRef]
- Gill, K.L.; Houston, J.B.; Galetin, A. Characterization of in vitro glucuronidation clearance of a range of drugs in human kidney microsomes: Comparison with liver and intestinal glucuronidation and impact of albumin. Drug Metab. Dispos. 2012, 40, 825–835. [Google Scholar] [CrossRef]
- Breyer-Pfaff, U.; Mey, U.; Green, M.D.; Tephly, T.R. Comparative N-glucuronidation kinetics of ketotifen and amitriptyline by expressed human UDP-glucuronosyltransferases and liver microsomes. Drug Metab. Dispos. 2000, 28, 869–872. [Google Scholar] [PubMed]
- Rowland, A.; Elliot, D.J.; Williams, J.A.; Mackenzie, P.I.; Dickinson, R.G.; Miners, J.O. In vitro characterization of lamotrigine N2-glucuronidation and the lamotrigine-valproic acid interaction. Drug Metab. Dispos. 2006, 34, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Hyland, R.; Osborne, T.; Payne, A.; Kempshall, S.; Logan, Y.R.; Ezzeddine, K.; Jones, B. In vitro and in vivo glucuronidation of midazolam in humans. Br. J. Clin. Pharmacol. 2009, 67, 445–454. [Google Scholar] [CrossRef]
- Haslemo, T.; Loryan, I.; Ueda, N.; Mannheimer, B.; Bertilsson, L.; Ingelman-Sundberg, M.; Molden, E.; Eliasson, E. UGT1A4*3 encodes significantly increased glucuronidation of olanzapine in patients on maintenance treatment and in recombinant systems. Clin. Pharmacol. Ther. 2012, 92, 221–227. [Google Scholar] [CrossRef]
- Uchaipichat, V.; Mackenzie, P.I.; Elliot, D.J.; Miners, J.O. Selectivity of substrate (trifluoperazine) and inhibitor (amitriptyline, androsterone, canrenoic acid, hecogenin, phenylbutazone, quinidine, quinine, and sulfinpyrazone) "probes" for human udp-glucuronosyltransferases. Drug Metab. Dispos. 2006, 34, 449–456. [Google Scholar] [CrossRef]
- Benoit-Biancamano, M.O.; Connelly, J.; Villeneuve, L.; Caron, P.; Guillemette, C. Deferiprone glucuronidation by human tissues and recombinant UDP glucuronosyltransferase 1A6: An in vitro investigation of genetic and splice variants. Drug Metab. Dispos. 2009, 37, 322–329. [Google Scholar] [CrossRef]
- Kostrubsky, S.E.; Sinclair, J.F.; Strom, S.C.; Wood, S.; Urda, E.; Stolz, D.B.; Wen, Y.H.; Kulkarni, S.; Mutlib, A. Phenobarbital and phenytoin increased acetaminophen hepatotoxicity due to inhibition of UDP-glucuronosyltransferases in cultured human hepatocytes. Toxicol. Sci. 2005, 87, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Matsumoto, Y.; Yamazaki, H. Effects of propofol analogs on glucuronidation of propofol, an anesthetic drug, by human liver microsomes. Drug Metab. Lett. 2007, 1, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Lautala, P.; Ethell, B.T.; Taskinen, J.; Burchell, B. The specificity of glucuronidation of entacapone and tolcapone by recombinant human UDP-glucuronosyltransferases. Drug Metab. Dispos. 2000, 28, 1385–1389. [Google Scholar]
- Kuehl, G.E.; Lampe, J.W.; Potter, J.D.; Bigler, J. Glucuronidation of nonsteroidal anti-inflammatory drugs: Identifying the enzymes responsible in human liver microsomes. Drug Metab. Dispos. 2005, 33, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Bernard, O.; Guillemette, C. The main role of UGT1A9 in the hepatic metabolism of mycophenolic acid and the effects of naturally occurring variants. Drug Metab. Dispos. 2004, 32, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Court, M.H.; Duan, S.X.; Guillemette, C.; Journault, K.; Krishnaswamy, S.; Von Moltke, L.L.; Greenblatt, D.J. Stereoselective conjugation of oxazepam by human UDP-glucuronosyltransferases (UGTs): S-oxazepam is glucuronidated by UGT2B15, while R-oxazepam is glucuronidated by UGT2B7 and UGT1A9. Drug Metab. Dispos. 2002, 30, 1257–1265. [Google Scholar] [CrossRef]
