Role of Long Non-Coding RNA Polymorphisms in Cancer Chemotherapeutic Response
Abstract
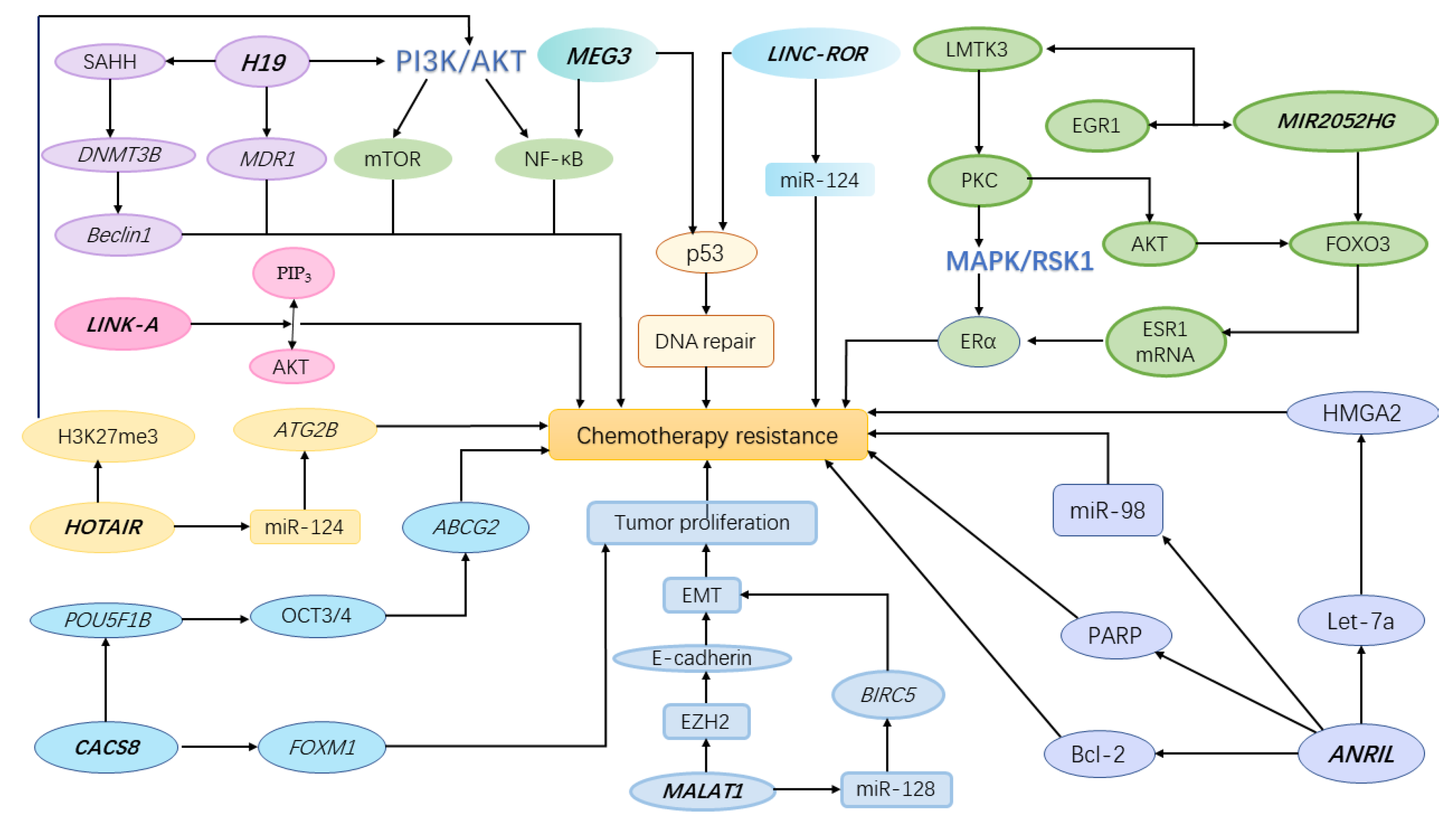
1. Background
2. lncRNA Polymorphisms in Cancer Chemotherapeutic Response
2.1. MIR2052HG
2.2. MEG3
2.3. H19
2.4. CASC8
2.5. LINK-A
2.6. Linc-ROR
2.7. MALAT1
2.8. ANRIL
2.9. HOTAIR
2.10. Other LncRNAs
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, F.F. Non-Coding RNAs, Meet Thy Masters. BioEssays 2010, 32, 599–608. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Mas, A.M.; Huarte, M. LncRNA-DNA Hybrids Regulate Distant Genes. EMBO Rep. 2020, 21, e50107. [Google Scholar] [CrossRef]
- Wang, K.; Sun, T.; Li, N.; Wang, Y.; Wang, J.-X.; Zhou, L.-Y.; Long, B.; Liu, C.-Y.; Liu, F.; Li, P.-F. MDRL LncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361. PLoS Genet. 2014, 10, e1004467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Han, C.; Song, K.; Chen, W.; Ungerleider, N.; Yao, L.; Ma, W.; Wu, T. The Long-Noncoding RNA MALAT1 Regulates TGF-Beta/Smad Signaling through Formation of a LncRNA-Protein Complex with Smads, SETD2 and PPM1A in Hepatic Cells. PLoS ONE 2020, 15, e0228160. [Google Scholar]
- Kanduri, C. Long Noncoding RNAs, Lessons from Genomic Imprinting. Biochim. Biophys. Acta 2016, 1859, 102–111. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. Starbase V2.0, Decoding MiRNA-CeRNA, MiRNA-NcRNA and Protein-RNA Interaction Networks from Large-Scale Clip-Seq Data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, C.; Chen, R.; Xiong, H.; Qiu, F.; Liu, S.; Zhang, M.; Wang, F.; Wang, Y.; Zhou, X.; et al. Disrupting MALAT1/miR-200c Sponge Decreases Invasion and Migration in Endometrioid Endometrial Carcinoma. Cancer Lett. 2016, 383, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yang, L. Long Noncoding RNA in Cancer, Wiring Signaling Circuitry. Trends Cell Biol. 2018, 28, 287–301. [Google Scholar] [CrossRef]
- Han, P.; Li, J.W.; Zhang, B.M.; Lv, J.C.; Li, Y.M.; Gu, X.Y.; Yu, Z.W.; Jia, Y.H.; Bai, X.F.; Li, L.; et al. The LncRNA CRNDE Promotes Colorectal Cancer Cell Proliferation and Chemoresistance Via miR-181a-5p-Mediated Regulation of Wnt/Beta-Catenin Signaling. Mol. Cancer 2017, 16, 9. [Google Scholar] [CrossRef]
- He, B.S.; Sun, H.L.; Xu, T.; Pan, Y.Q.; Lin, K.; Gao, T.Y.; Zhang, Z.Y.; Wang, S.K. Association of Genetic Polymorphisms in the LncRNAs with Gastric Cancer Risk in a Chinese Population. J. Cancer 2017, 8, 531–536. [Google Scholar] [CrossRef]
- Hurgobin, B.; Edwards, D. SNP Discovery Using a Pangenome, Has the Single Reference Approach Become Obsolete? Biology 2017, 6, 21. [Google Scholar] [CrossRef]
- Abdi, E.; Latifi-Navid, S.; Latifi-Navid, H.; Safaralizadeh, R. LncRNA Polymorphisms and Upper Gastrointestinal Cancer Risk. Pathol. Res. Pract. 2021, 218, 153324. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Taheri, M. Expressed Gene 3 (MEG3), A Tumor Suppressor Long Non Coding RNA. Biomed. Pharmacother. 2019, 118, 109129. [Google Scholar] [CrossRef]
- Fan, Z.; Chen, X.; Liu, L.; Zhu, C.; Xu, J.; Yin, X.; Sheng, Y.; Zhu, Z.; Wen, L.; Zuo, X.; et al. Association of the Polymorphism Rs13259960 in Slear with Predisposition to Systemic Lupus Erythematosus. Arthritis Rheumatol 2020, 72, 985–996. [Google Scholar] [CrossRef]
- Piskin, I.; Akcan, G.; Firat, A.; Tufan, A.C. A Single-Nucleotide Polymorphism (Rs8176070) of LncRNA Part1 May Reflect the Risk for Knee Osteoarthritis. Eur. J. Rheumatol. 2020, 7, 88–89. [Google Scholar] [CrossRef]
- Gong, W.-J.; Peng, J.-B.; Yin, J.-Y.; Li, X.-P.; Zheng, W.; Xiao, L.; Tan, L.-M.; Xiao, D.; Chen, Y.-X.; Li, X.; et al. Association between Well-Characterized Lung Cancer LncRNA Polymorphisms and Platinum-Based Chemotherapy Toxicity in Chinese Patients with Lung Cancer. Acta Pharmacol. Sin. 2017, 38, 581–590. [Google Scholar] [CrossRef]
