Genetic Variants Allegedly Linked to Antisocial Behaviour Are Equally Distributed Across Different Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Variants
2.2. Statistical Analysis
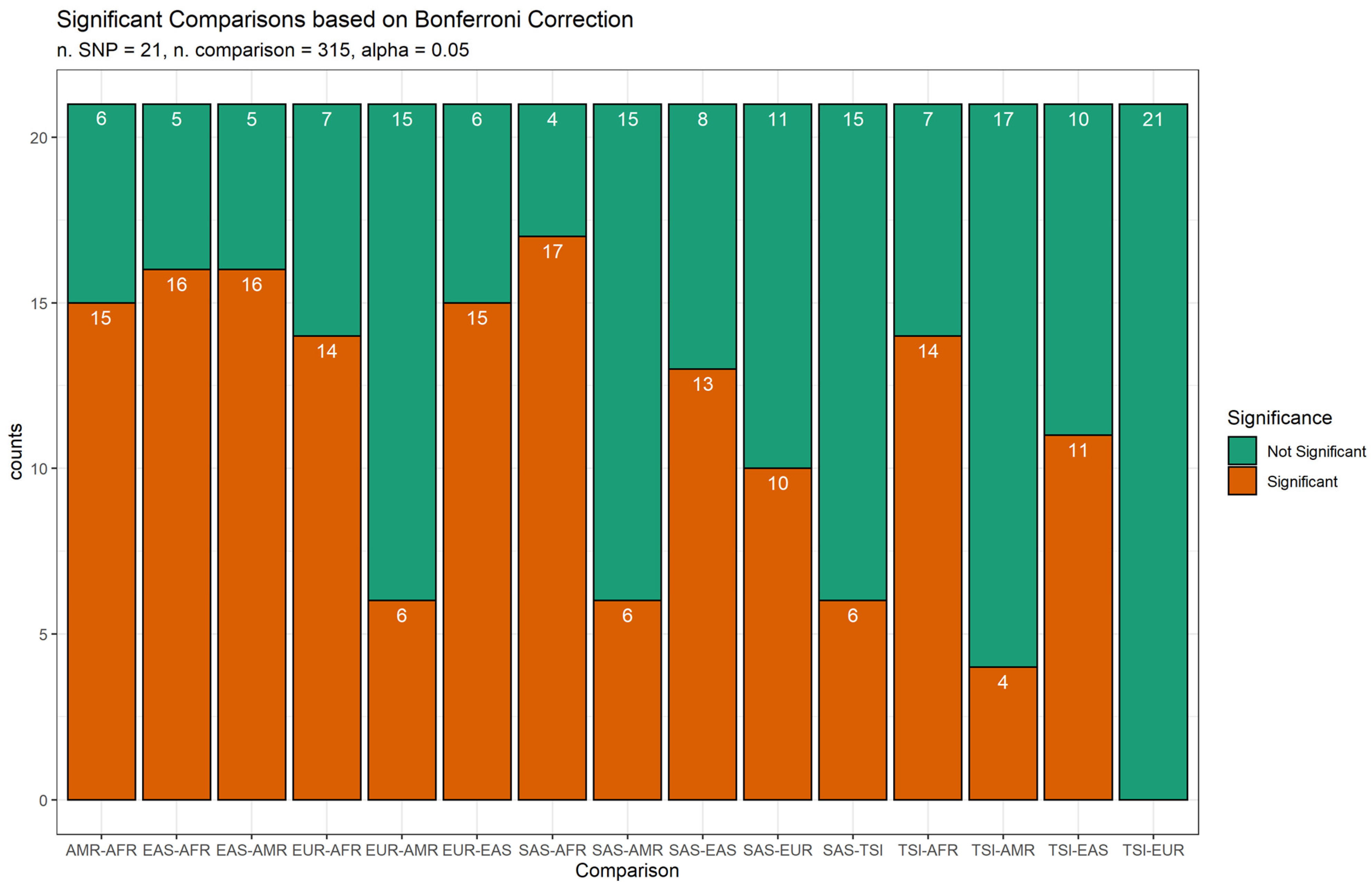
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ebstein, R.P.; Israel, S. Molecular Genetics of Personality: How Our Genes can Bring Us to a Better Understanding of Why We Act the Way We Do. In Handbook of Behavior Genetics, 1st ed.; Yong-Kyu, K., Ed.; Springer: New York, NY, USA, 2009; pp. 243–244. [Google Scholar] [CrossRef]
- Loehlin, J.C. History of Behavior Genetics. In Handbook of Behavior Genetics, 1st ed.; Yong-Kyu, K., Ed.; Springer: New York, NY, USA, 2009; pp. 3–11. [Google Scholar] [CrossRef]
- O’Mahony, C.; de Paor, A. The use of behavioural genetics in the criminal justice system: A disability & human rights perspective. Int. J. Law Psychiatry 2017, 54, 16–25. [Google Scholar] [CrossRef]
- van der Gronde, T.; Kempes, M.; van El, C.; Rinne, T.; Pieters, T. Neurobiological correlates in forensic assessment: A systematic review. PLoS ONE 2014, 9, e110672. [Google Scholar] [CrossRef]
- Manchia, M.; Comai, S.; Pinna, M.; Pinna, F.; Fanos, V.; Denovan-Wright, E.; Carpiniello, B. Biomarkers in aggression. Adv. Clin. Chem. 2019, 93, 169–237. [Google Scholar] [CrossRef]
- Baker, C. What Is Behavioral Genetics? In Behavioral Genetics an Introduction to How Genes and Environments Interact through Development to Shape Differences in Mood, Personality, and Intelligence, 1st ed.; American Association for the Advancement of Science Directorate for Science & Policy Programs: Washington, DC, USA, 2004; pp. 1–7. [Google Scholar]
- Assary, E.; Vincent, J.P.; Keers, R.; Pluess, M. Gene-environment interaction and psychiatric disorders: Review and future directions. Semin. Cell Dev. Biol. 2018, 77, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gainetdinov, R.R.; Beaulieu, J.-M.; Sotnikova, T.D.; Burch, L.H.; Williams, R.B.; Schwartz, D.A.; Krishnan, K.R.R.; Caron, M.G. Loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron 2005, 45, 11–16. [Google Scholar] [CrossRef] [PubMed]