- Court, M.H.; Krishnaswamy, S.; Hao, Q.; Duan, S.X.; Patten, C.J.; Von Moltke, L.L.; Greenblatt, D.J. Evaluation of 3’-azido-3’-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: Specificity and influence of the UGT2B7*2 polymorphism. Drug Metab. Dispos. 2003, 31, 1125–1133. [Google Scholar] [CrossRef]
- Innocenti, F.; Iyer, L.; Ramirez, J.; Green, M.D.; Ratain, M.J. Epirubicin glucuronidation is catalyzed by human UDP-glucuronosyltransferase 2B7. Drug Metab. Dispos. 2001, 29, 686–692. [Google Scholar]
- Coffman, B.L.; Rios, G.R.; King, C.D.; Tephly, T.R. Human UGT2B7 catalyzes morphine glucuronidation. Drug Metab. Dispos. 1997, 25, 1–4. [Google Scholar]
- Di Marco, A.; D’Antoni, M.; Attaccalite, S.; Carotenuto, P.; Laufer, R. Determination of drug glucuronidation and UDP-glucuronosyltransferase selectivity using a 96-well radiometric assay. Drug Metab. Dispos. 2005, 33, 812–819. [Google Scholar] [CrossRef]
- Barbier, O.; Turgeon, D.; Girard, C.; Green, M.D.; Tephly, T.R.; Hum, D.W.; Belanger, A. 3’-azido-3’-deoxythimidine (AZT) is glucuronidated by human UDP-glucuronosyltransferase 2B7 (UGT2B7). Drug Metab. Dispos. 2000, 28, 497–502. [Google Scholar]
- Uchaipichat, V.; Suthisisang, C.; Miners, J.O. The glucuronidation of R- and S-lorazepam: Human liver microsomal kinetics, UDP-glucuronosyltransferase enzyme selectivity, and inhibition by drugs. Drug Metab. Dispos. 2013, 41, 1273–1284. [Google Scholar] [CrossRef]
- Paulson, S.K.; Hribar, J.D.; Liu, N.W.; Hajdu, E.; Bible, R.H., Jr.; Piergies, A.; Karim, A. Metabolism and excretion of [(14)C]celecoxib in healthy male volunteers. Drug Metab. Dispos. 2000, 28, 308–314. [Google Scholar]
- Zhang, J.Y.; Zhan, J.; Cook, C.S.; Ings, R.M.; Breau, A.P. Involvement of human UGT2B7 and 2B15 in rofecoxib metabolism. Drug Metab. Dispos. 2003, 31, 652–658. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, J.; Chen, X.; Li, N.; Fu, F.; He, J.; Wang, G.; Zhang, L.; Zheng, Y.; Qiu, Z.; et al. Identification of human UDP-glucuronosyltransferase isoforms responsible for the glucuronidation of glycyrrhetinic acid. Drug Metab. Pharmacokinet. 2009, 24, 523–528. [Google Scholar] [CrossRef]
- He, Y.Q.; Liu, Y.; Zhang, B.F.; Liu, H.X.; Lu, Y.L.; Yang, L.; Xiong, A.Z.; Xu, L.L.; Wang, C.H.; Yang, L.; et al. Identification of the UDP-glucuronosyltransferase isozyme involved in senecionine glucuronidation in human liver microsomes. Drug Metab. Dispos. 2010, 38, 626–634. [Google Scholar] [CrossRef]
- Owens, K.H.; Murphy, P.G.; Medlicott, N.J.; Kennedy, J.; Zacharias, M.; Curran, N.; Sreebhavan, S.; Thompson-Fawcett, M.; Reith, D.M. Population pharmacokinetics of intravenous acetaminophen and its metabolites in major surgical patients. J. Pharmacokinet. Pharmacodyn. 2014, 41, 211–221. [Google Scholar] [CrossRef]
- Miyagi, S.J.; Collier, A.C. Pediatric development of glucuronidation: The ontogeny of hepatic UGT1A4. Drug Metab. Dispos. 2007, 35, 1587–1592. [Google Scholar] [CrossRef]
- Court, M.H. Interindividual variability in hepatic drug glucuronidation: Studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system. Drug Metab. Rev. 2009, 42, 209–224. [Google Scholar] [CrossRef]