- Lin, A.; Hu, Q.; Li, C.; Xing, Z.; Ma, G.; Wang, C.; Li, J.; Ye, Y.; Yao, J.; Liang, K.; et al. The Link—A LncRNA Interacts with Ptdins(3,4,5)P3 to Hyperactivate AKT and Confer Resistance to AKT Inhibitors. Nat. Cell Biol. 2017, 19, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liu, M.; Jia, D.; Tao, T.; Hao, D. LncRNA TINCR SNPS and Expression Levels Are Associated with Bladder Cancer Susceptibility. Genet. Test Mol. Biomarkers 2021, 25, 31–41. [Google Scholar] [CrossRef]
- Ingle, J.N.; Xie, F.; Ellis, M.J.; Goss, P.E.; Shepherd, L.E.; Chapman, J.-A.W.; Chen, B.E.; Kubo, M.; Furukawa, Y.; Momozawa, Y.; et al. Genetic Polymorphisms in the Long Noncoding RNA miR2052HG Offer a Pharmacogenomic Basis for the Response of Breast Cancer Patients to Aromatase Inhibitor Therapy. Cancer Res. 2016, 23, 7012. [Google Scholar] [CrossRef]
- Cairns, J.; Ingle, J.N.; Kalari, K.R.; Shepherd, L.E.; Kubo, M.; Goetz, M.P.; Weinshilboum, R.M.; Wang, L. The LncRNA miR2052HG Regulates Eralpha Levels and Aromatase Inhibitor Resistance through LMTK3 by Recruiting EGR1. Breast Cancer Res. 2019, 21, 47. [Google Scholar] [CrossRef] [PubMed]
- Bayarmaa, B.; Wu, Z.; Peng, J.; Wang, Y.; Xu, S.; Yan, T.; Yin, W.; Lu, J.; Zhou, L. Association of LncRNA MEG3 Polymorphisms with Efficacy of Neoadjuvant Chemotherapy in Breast Cancer. BMC Cancer 2019, 19, 877. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Z.; Zhao, Y.; Jin, Y.; An, L.; Wu, B.; Liu, Z.; Chen, X.; Chen, X.; Zhou, H.; et al. Genetic Polymorphisms of LncRNA-P53 Regulatory Network Genes Are Associated with Concurrent Chemoradiotherapy Toxicities and Efficacy in Nasopharyngeal Carcinoma Patients. Sci. Rep. 2017, 7, 8320. [Google Scholar] [CrossRef]
- Zeng, Y.; Li, T.-L.; Zhang, H.-B.; Deng, J.-L.; Zhang, R.; Sun, H.; Wan, Z.-R.; Liu, Y.-Z.; Zhu, Y.-S.; Wang, G. Polymorphisms in IGF2/H19 Gene Locus Are Associated with Platinum-Based Chemotherapeutic Response in Chinese Patients with Epithelial Ovarian Cancer. Pharmacogenomics 2019, 20, 179–188. [Google Scholar] [CrossRef]
- Lampropoulou, D.-I.; Aravantinos, G.; Katifelis, H.; Lazaris, F.; Laschos, K.; Theodosopoulos, T.; Papadimitriou, C.; Gazouli, M. Long Non-Coding RNA Polymorphisms and Prediction of Response to Chemotherapy Based on Irinotecan in Patients with Metastatic Colorectal Cancer. Cancer Biomarks 2019, 25, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Chen, S.-H.; Lv, Q.-L.; Sun, B.; Qu, Q.; Qin, C.-Z.; Fan, L.; Guo, Y.; Cheng, L.; Zhou, H.-H. Clinical Significance of Long Non-Coding RNA Casc8 Rs10505477 Polymorphism in Lung Cancer Susceptibility, Platinum-Based Chemotherapy Response, and Toxicity. Int. J. Environ. Res. Public Health 2016, 13, 545. [Google Scholar] [CrossRef]
- Melzer, D.; Perry, J.R.; Hernandez, D.; Corsi, A.M.; Stevens, K.; Rafferty, I.; Lauretani, F.; Murray, A.; Gibbs, J.R.; Paolisso, G.; et al. A Genome-Wide Association Study Identifies Protein Quantitative Trait Loci (PQTLS). PLoS Genet 2008, 4, e1000072. [Google Scholar] [CrossRef]
- Ma, C.X.; Reinert, T.; Chmielewska, I.; Ellis, M.J. Mechanisms of Aromatase Inhibitor Resistance. Nat. Rev. Cancer 2015, 15, 261–275. [Google Scholar] [CrossRef]
- Stebbing, J.; Filipovic, A.; Lit, L.C.; Blighe, K.; Grothey, A.; Xu, Y.; Miki, Y.; Chow, L.W.; Coombes, R.C.; Sasano, H.; et al. LMTK3 Is Implicated in Endocrine Resistance Via Multiple Signaling Pathways. Oncogene 2013, 32, 3371–3380. [Google Scholar] [CrossRef]
- Feng, S.Q.; Zhang, X.Y.; Fan, H.T.; Sun, Q.J.; Zhang, M. Up-Regulation of LncRNA MEG3 Inhibits Cell Migration and Invasion and Enhances Cisplatin Chemosensitivity in Bladder Cancer Cells. Neoplasma 2018, 65, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.H.; Wang, X. LncRNA MEG3 Inhibit Proliferation and Metastasis of Gastric Cancer via P53 Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3850–3856. [Google Scholar] [PubMed]
