The 3′-UTR Polymorphisms in the Thymidylate Synthase (TS) Gene Associated with the Risk of Ischemic Stroke and Silent Brain Infarction
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Study Population
2.3. Genotyping
2.4. Statistical Analyses
2.5. Cell Culture and Dual-Luciferase Activity Assay
3. Results
3.1. Population Characteristics
3.2. Association between TS Polymorphisms and Ischemic Stroke Prevalence
3.3. Gene-Environment Interaction Analyses and Haplotype Analysis
3.4. Difference of Plasma tHcy Level by Genotypes
3.5. Altered Gene Expression Level of TS According to TS 3′-UTR Polymorphisms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. The World Health Report 2002: Reducing Risks, Promoting Healthy Life. Available online: http://www.who.int/whr/2002/en/ (accessed on 4 October 2020).
- Goldstein, L.B.; Adams, R.; Becker, K.; Furberg, C.D.; Gorelick, P.B.; Hademenos, G.; Hill, M.; Howard, G.; Howard, V.J.; Jacobs, B.; et al. Primary Prevention of Ischemic Stroke. Circulation 2001, 103, 163–182. [Google Scholar] [CrossRef]
- Korea National Statistical Office. The Korea National Statistical Office Report 2009: Change in leading causes of death (1999–2009). Available online: http://www.kosis.kr/ups3/service/ch_file_down.jps?PUBCODE=YD&FILE_NAME=/ups3/upload/101/YD/VD0005.xls&SEQ=8 (accessed on 26 June 2020).
- Vermeer, S.E.; Koudstaal, P.J.; Oudkerk, M.; Hofman, A.; Breteler, M.M. Prevalence and Risk Factors of Silent Brain Infarcts in the Population-Based Rotterdam Scan Study. Stroke 2002, 33, 21–25. [Google Scholar] [CrossRef]
- Duell, P.B.; Malinow, M.R. Homocyst(e)ine: An important risk factor for atherosclerotic vascular disease. Curr. Opin. Lipidol. 1997, 8, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Kluijtmans, L.A.J.; Young, I.S.; Boreham, C.A.; Murray, L.; McMaster, D.; McNulty, H.; Strain, J.J.; McPartlin, J.; Scott, J.M.; Whitehead, A.S. Genetic and nutritional factors contributing to hyperhomocysteinemia in young adults. Blood 2003, 101, 2483–2488. [Google Scholar] [CrossRef] [PubMed]