- Furlan, V.; Demirdjian, S.; Bourdon, O.; Magdalou, J.; Taburet, A.M. Glucuronidation of drugs by hepatic microsomes derived from healthy and cirrhotic human livers. J. Pharmacol. Exp. Ther. 1999, 289, 1169–1175. [Google Scholar]
- Ye, L.; Yang, X.; Guo, E.; Chen, W.; Lu, L.; Wang, Y.; Peng, X.; Yan, T.; Zhou, F.; Liu, Z. Sorafenib metabolism is significantly altered in the liver tumor tissue of hepatocellular carcinoma patient. PLoS ONE 2014, 9, e96664. [Google Scholar] [CrossRef]
- Jarrar, Y.B.; Al-Essa, L.; Kilani, A.; Hasan, M.; Al-Qerem, W. Alterations in the gene expression of drug and arachidonic acid-metabolizing Cyp450 in the livers of controlled and uncontrolled insulin-dependent diabetic mice. Diabetes Metab. Syndr. Obes. 2018, 11, 483–492. [Google Scholar] [CrossRef]
- Dostalek, M.; Court, M.H.; Hazarika, S.; Akhlaghi, F. Diabetes mellitus reduces activity of human UDP-glucuronosyltransferase 2B7 in liver and kidney leading to decreased formation of mycophenolic acid acyl-glucuronide metabolite. Drug Metab. Dispos. 2011, 39, 448–455. [Google Scholar] [CrossRef]
- Gallagher, C.J.; Balliet, R.M.; Sun, D.; Chen, G.; Lazarus, P. Sex differences in UDP-glucuronosyltransferase 2B17 expression and activity. Drug Metab. Dispos. 2010, 38, 2204–2209. [Google Scholar] [CrossRef]
- Picard, N.; Ratanasavanh, D.; Premaud, A.; Le Meur, Y.; Marquet, P. Identification of the UDP-glucuronosyltransferase isoforms involved in mycophenolic acid phase II metabolism. Drug Metab. Dispos. 2005, 33, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Rae, J.M.; Drury, S.; Hayes, D.F.; Stearns, V.; Thibert, J.N.; Haynes, B.P.; Salter, J.; Sestak, I.; Cuzick, J.; Dowsett, M. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012, 104, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.K.; Aithal, G.P.; Leathart, J.B.; Swainsbury, R.A.; Dang, T.S.; Day, C.P. Genetic susceptibility to diclofenac-induced hepatotoxicity: Contribution of UGT2B7, CYP2C8, and ABCC2 genotypes. Gastroenterology 2007, 132, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.; Das, S.; Janisch, L.; Wen, M.; Ramirez, J.; Karrison, T.; Fleming, G.F.; Vokes, E.E.; Schilsky, R.L.; Ratain, M.J. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002, 2, 43–47. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, D.; Kuang, Q.; Liu, G.; Xu, W. Association between UGT1A1*28 polymorphisms and clinical outcomes of irinotecan-based chemotherapies in colorectal cancer: A meta-analysis in Caucasians. PLoS ONE 2013, 8, e58489. [Google Scholar]
- Jarrar, Y.B.; Kim, D.H.; Lee, S.J.; Shin, J.G. Inhibition of 20-hydroxyeicosatetraenoic acid (20-HETE) glucuronidation by non-steroidal anti-inflammatory drugs in human liver microsomes and recombinant UDP-glucuronosyltransferase enzymes. Prostaglandins Leukot. Essent. Fatty Acids 2020, 153, 102055. [Google Scholar] [CrossRef] [PubMed]
- Burger, D.M.; Meenhorst, P.L.; Koks, C.H.; Beijnen, J.H. Pharmacokinetic interaction between rifampin and zidovudine. Antimicrob. Agents Chemother. 1993, 37, 1426–1431. [Google Scholar] [CrossRef]
- Benowitz, N.L. Cigarette smoking and the personalization of irinotecan therapy. J. Clin. Oncol. 2007, 25, 2646–2647. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; McDonagh, E.M.; Hebert, J.M.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Altman, R.B.; Klein, T.E. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012, 92, 414–417. [Google Scholar] [CrossRef]