- McKinney, J.; Johansson, S.; Halmøy, A.; Dramsdahl, M.; Winge, I.; Knappskog, P.M.; Haavik, J. A loss-of-function mutation in tryptophan hydroxylase 2 segregating with attention-deficit/hyperactivity disorder. Mol. Psychiatry 2008, 13, 365–367. [Google Scholar] [CrossRef]
- Kamaluddin, M.; Shariff, N.S.; Nurfarliza, S.; Othman, A.; Ismail, K.H.; Mat Saat, G.A. Psychological traits underlying different killing methods among Malaysian male murderers. Malays. J. Pathol. 2014, 36, 41–50. [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association Publisher: Washington, DC, USA, 2013; ISBN 9780890425558/0890425558. [Google Scholar]
- Rosell, D.R.; Siever, L.J. The neurobiology of aggression and violence. CNS Spectr. 2015, 20, 254–279. [Google Scholar] [CrossRef] [PubMed]
- Waltes, R.; Chiocchetti, A.G.; Freitag, C.M. The neurobiological basis of human aggression: A review on genetic and epigenetic mechanisms. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2016, 171, 650–675. [Google Scholar] [CrossRef] [PubMed]
- Cupaioli, F.A.; Zucca, F.A.; Caporale, C.; Lesch, K.P.; Passamonti, L.; Zecca, L. The neurobiology of human aggressive behavior: Neuroimaging, genetic, and neurochemical aspects. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 106, 110059. [Google Scholar] [CrossRef] [PubMed]
- Kolla, N.J.; Bortolato, M. The role of monoamine oxidase A in the neurobiology of aggressive, antisocial, and violent behavior: A tale of mice and men. Prog. Neurobiol. 2020, 194, 101875. [Google Scholar] [CrossRef]
- Asherson, P.; Cormand, B. The genetics of aggression: Where are we now? Am. J. Med. Genet. B Neuropsychiatr. Genet. 2016, 171, 559–561. [Google Scholar] [CrossRef]