- Iso, H.; Naito, Y.; Sato, S.; Kitamura, A.; Okamura, T.; Sankai, T.; Shimamoto, T.; Iida, M.; Komachi, Y. Serum Triglycerides and Risk of Coronary Heart Disease among Japanese Men and Women. Am. J. Epidemiology 2001, 153, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Heinecke, A.; Kurnik, K.; Heller, C.; Nowak-Göttl, U.; Kosch, A.; Koch, H.G. For the Childhood Thrombophilia Study Group Increased fasting total homocysteine plasma levels as a risk factor for thromboembolism in children. Thromb. Haemost. 2004, 91, 308–314. [Google Scholar] [CrossRef]
- Matsui, T.; Arai, H.; Yuzuriha, T.; Yao, H.; Miura, M.; Hashimoto, S.; Higuchi, S.; Matsushita, S.; Morikawa, M.; Kato, A.; et al. Elevated Plasma Homocysteine Levels and Risk of Silent Brain Infarction in Elderly People. Stroke 2001, 32, 1116–1119. [Google Scholar] [CrossRef]
- Vermeer, S.E.; Van Dijk, E.J.; Koudstaal, P.J.; Oudkerk, M.; Hofman, A.; Clarke, R.; Breteler, M.M. Homocysteine, silent brain infarcts, and white matter lesions: The Rotterdam Scan Study. Ann. Neurol. 2002, 51, 285–289. [Google Scholar] [CrossRef]
- Kim, N.K.; Choi, B.O.; Jung, W.S.; Choi, Y.J.; Choi, K.G. Hyperhomocysteinemia as an independent risk factor for silent brain infarction. Neurology 2003, 61, 1595–1599. [Google Scholar] [CrossRef]
- Howard, G.; Wagenknecht, L.E.; Burke, G.L.; Diez-Roux, A.; Evans, G.W.; McGovern, P.; Nieto, F.J.; Tell, G.S. For the ARIC Investigators Cigarette Smoking and Progression of Atherosclerosis. JAMA 1998, 279, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Corbin, J.M.; Ruiz-Echevarría, M.J. One-Carbon Metabolism in Prostate Cancer: The Role of Androgen Signaling. Int. J. Mol. Sci. 2016, 17, 1208. [Google Scholar] [CrossRef]
- Lehotský, J.; Tothová, B.; Kovalská, M.; Dobrota, D.; Beňová, A.; Kalenská, D.; Kaplán, P. Role of Homocysteine in the Ischemic Stroke and Development of Ischemic Tolerance. Front. Neurosci. 2016, 10, 538. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Li, H.; Zuo, Z.; Lin, J.; Wang, A.; Zhao, X.; Liu, L.; Wang, Y.; on behalf of the CHANCE Investigators. Homocysteine Level Predicts Response to Dual Antiplatelet in Women With Minor Stroke or Transient Ischemic Attack. Arter. Thromb. Vasc. Biol. 2020, 40, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Pirouzpanah, S.; Varshosaz, P.; Fakhrjou, A.; Montazeri, V. The contribution of dietary and plasma folate and cobalamin to levels of angiopoietin-1, angiopoietin-2 and Tie-2 receptors depend on vascular endothelial growth factor status of primary breast cancer patients. Sci. Rep. 2019, 9, 14851–14918. [Google Scholar] [CrossRef] [PubMed]