- Choe, P.G.; Park, W.B.; Song, J.S.; Kim, N.H.; Song, K.H.; Park, S.W.; Kim, H.B.; Kim, N.J.; Oh, M.D. Incidence of atazanavir-associated hyperbilirubinemia in Korean HIV patients: 30 months follow-up results in a population with low UDP-glucuronosyltransferase1A1*28 allele frequency. J. Korean Med. Sci. 2010, 25, 1427–1430. [Google Scholar] [CrossRef]
- Onoue, M.; Terada, T.; Kobayashi, M.; Katsura, T.; Matsumoto, S.; Yanagihara, K.; Nishimura, T.; Kanai, M.; Teramukai, S.; Shimizu, A.; et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int. J. Clin. Oncol. 2009, 14, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.B.; Smith, D.M.; Shaak, T.L. Genomic sequencing of uric acid metabolizing and clearing genes in relationship to xanthine oxidase inhibitor dose. Rheumatol. Int. 2017, 37, 445–453. [Google Scholar] [CrossRef]
- Takekuma, Y.; Takenaka, T.; Kiyokawa, M.; Yamazaki, K.; Okamoto, H.; Kitabatake, A.; Tsutsui, H.; Sugawara, M. Contribution of polymorphisms in UDP-glucuronosyltransferase and CYP2D6 to the individual variation in disposition of carvedilol. J. Pharm. Pharm. Sci. 2006, 9, 101–112. [Google Scholar]
- Lankisch, T.O.; Moebius, U.; Wehmeier, M.; Behrens, G.; Manns, M.P.; Schmidt, R.E.; Strassburg, C.P. Gilbert’s disease and atazanavir: From phenotype to UDP-glucuronosyltransferase haplotype. Hepatology 2006, 44, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Hirvensalo, P.; Tornio, A.; Neuvonen, M.; Tapaninen, T.; Paile-Hyvarinen, M.; Karja, V.; Mannisto, V.T.; Pihlajamaki, J.; Backman, J.T.; Niemi, M. Comprehensive Pharmacogenomic Study Reveals an Important Role of UGT1A3 in Montelukast Pharmacokinetics. Clin. Pharmacol. Ther. 2018, 104, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Oh, E.S.; Park, K.; Park, M.S.; Chung, J.Y. The UGT1A3*2 polymorphism affects atorvastatin lactonization and lipid-lowering effect in healthy volunteers. Pharmacogenet Genomics 2012, 22, 598–605. [Google Scholar] [CrossRef]
- Allegra, S.; Cusato, J.; De Francia, S.; Longo, F.; Pirro, E.; Massano, D.; Avataneo, V.; De Nicolo, A.; Piga, A.; D’Avolio, A. The effect of vitamin D pathway genes and deferasirox pharmacogenetics on liver iron in thalassaemia major patients. Pharmacogenomics J. 2019, 19, 417–427. [Google Scholar] [CrossRef]
- Levesque, E.; Belanger, A.S.; Harvey, M.; Couture, F.; Jonker, D.; Innocenti, F.; Cecchin, E.; Toffoli, G.; Guillemette, C. Refining the UGT1A haplotype associated with irinotecan-induced hematological toxicity in metastatic colorectal cancer patients treated with 5-fluorouracil/irinotecan-based regimens. J. Pharmacol. Exp. Ther. 2013, 345, 95–101. [Google Scholar] [CrossRef]
- Visscher, H.; Ross, C.J.; Rassekh, S.R.; Sandor, G.S.; Caron, H.N.; van Dalen, E.C.; Kremer, L.C.; van der Pal, H.J.; Rogers, P.C.; Rieder, M.J.; et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr. Blood Cancer 2013, 60, 1375–1381. [Google Scholar] [CrossRef]
- Dadheech, S.; Rao, A.V.; Shaheen, U.; Hussien, M.D.; Jain, S.; Jyothy, A.; Munshi, A. Three most common nonsynonymous UGT1A6*2 polymorphisms (Thr181Ala, Arg184Ser and Ser7Ala) and therapeutic response to deferiprone in beta-thalassemia major patients. Gene 2013, 531, 301–305. [Google Scholar] [CrossRef]