- Ragazzo, M.; Puleri, G.; Errichiello, V.; Manzo, L.; Luzzi, L.; Potenza, S.; Strafella, C.; Peconi, C.; Nicastro, F.; Caputo, V.; et al. Evaluation of OpenArray™ as a Genotyping Method for Forensic DNA Phenotyping and Human Identification. Genes 2021, 12, 221. [Google Scholar] [CrossRef]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Strafella, C.; Caputo, V.; Termine, A.; Barati, S.; Gambardella, S.; Borgiani, P.; Caltagirone, C.; Novelli, G.; Giardina, E.; Cascella, R. Analysis of ACE2 Genetic Variability among Populations Highlights a Possible Link with COVID-19-Related Neurological Complications. Genes 2020, 11, 741. [Google Scholar] [CrossRef]
- Strafella, C.; Caputo, V.; Termine, A.; Barati, S.; Caltagirone, C.; Giardina, E.; Cascella, R. Investigation of Genetic Variations of IL6 and IL6R as Potential Prognostic and Pharmacogenetics Biomarkers: Implications for COVID-19 and Neuroinflammatory Disorders. Life 2020, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.; Zheng-Bradley, X.; Smith, R.; Kulesha, E.; Xiao, C.; Toneva, I.; Vaughan, B.; Preuss, D.; Leinonen, R.; Shumway, M.; et al. 1000 Genomes Project Consortium. The 1000 Genomes Project: Data management and community access. Nat. Methods 2012, 9, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Abdi, H. The Bonferroni and Sidak corrections for multiple comparisons. In Encyclopedia of Measurement and Statistics; Salkind, N.J., Ed.; Sage: Thousand Oaks, CA, USA, 2007. [Google Scholar]
- Simes, R.J. An improved Bonferroni procedure for multiple tests of significance. Biometrika 1986, 50, 813–820. [Google Scholar] [CrossRef]
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D. qvalue: Q-value Estimation for False Discovery Rate Control. R package version 2.18.0. 2019. Available online: http://github.com/jdstorey/qvalue (accessed on 2 February 2021).
- Thissen, D.; Steinberg, L.; Kuang, D. Quick and easy implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J. Educ. Behav. Stat. 2002, 27, 77–83. [Google Scholar] [CrossRef]
- Ruxton, G.D. The unequal variance t-test is an underused alternative to Student’s t-test and the Mann–Whitney U test. Behav. Ecol. 2006, 17, 688–690. [Google Scholar] [CrossRef]
- Wilcox, R.R. Introduction to Robust Estimation and Hypothesis Testing, 3rd ed.; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 2 February 2021).