- Wang, C.; Nie, H.; Li, Y.; Liu, G.; Wang, X.; Xing, S.; Zhang, L.; Chen, X.; Chen, Y.; Li, Y. The Study of the Relation of DNA Repair Pathway Genes SNPS and the Sensitivity to Radiotherapy and Chemotherapy of NSCLC. Sci. Rep. 2016, 6, 26526. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhang, L.; Yuan, C.; Zhou, L.; Xu, S.; Lin, Y.; Zhang, J.; Yin, W.; Lu, J. Expression Profile Analysis of Long Noncoding RNA in Er-Positive Subtype Breast Cancer Using Microarray Technique and Bioinformatics. Cancer. Manag. Res. 2017, 9, 891–901. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cao, X.; Zhuang, S.; Hu, Y.; Xi, L.; Deng, L.; Sheng, H.; Shen, W. Associations between Polymorphisms of Long Non-Coding RNA MEG3 and Risk of Colorectal Cancer in Chinese. Oncotarget 2016, 7, 19054–19059. [Google Scholar] [CrossRef]
- Liu, J.; Wan, L.; Lu, K.; Sun, M.; Pan, X.; Zhang, P.; Lu, B.; Liu, G.; Wang, Z. The Long Noncoding RNA MEG3 Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma. PLoS ONE 2015, 10, e0114586. [Google Scholar] [CrossRef]
- Xia, Y.; He, Z.; Liu, B.; Wang, P.; Chen, Y. Downregulation of MEG3 Enhances Cisplatin Resistance of Lung Cancer Cells through Activation of the Wnt/Beta-Catenin Signaling Pathway. Mol. Med. Rep. 2015, 12, 4530–4537. [Google Scholar] [CrossRef]
- Zhang, A.; Xu, M.; Mo, Y.Y. Role of the LncRNA-P53 Regulatory Network in Cancer. J. Mol. Cell. Biol. 2014, 6, 181–191. [Google Scholar] [CrossRef]
- Gong, W.J.; Yin, J.Y.; Li, X.P.; Fang, C.; Xiao, D.; Zhang, W.; Zhou, H.H.; Li, X.; Liu, Z.Q. Association of Well-Characterized Lung Cancer LncRNA Polymorphisms with Lung Cancer Susceptibility and Platinum-Based Chemotherapy Response. Tumour Biol. 2016, 37, 8349–8358. [Google Scholar] [CrossRef]
- Lottin, S.; Adriaenssens, E.; Dupressoir, T.; Berteaux, N.; Montpellier, C.; Coll, J.; Dugimont, T.; Curgy, J.J. Overexpression of an Ectopic H19 Gene Enhances the Tumorigenic Properties of Breast Cancer Cells. Carcinogenesis 2002, 23, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Bi, J.; Xue, X.; Zheng, L.; Zhi, K.; Hua, J.; Fang, G. Up-Regulated Long Non-Coding RNA H19 Contributes to Proliferation of Gastric Cancer Cells. FEBS J. 2012, 279, 3159–3165. [Google Scholar] [CrossRef]
- Tsang, W.P.; Kwok, T.T. Riboregulator H19 Induction of Mdr1-Associated Drug Resistance in Human Hepatocellular Carcinoma Cells. Oncogene 2007, 26, 4877–4881. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhu, Z.; Li, J.; Yao, J.; Jiang, H.; Ran, R.; Li, X.; Li, Z. Expression and Prognostic Value of Long Non-Coding RNA H19 in Glioma Via Integrated Bioinformatics Analyses. Aging 2020, 12, 3407–3430. [Google Scholar] [CrossRef]
- Guo, Q.-Y.; Wang, H.; Wang, Y. LncRNA H19 Polymorphisms Associated with the Risk of Oscc in Chinese Population. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3770–3774. [Google Scholar]
- He, T.-D.; Xu, D.; Sui, T.; Zhu, J.-K.; Wei, Z.-X.; Wang, Y.-M. Association between H19 Polymorphisms and Osteosarcoma Risk. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3775–3780. [Google Scholar] [PubMed]
- Hua, Q.; Lv, X.; Gu, X.; Chen, Y.; Chu, H.; Du, M.; Gong, W.; Wang, M.; Zhang, Z. Genetic Variants in LncRNA H19 Are Associated with the Risk of Bladder Cancer in a Chinese Population. Mutagenesis 2016, 31, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Tang, R.; Ma, X.; Wang, Y.; Luo, D.; Xu, Z.; Zhu, Y.; Yang, L. Tag SNPs in Long Non-Coding RNA H19 Contribute to Susceptibility to Gastric Cancer in the Chinese Han Population. Oncotarget 2015, 6, 15311–15320. [Google Scholar] [CrossRef]
- Yang, M.L.; Huang, Z.; Wang, Q.; Chen, H.H.; Ma, S.N.; Wu, R.; Cai, W.S. The Association of Polymorphisms in LncRNA-H19 with Hepatocellular Cancer Risk and Prognosis. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Zhu, Q.-N.; Wang, G.; Guo, Y.; Peng, Y.; Zhang, R.; Deng, J.-L.; Li, Z.-X.; Zhu, Y.-S. LncRNA H19 Is a Major Mediator of Doxorubicin Chemoresistance in Breast Cancer Cells through a Cullin4a-MDR1 Pathway. Oncotarget 2017, 8, 91990–92003. [Google Scholar] [CrossRef]
- Yu, S.; Wu, C.; Tan, Q.; Liu, H. Long Noncoding RNA H19 Promotes Chemotherapy Resistance in Choriocarcinoma Cells. J. Cell. Biochem. 2019, 120, 15131–15144. [Google Scholar] [CrossRef]