- Kim, S.Y.; Hong, Y.S.; Shim, E.K.; Kong, S.Y.; Shin, A.; Baek, J.Y.; Jung, K.H. S-1 plus irinotecan and oxaliplatin for the first-line treatment of patients with metastatic colorectal cancer: A prospective phase II study and pharmacogenetic analysis. Br. J. Cancer 2013, 109, 1420–1427. [Google Scholar] [CrossRef] [PubMed]
- Woillard, J.B.; Rerolle, J.P.; Picard, N.; Rousseau, A.; Drouet, M.; Munteanu, E.; Essig, M.; Marquet, P.; Le Meur, Y. Risk of diarrhoea in a long-term cohort of renal transplant patients given mycophenolate mofetil: The significant role of the UGT1A8 2 variant allele. Br. J. Clin. Pharmacol. 2010, 69, 675–683. [Google Scholar] [CrossRef]
- Lu, Y.; Fang, Y.; Wu, X.; Ma, C.; Wang, Y.; Xu, L. Effects of UGT1A9 genetic polymorphisms on monohydroxylated derivative of oxcarbazepine concentrations and oxcarbazepine monotherapeutic efficacy in Chinese patients with epilepsy. Eur. J. Clin. Pharmacol. 2017, 73, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Lim, H.S.; Park, Y.H.; Lee, S.Y.; Lee, J.S. Integrated pharmacogenetic prediction of irinotecan pharmacokinetics and toxicity in patients with advanced non-small cell lung cancer. Lung Cancer 2009, 63, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Court, M.H.; Freytsis, M.; Wang, X.; Peter, I.; Guillemette, C.; Hazarika, S.; Duan, S.X.; Greenblatt, D.J.; Lee, W.M.; Acute Liver Failure Study Group. The UDP-glucuronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A exon 5a/5b splice variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure. J. Pharmacol. Exp. Ther. 2013, 345, 297–307. [Google Scholar]
- Li, J.; Peng, P.; Mei, Q.; Xia, S.; Tian, Y.; Hu, L.; Chen, Y. The impact of UGT2B7 C802T and CYP3A4*1G polymorphisms on pain relief in cancer patients receiving oxycontin. Support Care Cancer 2018, 26, 2763–2767. [Google Scholar] [CrossRef] [PubMed]
- Ning, M.; Tao, Y.; Hu, X.; Guo, L.; Ni, J.; Hu, J.; Shen, H.; Chen, Y. Roles of UGT2B7 C802T gene polymorphism on the efficacy of morphine treatment on cancer pain among the Chinese han population. Niger. J. Clin. Pract. 2019, 22, 1319–1323. [Google Scholar]
- Darbari, D.S.; van Schaik, R.H.; Capparelli, E.V.; Rana, S.; McCarter, R.; van den Anker, J. UGT2B7 promoter variant -840G>A contributes to the variability in hepatic clearance of morphine in patients with sickle cell disease. Am. J. Hematol. 2008, 83, 200–202. [Google Scholar] [CrossRef]
- Tian, J.N.; Ho, I.K.; Tsou, H.H.; Fang, C.P.; Hsiao, C.F.; Chen, C.H.; Tan, H.K.; Lin, L.; Wu, C.S.; Su, L.W.; et al. UGT2B7 genetic polymorphisms are associated with the withdrawal symptoms in methadone maintenance patients. Pharmacogenomics 2012, 13, 879–888. [Google Scholar] [CrossRef]
- Singkham, N.; Towanabut, S.; Lertkachatarn, S.; Punyawudho, B. Influence of the UGT2B7 -161C>T polymorphism on the population pharmacokinetics of lamotrigine in Thai patients. Eur. J. Clin. Pharmacol. 2013, 69, 1285–1291. [Google Scholar] [CrossRef]