- Forzano, F.; Borry, P.; Cambon-Thomsen, A.; Hodgson, S.V.; Tibben, A.; de Vries, P.; van El, C.; Cornel, M. Italian appeal court: A genetic predisposition to commit murder? Eur. J. Hum. Genet. 2010, 18, 519–521. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barbujani, G.; Ghirotto, S.; Tassi, F. Nine things to remember about human genome diversity. Tissue Antigens. 2013, 82, 155–164. [Google Scholar] [CrossRef] [PubMed]
| Frequency Data | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Variant | Associated Allele | AFR | AMR | EAS | EUR | TSI | SAS |
| TPH1 | rs1800532 | G | G: 84% T: 16% | G: 63% T: 37% | G: 52% T: 48% | G: 61% T: 39% | G: 62% T: 38% | G: 73% T: 27 |
| TPH1 | rs1799913 | T | G: 84% T: 16% | G: 63% T: 37% | G: 52% T: 48% | G: 61% T: 39% | G: 62% T: 38% | G: 73% T: 27% |
| TPH2 | rs4570625 | T | G: 63% T: 37% | G: 66% T: 34% | G: 45% T: 55% | G: 79% T: 21% | G: 77% T: 23% | G: 72% T: 28% |
| TPH2 | rs6582071 | A | G: 47% A: 53% | G: 64% A: 36% | G: 45% A: 55% | G: 78% A: 22% | G: 77% A: 23%) | G: 72% A: 28% |
| SLC6A4 | rs25531 | C | T: 78% C: 22% | T: 95% C: 5% | T: 87% C: 13% | T: 91% C: 9% | T: 93% C: 7% | T: 86% C: 14% |
| COMT | rs4680 | A | G: 72% A: 28% | G: 62% A: 38% | G: 72% A: 28% | G: 50% A: 50% | G: 55% A: 45% | G: 56% A: 44% |
| COMT | rs6269 | G | A: 63% G: 37% | A: 69% G: 31% | A: 66% G: 34% | A: 59% G: 41% | A: 51% G: 49% | A: 67% G: 33% |
| COMT | rs4818 | G | C: 83% G: 17% | C: 70% G: 30% | C: 66% G: 34% | C: 60% G: 40% | C: 53% G: 47% | C: 69% G: 31% |
| MAOA | rs1346551029 | n.c. | ACCG…: 99% ACCG…: 1% | ACCG…: 100% ACCG…: 0% | ACCG…: 100% ACCG…: 0% | ACCG…: 99% ACCG…: 1% | NA | ACCG…: 100% ACCG…: 0% |
| DRD4 | rs761010487 | n.c. | CGCC…: 100% CGCC…: 0% | CGCC…: 100% CGCC…: 0% | CGCC…: 100% CGCC…: 0% | CGCC…: 100% CGCC…: 0% | NA | CGCC…: 100% CGCC…: 0% |
| HTR1B | rs6296 | G | C: 76% G: 24% | C: 60% G: 40% | C: 49% G: 51% | C: 74% G: 26% | C: 78% G: 22% | C: 68% G: 32% |
| HTR1B | rs130058 | A | T: 97% A: 3% | T: 72% A: 28% | T: 91% A: 9% | T: 66% A: 34% | T: 63% A: 37% | T: 74% A: 26% |
| HTR1B | rs13212041 | C | C: 56% T: 44% | C: 17% T: 83% | C: 23% T: 77% | C: 19% T: 81% | C: 13% T: 87% | C: 16% T: 84% |
| HTR2B | rs79874540 | A | G: 100% | G: 100% | G: 100% | G: 100% A: 0% | G: 100% | G: 100% |
| HTR2A | rs6313 | G | G: 61% A: 39% | G: 65% A: 35% | G: 41% A: 59% | G: 56% A: 44% | G: 50% A: 50% | G: 58% A: 42% |
| HTR2A | rs6311 | C | C: 59% T: 41% | C: 64% T: 36% | C: 41% T: 59% | C: 56% T: 44% | C: 50% T: 50% | C: 60% T: 40% |
| HTR2A | rs7322347 | A | T: 32% A: 68% | T: 60% A: 40% | T: 79% A: 21% | T: 56% A: 44% | C: 49% T: 51% | T: 67% A: 33 |
| SLC6A3 | rs28363170 | n.c. | NA | |||||