- Duthie, S.J. Epigenetic modifications and human pathologies: Cancer and CVD. Proc. Nutr. Soc. 2010, 70, 47–56. [Google Scholar] [CrossRef]
- Choi, S.-W.; Mason, J.B. Folate Status: Effects on Pathways of Colorectal Carcinogenesis. J. Nutr. 2002, 132, 2413S–2418S. [Google Scholar] [CrossRef]
- Slattery, M.L.; Potter, J.D.; Samowitz, W.; Schaffer, D.; Leppert, M. Methylenetetrahydrofolate reductase, diet, and risk of colon cancer. Cancer Epidemiol. Biomark. Prev. 1999, 8, 513–518. [Google Scholar]
- Kawakami, K.; Omura, K.; Kanehira, E.; Watanabe, Y. Polymorphic tandem repeats in the thymidylate synthase gene is associated with its protein expression in human gastrointestinal cancers. Anticancer Res. 2000, 19, 3249–3252. [Google Scholar]
- Kim, H.C.; Choi, D.P.; Ahn, S.V.; Nam, C.M.; Suh, I. Six-Year Survival and Causes of Death among Stroke Patients in Korea. Neuroepidemiology 2008, 32, 94–100. [Google Scholar] [CrossRef]
- Adams, H.P.; Biller, J. Classification of Subtypes of Ischemic Stroke. Stroke 2015, 46, e114–e117. [Google Scholar] [CrossRef]
- Lee, B.-C.; Roh, J.K. International Experience in Stroke Registries. Am. J. Prev. Med. 2006, 31, S243–S245. [Google Scholar] [CrossRef]
- Papazoglou, D.; Galazios, G.; Koukourakis, M.I.; Panagopoulos, I.; Kontomanolis, E.N.; Papatheodorou, K.; Maltezos, E. Vascular endothelial growth factor gene polymorphisms and pre-eclampsia. Mol. Hum. Reprod. 2004, 10, 321–324. [Google Scholar] [CrossRef]
- Ryu, C.S.; Sakong, J.H.; Ahn, E.H.; Kim, J.O.; Ko, D.; Kim, J.H.; Lee, W.S.; Kim, N.K. Association study of the three functional polymorphisms (TAS2R46G>A, OR4C16G>A, and OR4X1A>T) with recurrent pregnancy loss. Genes Genom. 2018, 41, 61–70. [Google Scholar] [CrossRef]