- Ma, C.L.; Wu, X.Y.; Jiao, Z.; Hong, Z.; Wu, Z.Y.; Zhong, M.K. SCN1A, ABCC2 and UGT2B7 gene polymorphisms in association with individualized oxcarbazepine therapy. Pharmacogenomics 2015, 16, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lin, X.Q.; Cai, W.K.; Xu, G.L.; Zhou, M.D.; Yang, M.; He, G.H. Effect of UGT2B7 genotypes on plasma concentration of valproic acid: A meta-analysis. Eur. J. Clin. Pharmacol. 2018, 74, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Woillard, J.B.; Picard, N.; Thierry, A.; Touchard, G.; Marquet, P. Associations between polymorphisms in target, metabolism, or transport proteins of mycophenolate sodium and therapeutic or adverse effects in kidney transplant patients. Pharmacogenet Genomics 2014, 24, 256–262. [Google Scholar] [CrossRef][Green Version]
- Knights, K.M.; Miners, J.O. Renal UDP-glucuronosyltransferases and the glucuronidation of xenobiotics and endogenous mediators. Drug Metab. Rev. 2010, 42, 63–73. [Google Scholar] [CrossRef]
- Bowalgaha, K.; Elliot, D.J.; Mackenzie, P.I.; Knights, K.M.; Miners, J.O. The glucuronidation of Delta4-3-Keto C19- and C21-hydroxysteroids by human liver microsomal and recombinant UDP-glucuronosyltransferases (UGTs): 6alpha- and 21-hydroxyprogesterone are selective substrates for UGT2B7. Drug Metab. Dispos. 2007, 35, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Tsoutsikos, P.; Miners, J.O.; Stapleton, A.; Thomas, A.; Sallustio, B.C.; Knights, K.M. Evidence that unsaturated fatty acids are potent inhibitors of renal UDP-glucuronosyltransferases (UGT): Kinetic studies using human kidney cortical microsomes and recombinant UGT1A9 and UGT2B7. Biochem. Pharmacol. 2004, 67, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Little, J.M.; Williams, L.; Xu, J.; Radominska-Pandya, A. Glucuronidation of the dietary fatty acids, phytanic acid and docosahexaenoic acid, by human UDP-glucuronosyltransferases. Drug Metab. Dispos. 2002, 30, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Trottier, J.; Perreault, M.; Rudkowska, I.; Levy, C.; Dallaire-Theroux, A.; Verreault, M.; Caron, P.; Staels, B.; Vohl, M.C.; Straka, R.J.; et al. Profiling serum bile acid glucuronides in humans: Gender divergences, genetic determinants, and response to fenofibrate. Clin. Pharmacol. Ther. 2013, 94, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, F.; Radominska, A.; Zimniak, P.; Little, J.M.; Havinga, R.; Vonk, R.J.; Lester, R. Defective biliary secretion of bile acid 3-O-glucuronides in rats with hereditary conjugated hyperbilirubinemia. J. Lipid Res. 1989, 30, 1835–1845. [Google Scholar] [CrossRef]
- Czernik, P.J.; Little, J.M.; Barone, G.W.; Raufman, J.P.; Radominska-Pandya, A. Glucuronidation of estrogens and retinoic acid and expression of UDP-glucuronosyltransferase 2B7 in human intestinal mucosa. Drug Metab. Dispos. 2000, 28, 1210–1216. [Google Scholar]
- Sneitz, N.; Krishnan, K.; Covey, D.F.; Finel, M. Glucuronidation of the steroid enantiomers ent-17beta-estradiol, ent-androsterone and ent-etiocholanolone by the human UDP-glucuronosyltransferases. Mol. Biol. 2012, 127, 282–288. [Google Scholar]
- Shatalova, E.G.; Loginov, V.I.; Braga, E.A.; Kazubskaia, T.P.; Sudomoina, M.A.; Blanchard, R.L.; Favorova, O.O. Association of polymorphisms in SULT1A1 and UGT1A1 Genes with breast cancer risk and phenotypes in Russian women. Mol. Biol. 2006, 40, 263–270. [Google Scholar] [CrossRef]
- Turgeon, D.; Chouinard, S.; Belanger, P.; Picard, S.; Labbe, J.F.; Borgeat, P.; Belanger, A. Glucuronidation of arachidonic and linoleic acid metabolites by human UDP-glucuronosyltransferases. J. Lipid Res. 2003, 44, 1182–1191. [Google Scholar] [CrossRef]