- Chen, S.; Huo, X.; Lin, Y.; Ban, H.; Lin, Y.; Li, W.; Zhang, B.; Au, W.W.; Xu, X. Association of Mdr1 and Ercc1 Polymorphisms with Response and Toxicity to Cisplatin-Based Chemotherapy in Non-Small-Cell Lung Cancer Patients. Int. J. Hyg. Environ. Health 2010, 213, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The Long Noncoding RNA H19 Promotes Tamoxifen Resistance in Breast Cancer Via Autophagy. J. Hematol. Oncol. 2019, 12, 81. [Google Scholar] [CrossRef]
- Wu, E.-R.; Hsieh, M.-J.; Chiang, W.-L.; Hsueh, K.-C.; Yang, S.-F.; Su, S.-C. Association of LncRNA CCAT2 and CASC8 Gene Polymorphisms with Hepatocellular Carcinoma. Int. J. Environ. Res. Public. Health 2019, 16, 2833. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Thyagarajan, B.; Gross, M.D.; Goodman, M.; Sun, Y.V.; Bostick, R.M. Genetic Variants at Chromosome 8q24, Colorectal Epithelial Cell Proliferation, and Risk for Incident, Sporadic Colorectal Adenomas. Mol. Carcinog. 2014, 53, E187–E192. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Jia, Z.; Cao, D.; Yang, N.; Wang, Y.; Cao, X.; Jiang, J. Long Non-Coding RNA Polymorphisms on 8q24 Are Associated with the Prognosis of Gastric Cancer in a Chinese Population. PeerJ 2020, 8, e8600. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Gao, M.; Yin, Z.; Yan, L.; Cui, L. Association between LncRNA Casc8 Polymorphisms and the Risk of Cancer, A Meta-Analysis. Cancer Manag. Res. 2018, 10, 3141–3148. [Google Scholar] [CrossRef] [PubMed]
- Gargallo, C.J.; Lanas, Á.; Carrera-Lasfuentes, P.; Ferrandez, Á.; Quintero, E.; Carrillo, M.; Alonso-Abreu, I.; García-Gonzalez, M.A. Genetic Susceptibility in the Development of Colorectal Adenomas According to Family History of Colorectal Cancer. Int. J. Cancer 2019, 144, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Du, M.; Wang, C.; Gu, D.; Wang, M.; Zhang, Q.; Zhao, T.; Zhang, X.; Tan, Y.; Huo, X.; et al. Clinical Significance of POU5F1P1 Rs10505477 Polymorphism in Chinese Gastric Cancer Patients Receving Cisplatin-Based Chemotherapy after Surgical Resection. Int. J. Mol. Sci. 2014, 15, 12764–12777. [Google Scholar] [CrossRef]
- Ma, G.; Gu, D.; Lv, C.; Chu, H.; Xu, Z.; Tong, N.; Wang, M.; Tang, C.; Xu, Y.; Zhang, Z.; et al. Genetic Variant in 8q24 Is Associated with Prognosis for Gastric Cancer in a Chinese Population. J. Gastroenterol. Hepatol. 2015, 30, 689–695. [Google Scholar] [CrossRef]
- Panagopoulos, I.; Möller, E.; Collin, A.; Mertens, F. The Pou5f1p1 Pseudogene Encodes a Putative Protein Similar to Pou5f1 Isoform 1. Oncol. Rep. 2008, 20, 1029–1033. [Google Scholar] [CrossRef]
- Hosokawa, Y.; Takahashi, H.; Inoue, A.; Kawabe, Y.; Funahashi, Y.; Kameda, K.; Sugimoto, K.; Yano, H.; Harada, H.; Kohno, S.; et al. Oct-3/4 Modulates the Drug-Resistant Phenotype of Glioblastoma Cells through Expression of ATP Binding Cassette Transporter G2. Biochim. Biophys. Acta 2015, 1850, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Ota, S.; Ishii, G.; Goto, K.; Kubota, K.; Kim, Y.H.; Kojika, M.; Murata, Y.; Yamazaki, M.; Nishiwaki, Y.; Eguchi, K.; et al. Immunohistochemical Expression of BCRP and ERCC1 in Biopsy Specimen Predicts Survival in Advanced Non-Small-Cell Lung Cancer Treated with Cisplatin-Based Chemotherapy. Lung Cancer 2009, 64, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Guan, J.; Xu, Y.; Ren, H.; Jiang, J.; Wudu, M.; Wang, Q.; Su, H.; Zhang, Y.; Zhang, B.; et al. Silencing of CASC8 Inhibits Non-Small Cell Lung Cancer Cells Function and Promotes Sensitivity to Osimertinib Via FOXM1. J. Cancer 2021, 12, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Bella, L.; Zona, S.; Nestal, G.; de Moraes, G.N.; Lam, E.W.-F. FOXM1: A Key Oncofoetal Transcription Factor in Health and Disease. Semin. Cancer Biol. 2014, 29, 32–39. [Google Scholar] [CrossRef]
- Luo, J.; Manning, B.D.; Cantley, L.C. Targeting the Pi3k-Akt Pathway in Human Cancer, Rationale and Promise. Cancer Cell 2003, 4, 257–262. [Google Scholar] [CrossRef]
- Vivanco, I.; Sawyers, C.L. The Phosphatidylinositol 3-Kinase Akt Pathway in Human Cancer. Nat. Rev. Cancer 2002, 2, 489–501. [Google Scholar] [CrossRef]