- Kim, O.J.; Hong, S.H.; Jeon, Y.J.; Oh, S.H.; Kim, H.S.; Park, Y.S.; Kim, E.J.; Kim, N.K. Gene–environment interactions between methylenetetrahydrofolate reductase (MTHFR) 677C>T and metabolic syndrome for the prevalence of ischemic stroke in Koreans. Neurosci. Lett. 2013, 533, 11–16. [Google Scholar] [CrossRef]
- Fekih-Mrissa, N.; Mrad, M.; Klai, S.; Mansour, M.; Nsiri, B.; Gritli, N.; Mrissa, R. Methylenetetrahydrofolate Reductase (C677T and A1298C) Polymorphisms, Hyperhomocysteinemia, and Ischemic Stroke in Tunisian Patients. J. Stroke Cerebrovasc. Dis. 2013, 22, 465–469. [Google Scholar] [CrossRef]
- Holmes, M.V.; Newcombe, P.A.; Hubacek, J.; Sofat, R.; Ricketts, S.L.; Cooper, J.; Breteler, M.M.B.E.; Bautista, L.; Sharma, P.; Whittaker, J.C.; et al. Effect modification by population dietary folate on the association between MTHFR genotype, homocysteine, and stroke risk: A meta-analysis of genetic studies and randomised trials. Lancet 2011, 378, 584–594. [Google Scholar] [CrossRef]
- Bersano, A.; Ballabio, E.; Bresolin, N.; Candelise, L. Genetic polymorphisms for the study of multifactorial stroke. Hum. Mutat. 2008, 29, 776–795. [Google Scholar] [CrossRef] [PubMed]
- Van Der Put, N.M.; Gabreëls, F.; Stevens, E.M.; Smeitink, J.A.; Trijbels, F.J.; Eskes, T.K.; Heuvel, L.P.V.D.; Blom, H.J. A Second Common Mutation in the Methylenetetrahydrofolate Reductase Gene: An Additional Risk Factor for Neural-Tube Defects? Am. J. Hum. Genet. 1998, 62, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Subbarayan, P.R.; Hammad, H.; Sarkar, M.; Ardalan, B.; Gupta, N. The presence of three repeats in the 5′ UTR region of thymidylate synthase (TS) is associated with increased TS mRNA expression in cultured human cancer cell lines in vitro. Oncol. Rep. 2011, 27, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Ho, V.; Massey, T.E.; King, W.D. Effects of methionine synthase and methylenetetrahydrofolate reductase gene polymorphisms on markers of one-carbon metabolism. Genes Nutr. 2013, 8, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Brevik, A.; Vollset, S.E.; Tell, G.S.; Refsum, H.; Ueland, P.M.; Loeken, E.B.; Drevon, C.A.; Andersen, L.F. Plasma concentration of folate as a biomarker for the intake of fruit and vegetables: The Hordaland Homocysteine Study. Am. J. Clin. Nutr. 2005, 81, 434–439. [Google Scholar] [CrossRef]