- Little, J.M.; Kurkela, M.; Sonka, J.; Jantti, S.; Ketola, R.; Bratton, S.; Finel, M.; Radominska-Pandya, A. Glucuronidation of oxidized fatty acids and prostaglandins B1 and E2 by human hepatic and recombinant UDP-glucuronosyltransferases. J. Lipid Res. 2004, 45, 1694–1703. [Google Scholar] [CrossRef]
- van Erk, M.J.; Wopereis, S.; Rubingh, C.; van Vliet, T.; Verheij, E.; Cnubben, N.H.; Pedersen, T.L.; Newman, J.W.; Smilde, A.K.; van der Greef, J.; et al. Insight in modulation of inflammation in response to diclofenac intervention: A human intervention study. BMC Med. Genomics 2010, 3, 5. [Google Scholar] [CrossRef]
- Faletti, A.; Bassi, D.; Franchi, A.M.; Gimeno, A.L.; Gimeno, M.A. Effects of morphine on arachidonic acid metabolism, on Ca2(+)-uptake and on cAMP synthesis in uterine strips from spayed rats. Prostaglandins Leukot. Essent Fatty Acids 1990, 41, 151–155. [Google Scholar] [CrossRef]
- Rivera, J.; Ward, N.; Hodgson, J.; Puddey, I.B.; Falck, J.R.; Croft, K.D. Measurement of 20-hydroxyeicosatetraenoic acid in human urine by gas chromatography-mass spectrometry. Clin. Chem. 2004, 50, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Watzer, B.; Reinalter, S.; Seyberth, H.W.; Schweer, H. Determination of free and glucuronide conjugated 20-hydroxyarachidonic acid (20-HETE) in urine by gas chromatography/negative ion chemical ionization mass spectrometry. Prostaglandins Leukot. Essent Fatty Acids. 2000, 62, 175–181. [Google Scholar] [CrossRef]
- Knights, K.M.; Tsoutsikos, P.; Miners, J.O. Novel mechanisms of nonsteroidal anti-inflammatory drug-induced renal toxicity. Expert Opin. Drug Metab. Toxicol. 2005, 1, 399–408. [Google Scholar] [CrossRef]
- Knights, K.M.; Mangoni, A.A.; Miners, J.O. Non-selective nonsteroidal anti-inflammatory drugs and cardiovascular events: Is aldosterone the silent partner in crime? Br. J. Clin. Pharmacol. 2006, 61, 738–740. [Google Scholar] [CrossRef] [PubMed]
- Knights, K.M.; Winner, L.K.; Elliot, D.J.; Bowalgaha, K.; Miners, J.O. Aldosterone glucuronidation by human liver and kidney microsomes and recombinant UDP-glucuronosyltransferases: Inhibition by NSAIDs. Br. J. Clin. Pharmacol. 2009, 68, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Crilly, M.A.; Mangoni, A.A. Non-steroidal anti-inflammatory drug (NSAID) related inhibition of aldosterone glucuronidation and arterial dysfunction in patients with rheumatoid arthritis: A cross-sectional clinical study. BMJ Open 2011, 1, e000076. [Google Scholar] [CrossRef]
- Crilly, M.A.; Mangoni, A.A.; Knights, K.M. Aldosterone glucuronidation inhibition as a potential mechanism for arterial dysfunction associated with chronic celecoxib and diclofenac use in patients with rheumatoid arthritis. Clin. Exp. Rheumatol. 2013, 31, 691–698. [Google Scholar]
- Sten, T.; Finel, M.; Ask, B.; Rane, A.; Ekstrom, L. Non-steroidal anti-inflammatory drugs interact with testosterone glucuronidation. Steroids 2009, 74, 971–977. [Google Scholar] [CrossRef]
- Graziani, G.; Tentori, L.; Portarena, I.; Vergati, M.; Navarra, P. Valproic acid increases the stimulatory effect of estrogens on proliferation of human endometrial adenocarcinoma cells. Endocrinology 2003, 144, 2822–2828. [Google Scholar] [CrossRef]
- Ethell, B.T.; Anderson, G.D.; Burchell, B. The effect of valproic acid on drug and steroid glucuronidation by expressed human UDP-glucuronosyltransferases. Biochem. Pharmacol. 2003, 65, 1441–1449. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).