| BDNF | rs6265 | C | C: 99% T: 1% | C: 85% T: 15% | C: 51% T: 49% | C: 80% T: 20% | C: 76% T: 24% | C: 80% T: 20% |
| ApoE | rs7412 (A > G ApoE epsylon4 Variant) | T | C: 90% T: 10% | C: 95% T: 5% | C: 90% T: 10% | C: 94% T: 6% | C: 95% T: 5% | C: 96% T: 4% |
| ApoE | rs429358 (A > G ApoE epsylon4 Variant) | T | T: 73% C: 27% | T: 90% C: 10% | T: 91% C: 9% | T: 84% C: 16% | T: 90% C: 10% | T: 91% C: 9% |
| NR3C2 | rs2070951 | C | G: 84% C: 16% | G: 45% C: 55% | G: 24% C: 76% | G: 51% C: 49% | G: 57% C: 43% | G: 32% C: 68% |
| MAOA | rs6323 | G | G: 14% T: 86% | G: 29% T: 71% | G: 57% T: 43% | G: 29% T: 71% | G: 27% T: 73% | G: 65% T: 35% |
| MAOA | rs1137070 | T | T: 36% C: 64% | T: 39% C: 61% | T: 58% C: 42% | T: 29% C: 71% | T: 28 C: 72% | T: 65% C: 35% |
| SNP (Count) | Comparison | p-Value (t-Test) | p-Value (Wilcoxon) |
|---|---|---|---|
| 21 | AMR–AFR | 0.95 | 0.67 |
| 21 | EAS–AFR | 0.63 | 0.61 |
| 21 | EUR–AFR | 0.96 | 0.74 |
| 21 | TSI–AFR | 0.97 | 0.84 |
| 21 | SAS–AFR | 0.67 | 0.57 |
| 21 | EAS–AMR | 0.64 | 0.55 |
| 21 | EUR–AMR | 0.90 | 0.97 |
| 21 | TSI–AMR | 0.91 | 0.99 |
| 21 | SAS–AMR | 0.69 | 1.00 |
| 21 | EUR–EAS | 0.54 | 0.40 |
| 21 | TSI–EAS | 0.56 | 0.36 |
| 21 | SAS–EAS | 0.97 | 0.96 |
| 21 | TSI–EUR | 0.98 | 0.90 |
| 21 | SAS–EUR | 0.59 | 0.76 |
| 21 | SAS–TSI | 0.61 | 0.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zampatti, S.; Ragazzo, M.; Fabrizio, C.; Termine, A.; Campoli, G.; Caputo, V.; Strafella, C.; Cascella, R.; Caltagirone, C.; Giardina, E. Genetic Variants Allegedly Linked to Antisocial Behaviour Are Equally Distributed Across Different Populations. J. Pers. Med. 2021, 11, 213. https://doi.org/10.3390/jpm11030213
Zampatti S, Ragazzo M, Fabrizio C, Termine A, Campoli G, Caputo V, Strafella C, Cascella R, Caltagirone C, Giardina E. Genetic Variants Allegedly Linked to Antisocial Behaviour Are Equally Distributed Across Different Populations. Journal of Personalized Medicine. 2021; 11(3):213. https://doi.org/10.3390/jpm11030213
Chicago/Turabian StyleZampatti, Stefania, Michele Ragazzo, Carlo Fabrizio, Andrea Termine, Giulia Campoli, Valerio Caputo, Claudia Strafella, Raffaella Cascella, Carlo Caltagirone, and Emiliano Giardina. 2021. "Genetic Variants Allegedly Linked to Antisocial Behaviour Are Equally Distributed Across Different Populations" Journal of Personalized Medicine 11, no. 3: 213. https://doi.org/10.3390/jpm11030213
APA StyleZampatti, S., Ragazzo, M., Fabrizio, C., Termine, A., Campoli, G., Caputo, V., Strafella, C., Cascella, R., Caltagirone, C., & Giardina, E. (2021). Genetic Variants Allegedly Linked to Antisocial Behaviour Are Equally Distributed Across Different Populations. Journal of Personalized Medicine, 11(3), 213. https://doi.org/10.3390/jpm11030213