- Stokoe, D.; Stephens, L.R.; Copeland, T.; Gaffney, P.R.J.; Reese, C.B.; Painter, G.F.; Holmes, A.B.; McCormick, F.; Hawkins, P.T. Dual Role of Phosphatidylinositol-3,4,5-Trisphosphate in the Activation of Protein Kinase B. Science 1997, 277, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant P53 in Breast Cancer, Potential as a Therapeutic Target and Biomarker. Breast Cancer Res. Treat 2018, 170, 213–219. [Google Scholar] [CrossRef]
- Li, C.; Zhao, Z.; Zhou, Z.; Liu, R. Linc-ROR Confers Gemcitabine Resistance to Pancreatic Cancer Cells Via Inducing Autophagy and Modulating the miR-124/PTBP1/PKM2 Axis. Cancer Chemother. Pharmacol. 2016, 78, 1199–1207. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, W.; Sun, Q.; Ye, L.; Zhou, D.; Wang, W. Linc-ROR Facilitates Hepatocellular Carcinoma Resistance to Doxorubicin by Regulating Twist1-Mediated Epithelial-Mesenchymal Transition. Mol. Med. Rep. 2021, 23, 340. [Google Scholar] [CrossRef]
- Luo, C.; Cao, J.; Peng, R.; Guo, Q.; Ye, H.; Wang, P.; Wang, K.; Song, C. Functional Variants in Linc-ROR Are Associated with MRNA Expression of Linc-ROR and Breast Cancer Susceptibility. Sci. Rep. 2018, 8, 4680. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a Novel Noncoding RNA, and Thymosin Beta4 Predict Metastasis and Survival in Early-Stage Non-Small Cell Lung Cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Wu, S.; Sun, H.; Wang, Y.; Yang, X.; Meng, Q.; Yang, H.; Zhu, H.; Tang, W.; Li, X.; Aschner, M.; et al. MALAT1 Rs664589 Polymorphism Inhibits Binding to miR-194-5p, Contributing to Colorectal Cancer Risk, Growth, and Metastasis. Cancer Res. 2019, 79, 5432–5441. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through Ezh2. Mol. Cancer Ther. 2017, 16, 739–751. [Google Scholar] [CrossRef]
- Ding, H.H.; Wu, W.D.; Jiang, T.; Cao, J.; Ji, Z.Y.; Jin, J.H.; Wang, J.J.; Song, W.F.; Wang, L.W. Meta-Analysis Comparing the Safety and Efficacy of Metastatic Colorectal Cancer Treatment Regimens, Capecitabine Plus Irinotecan (CAPIRI) and 5-Fluorouracil/Leucovorin Plus Irinotecan (FOLFIRI). Tumour. Biol. 2015, 36, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Ratain, M.J. Irinotecan Dosing, Does the Cpt in Cpt-11 Stand for Can’t Predict Toxicity? J. Clin. Oncol. 2002, 20, 7–8. [Google Scholar] [CrossRef]
- Fujita, K.-I.; Kubota, Y.; Ishida, H.; Sasaki, Y. Irinotecan, a Key Chemotherapeutic Drug for Metastatic Colorectal Cancer. World J. Gastroenterol. 2015, 21, 1223–1248. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Yu, J.; Dhanasekaran, S.M.; Kim, J.H.; Mani, R.-S.; Tomlins, S.A.; Mehra, R.; Laxman, B.; Cao, X.; Kleer, C.G.; et al. Repression of E-Cadherin by the Polycomb Group Protein Ezh2 in Cancer. Oncogene 2008, 27, 7274–7284. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Li, Z.; Wang, W.; Zeng, Y.; Liu, Z.; Qiu, J. Long Non-Coding RNA H19 Increases Bladder Cancer Metastasis by Associating with Ezh2 and Inhibiting E-Cadherin Expression. Cancer Lett. 2013, 333, 213–221. [Google Scholar] [CrossRef]
- Yamasaki, T.; Seki, N.; Yoshino, H.; Itesako, T.; Hidaka, H.; Yamada, Y.; Tatarano, S.; Yonezawa, T.; Kinoshita, T.; Nakagawa, M.; et al. MicroRNA-218 Inhibits Cell Migration and Invasion in Renal Cell Carcinoma through Targeting Caveolin-2 Involved in Focal Adhesion Pathway. J. Urol. 2013, 190, 1059–1068. [Google Scholar] [CrossRef]
- Li, P.L.; Zhang, X.; Wang, L.L.; Du, L.T.; Yang, Y.M.; Li, J.; Wang, C.X. MicroRNA-218 is a Prognostic Indicator in Colorectal Cancer and Enhances 5-Fluorouracil-Induced Apoptosis by Targeting Birc5. Carcinogenesis 2015, 36, 1484–1493. [Google Scholar]
- Fang, Z.; Chen, W.; Yuan, Z.; Liu, X.; Jiang, H. LncRNA-MALAT1 Contributes to the Cisplatin-Resistance of Lung Cancer by Upregulating Mrp1 and Mdr1 Via Stat3 Activation. Biomed. Pharmacother. 2018, 101, 536–542. [Google Scholar] [CrossRef]
- Kong, Y.; Hsieh, C.-H.; Alonso, L.C. ANRIL: A LncRNA at the CDKN2A/B Locus with Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.-Q.; Sun, M.; Yang, J.-S.; Xie, M.; Xu, T.-P.; Xia, R.; Liu, Y.-W.; Liu, X.-H.; Zhang, E.-B.; Lu, K.-H.; et al. Long Noncoding RNA Anril Promotes Non-Small Cell Lung Cancer Cell Proliferation and Inhibits Apoptosis by Silencing KLF2 and P21 Expression. Mol. Cancer Ther. 2015, 14, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Gu, Z.-T.; Chen, W.-H.; Cao, K.-J. Increased Expression of the Long Non-Coding RNA ANRIL Promotes Lung Cancer Cell Metastasis and Correlates with Poor Prognosis. Diagn. Pathol. 2015, 10, 14. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, W.; Shao, Z. Association between Long Non-Coding RNA Polymorphisms and Cancer Risk, A Meta-Analysis. Biosci. Rep. 2018, 38, BSR20180365. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular Interplay of the Noncoding RNA Anril and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of Ink4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.W.; Zhu, X.F.; Huang, X.F.; Sheng, P.Y.; He, A.S.; Yang, Z.B.; Deng, R.; Feng, G.K.; Liao, W.M. P14arf Sensitizes Human Osteosarcoma Cells to Cisplatin-Induced Apoptosis in a P53-Independent Manner. Cancer Biol. Ther. 2007, 6, 1074–1080. [Google Scholar] [CrossRef]
- Al-Mohanna, M.A.; Manogaran, P.S.; Al-Mukhalafi, Z.; Al-Hussein, K.A.; Aboussekhra, A. The Tumor Suppressor P16(Ink4a) Gene Is a Regulator of Apoptosis Induced by Ultraviolet Light and Cisplatin. Oncogene 2004, 23, 201–212. [Google Scholar] [CrossRef]
- Xu, R.; Mao, Y.; Chen, K.; He, W.; Shi, W.; Han, Y. The Long Noncoding RNA Anril Acts as an Oncogene and Contributes to Paclitaxel Resistance of Lung Adenocarcinoma A549 Cells. Oncotarget 2017, 8, 39177–39184. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.-T.; Gao, J.-H.; Chen, Y.-Q.; Chen, H.; Meng, H.-Y.; Lou, G. LncRNA Anril Affects the Sensitivity of Ovarian Cancer to Cisplatin Via Regulation of Let-7a/HMGA2 Axis. Biosci. Rep. 2019, 39, BSR20182101. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, G.; Cheng, Z.; Dai, L.; Jia, L.; Jing, X.; Wang, H.; Zhang, R.; Liu, M.; Jiang, T.; et al. Knockdown of LncRNA Anril Inhibits the Development of Cisplatin Resistance by Upregulating miR98 in Lung Cancer Cells. Oncol. Rep. 2020, 44, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Cunnington, M.S.; Koref, M.S.; Mayosi, B.M.; Burn, J.; Keavney, B. Chromosome 9p21 Snps Associated with Multiple Disease Phenotypes Correlate with Anril Expression. PLoS Genet. 2010, 6, e1000899. [Google Scholar] [CrossRef]
- Nakagawa, T.; Endo, H.; Yokoyama, M.; Abe, J.; Tamai, K.; Tanaka, N.; Sato, I.; Takahashi, S.; Kondo, T.; Satoh, K. Large Noncoding RNA Hotair Enhances Aggressive Biological Behavior and is Associated with Short Disease-Free Survival in Human Non-Small Cell Lung Cancer. Biochem. Biophys. Res. Commun. 2013, 436, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Moazeni-Roodi, A.; Aftabi, S.; Sarabandi, S.; Karami, S.; Hashemi, M.; Ghavami, S. Genetic Association between Hotair Gene and the Risk of Cancer, An Updated Meta-Analysis. J. Genet. 2020, 99, 1–16. [Google Scholar] [CrossRef]
- Senousy, M.A.; El-Abd, A.M.; Abdel-Malek, R.R.; Rizk, S.M. Circulating Long Non-Coding RNAs Hotair, Linc-P21, GAS5 and Xist Expression Profiles in Diffuse Large B-Cell Lymphoma, Association with R-Chop Responsiveness. Sci. Rep. 2021, 11, 2095. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qian, W.; Ye, X. Long Noncoding RNAS in Diffuse Large B-Cell Lymphoma, Current Advances and Perspectives. Onco Targets Ther. 2020, 13, 4295–4303. [Google Scholar] [CrossRef]
- Coiffier, B.; Sarkozy, C. Diffuse Large B-Cell Lymphoma: R-Chop Failure—What to Do? Hematol. Am. Soc. Hematol. Educ. Program. 2016, 2016, 366–378. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, K.; Tang, Y.; Luan, X.; Zheng, X.; Lu, X.; Mao, J.; Hu, L.; Zhang, S.; Zhang, X.; et al. LncRNA-HOTAIR Activates Autophagy and Promotes the Imatinib Resistance of Gastrointestinal Stromal Tumor Cells through a Mechanism Involving the miR-130a/Atg2b Pathway. Cell Death Dis. 2021, 12, 367. [Google Scholar] [CrossRef]
- Kovaleva, V.; Mora, R.; Park, Y.J.; Plass, C.; Chiramel, A.I.; Bartenschlager, R.; Dohner, H.; Stilgenbauer, S.; Pscherer, A.; Lichter, P.; et al. MiRNA-130a Targets ATG2B and DICER1 to Inhibit Autophagy and Trigger Killing of Chronic Lymphocytic Leukemia Cells. Cancer Res. 2012, 72, 1763–1772. [Google Scholar] [CrossRef]
- Jiang, B.; Xue, M.; Xu, D.; Song, J.; Zhu, S. Down-Regulated LncRNA Hotair Alleviates Polycystic Ovaries Syndrome in Rats by Reducing Expression of Insulin-Like Growth Factor 1 Via MicroRNA-130a. J. Cell. Mol. Med. 2020, 24, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Mazor, G.; Levin, L.; Picard, D.; Ahmadov, U.; Caren, H.; Borkhardt, A.; Reifenberger, G.; Leprivier, G.; Remke, M.; Rotblat, B. The LncRNA Tp73-As1 Is Linked to Aggressiveness in Glioblastoma and Promotes Temozolomide Resistance in Glioblastoma Cancer Stem Cells. Cell Death Dis. 2019, 10, 246. [Google Scholar] [CrossRef]
- Chen, W.; Xiao, J.; Shi, L.; Lin, L.; Jiang, M.; Ge, Y.; Li, Z.; Fan, H.; Yang, L.; Xu, Z. Association of Tp73-As1 Gene Polymorphisms with the Risk and Survival of Gastric Cancer in a Chinese Han Population. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3814–3822. [Google Scholar] [CrossRef] [PubMed]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a Non-Protein-Coding RNA, Controls Apoptosis and Is Downregulated in Breast Cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.-P.; Gao, W.-S.; Huo, J.-X.; Yang, Z.-S. Long Non-Coding RNA GAS5 Functions as a Tumor Suppressor in Renal Cell Carcinoma. Asian Pac. J. Cancer Prev. 2013, 14, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Kong, R.; Chen, F.; Song, Y. A Critical Role for the Long Non-Coding RNA GAS5 in Proliferation and Apoptosis in Non-Small-Cell Lung Cancer. Mol. Carcinog. 2015, 54, E1–E12. [Google Scholar] [CrossRef]
- Sun, M.; Jin, F.-Y.; Xia, R.; Kong, R.; Li, J.-H.; Xu, T.-P.; Liu, Y.-W.; Zhang, E.-B.; Liu, X.-H.; De, W. Decreased Expression of Long Noncoding RNA GAS5 Indicates a Poor Prognosis and Promotes Cell Proliferation in Gastric Cancer. BMC Cancer 2014, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zheng, W.; Li, C. Association of Long-Chain Non-Coding RNA GAS5 Gene Polymorphisms with Prostate Cancer Risk and Prognosis in Chinese Han Population. Medicine 2020, 99, e21790. [Google Scholar] [CrossRef]
- Yan, H.; Zhang, D.-Y.; Li, X.; Yuan, X.-Q.; Yang, Y.-L.; Zhu, K.-W.; Zeng, H.; Li, X.-L.; Cao, S.; Zhou, H.-H.; et al. Long Non-Coding RNA GAS5 Polymorphism Predicts a Poor Prognosis of Acute Myeloid Leukemia in Chinese Patients Via Affecting Hematopoietic Reconstitution. Leuk. Lymphoma 2017, 58, 1948–1957. [Google Scholar] [CrossRef]
- Chen, Z.; Pan, T.; Jiang, D.; Jin, L.; Geng, Y.; Feng, X.; Shen, A.; Zhang, L. The LncRNA-GAS5/miR-221-3p/DKK2 Axis Modulates ABCB1-Mediated Adriamycin Resistance of Breast Cancer Via the Wnt/Beta-Catenin Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 19, 1434–1448. [Google Scholar] [CrossRef]
- Xue, F.; Cheng, Y.; Xu, L.; Tian, C.; Jiao, H.; Wang, R.; Gao, X. LncRNA Neat1/miR-129/Bcl-2 Signaling Axis Contributes to Hdac Inhibitor Tolerance in Nasopharyngeal Cancer. Aging 2020, 12, 1417–1488. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Yan, Y.; Ma, N.; He, G.; Wang, K.; Zhang, Y.; Yin, J.; Song, C.; Wang, P.; Ye, H.; et al. Variant of SNPs at LncRNA Neat1 Contributes to Gastric Cancer Susceptibility in Chinese Han Population. Int. J. Clin. Oncol. 2021, 26, 694–700. [Google Scholar] [CrossRef]
- Yang, Z.; Yuan, L.; Yang, L.; Peng, S.; Yang, P.; He, X.; Bao, G. Association Study between Kcnq1 and Kcnq1ot1 Genetic Polymorphisms and Gastric Cancer Susceptibility and Survival in a Chinese Han Population, A Case-Control Study. Ann. Transl. Med. 2021, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Chang, A. Chemotherapy, Chemoresistance and the Changing Treatment Landscape for NSCLC. Lung Cancer 2011, 71, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 605. [Google Scholar] [CrossRef]
| LncRNA | Polymorphisms | Cancer Type | Patient Population | Drug | Effect | Reference |
|---|---|---|---|---|---|---|
| MIR2052 Host Gene (MIR2052HG) | rs4476990 and rs3802201 | Breast cancer | 4658 women with breast cancer, including 252 women experiencing a breast cancer recurrence | Aromatase Inhibitor (AIs) | Regulated ERα expression in the presence of AIs | [21] |
| 4406 controls without recurrence of breast cancer and 252 cases with recurrence | [22] | |||||
| Maternally Expressed 3(MEG3) | rs10132552 | Breast cancer | 144 women with locally advanced invasive breast cancer | Paclitaxel and cisplatin | Associated with good DFS and PCR rate | [23] |
| Nasopharyngeal carcinoma | 505 newly diagnosed nasopharyngeal carcinoma patients | Platinum-based chemotherapy drug | Associated with treatment response and risk of developing anemia | [24] | ||
| rs941576 | Breast cancer | 144 women with locally advanced invasive breast cancer | Paclitaxel and cisplatin | Associated with good DFS | [23] | |
| rs116907618 | Lung cancer | 467 lung cancer patients | Platinum-based chemotherapy drug | Associated with severe gastrointestinal toxicity | [18] | |
| H19 Imprinted Maternally Expressed Transcript(H19) | rs2839698, rs3842761, rs4244809, rs7924316, rs4244809 | Epithelial ovarian cancer (EOC) | 43 platinum-resistant and 138 platinum-sensitive EOC patients | Platinum-based chemotherapy drug | Associated with platinum-based chemoresistance | [25] |
| rs2104725 | Lung cancer | 467 lung cancer patients | Platinum-based chemotherapy drug | Associated with severe gastrointestinal toxicity | [18] | |
| rs2839698 | Associated with severe gastrointestinal or hematologic toxicities | |||||
| antisense non-coding RNA in the INK4 locus (ANRIL) | rs1333049 | Lung cancer | 467 lung cancer patients | Platinum-based chemotherapy drug | Associated with the incidence of severe gastrointestinal toxicity | [18] |
| rs10120688 | Associated with severe hematologic toxicity | |||||
| HOX Transcript Antisense RNA (HOTAIR) | rs7958904 | Lung cancer | 467 lung cancer patients | Platinum-based chemotherapy drug | Associated with the incidence of severe gastrointestinal toxicity | [18] |
| rs1899663 | Associated with severe gastrointestinal toxicity in age ≥ 57 | |||||
| metastasis-associated with lung adenocarcinoma transcript-1 (MALAT1) | rs619586 | Lung cancer | 467 lung cancer patients | Platinum-based chemotherapy drug | Associated with gastrointestinal toxicity | [18] |
| rs3200401 | Metastatic colorectal cancer | 98 colorectal cancer patients | Irinotecan | Associated with Pb derived toxicity and tumor resistance to irinotecan | [26] | |
| cancer susceptibility candidate 8 (CASC8) | rs10505477 | Lung Cancer | 498 lung cancer patients and healthy controls | Platinum-based chemotherapy drug | Associated with platinum-based chemotherapy response and toxicity | [27] |
| Long Intergenic Non-Protein Coding RNA 1139(LINK-A) | rs12095274 | Breast cancer | Breast cancer patients | AKT inhibitors | Leading to resistance to AKT inhibitors | [19] |
| Long intergenic non-protein coding RNA-regulator of reprogramming (Linc-ROR) | rs2027701 | Nasopharyngeal carcinoma | 505 newly diagnosed nasopharyngeal carcinoma patients | Platinum-based chemotherapy drug | Associated with chemoresistance and toxicity | [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Gu, M.; Gu, Z.; Lou, Y.-R. Role of Long Non-Coding RNA Polymorphisms in Cancer Chemotherapeutic Response. J. Pers. Med. 2021, 11, 513. https://doi.org/10.3390/jpm11060513
Zhang Z, Gu M, Gu Z, Lou Y-R. Role of Long Non-Coding RNA Polymorphisms in Cancer Chemotherapeutic Response. Journal of Personalized Medicine. 2021; 11(6):513. https://doi.org/10.3390/jpm11060513
Chicago/Turabian StyleZhang, Zheng, Meng Gu, Zhongze Gu, and Yan-Ru Lou. 2021. "Role of Long Non-Coding RNA Polymorphisms in Cancer Chemotherapeutic Response" Journal of Personalized Medicine 11, no. 6: 513. https://doi.org/10.3390/jpm11060513
APA StyleZhang, Z., Gu, M., Gu, Z., & Lou, Y.-R. (2021). Role of Long Non-Coding RNA Polymorphisms in Cancer Chemotherapeutic Response. Journal of Personalized Medicine, 11(6), 513. https://doi.org/10.3390/jpm11060513