- McCully, K.S. Vascular pathology of homocysteinemia: Implications for the pathogenesis of arteriosclerosis. Am. J. Pathol. 1969, 56, 111–128. [Google Scholar]
- Joachim, E.; Goldenberg, N.A.; Bernard, T.J.; Armstrong-Wells, J.; Stabler, S.P.; Manco-Johnson, M.J. The Methylenetetrahydrofolate Reductase Polymorphism (MTHFR c.677C>T) and Elevated Plasma Homocysteine Levels in a U.S. Pediatric Population with Incident Thromboembolism. Thromb. Res. 2013, 132, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; An, S.-A.; Lee, H.-B.; Kim, Y.; Kim, N.-K.; Kim, S.-H.; Kim, O.-J.; Oh, S.-H. Different impact of hyperhomocysteinemia on cerebral small vessel ischemia and cervico-cerebral atherosclerosis in non-stroke individuals. Thromb. Res. 2013, 131, e12–e16. [Google Scholar] [CrossRef] [PubMed]
- Blom, H.J.; Smulders, Y. Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects. J. Inherit. Metab. Dis. 2010, 34, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Liu, H.; Ju, J.; Li, Y.; Li, P.; Wang, L.-E.; Brewster, A.M.; Buchholz, T.A.; Arun, B.K.; Wei, Q.; et al. Genetic variant rs16430 6bp > 0bp at the microRNA-binding site inTYMSand risk of sporadic breast cancer risk in non-hispanic white women aged ≤55 years. Mol. Carcinog. 2013, 54, 281–290. [Google Scholar] [CrossRef]
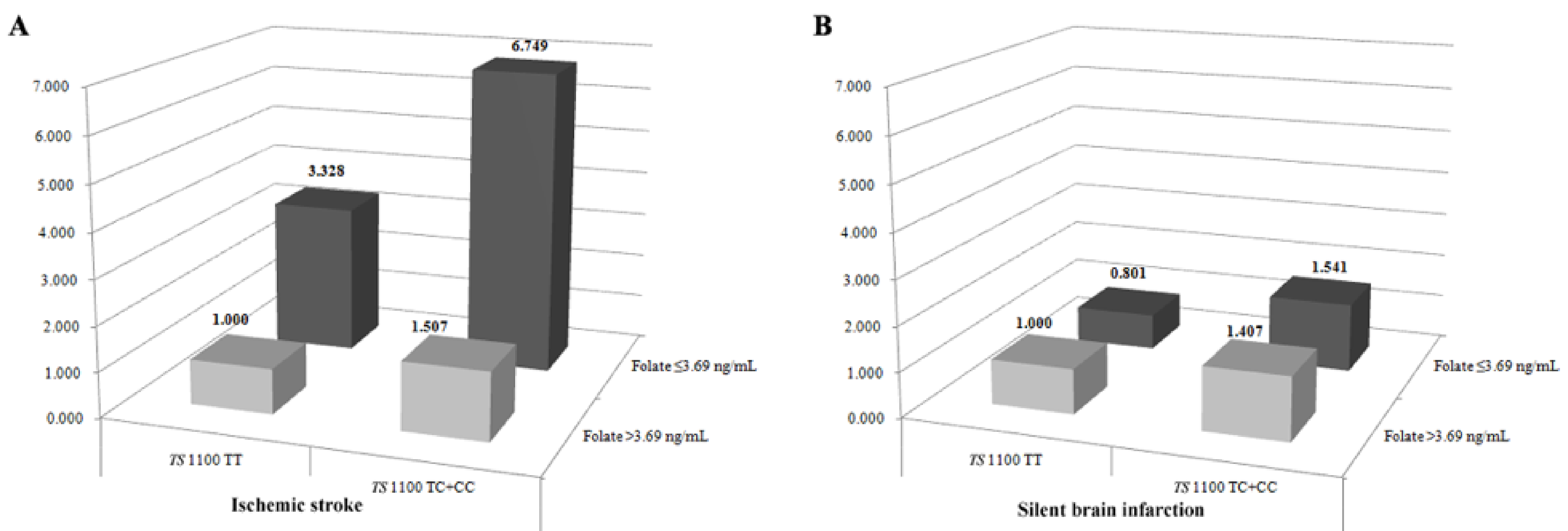
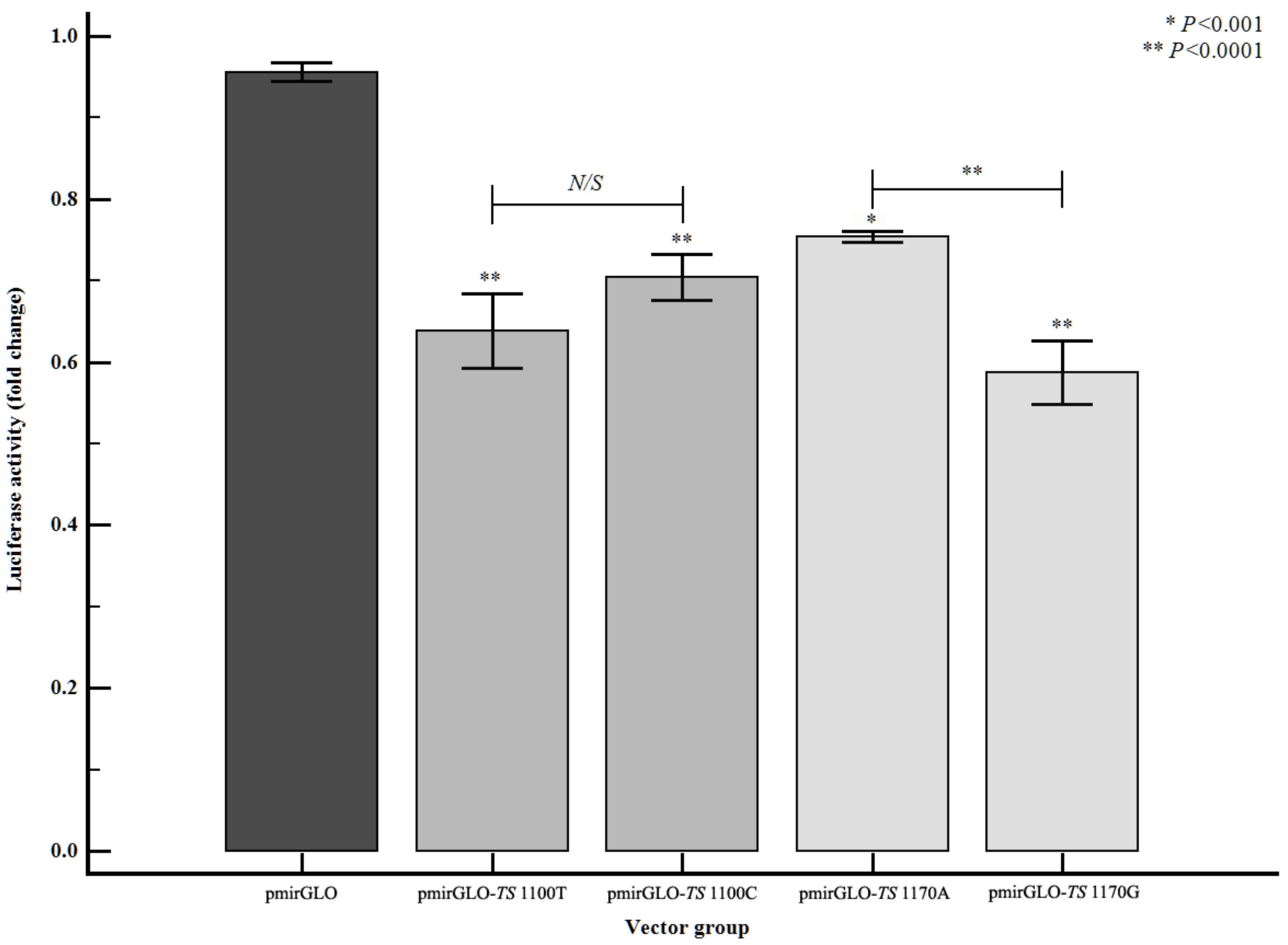
| Characteristics | Controls (n = 409) | Stroke (n = 507) | P | LAD (n = 201) | P | SVD (n = 149) | P | CE (n = 54) | P | SBI (n = 383) | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Male (n, %) | 173 (42.3) | 217 (42.8) | 0.952 | 83 (41.3) | 0.937 | 72 (48.3) | 0.441 | 22 (40.7) | 1.000 | 161 (42.0) | 0.795 |
| Age (years, mean ± SD) | 62.77 ± 10.61 | 62.95 ± 10.93 | 0.806 | 64.04 ± 10.43 | 0.165 | 60.89 ± 10.87 | 0.097 | 65.78 ± 11.98 | 0.055 | 64.02 ± 10.67 | 0.828 |
| HDL-C (mg/dL, mean ± SD) | 46.39 ± 13.75 | 44.50 ± 15.60 | 0.159 | 43.22 ± 13.15 | 0.025 | 44.50 ± 13.61 | 0.287 | 45.87 ± 13.75 | 0.810 | 45.22 ± 10.27 | 0.629 |
| LDL-C (mg/dL, mean ± SD) | 118.34 ± 42.14 | 120.99 ± 33.50 | 0.409 | 126.09 ± 38.55 | 0.069 | 116.51 ± 29.15 | 0.788 | 114.79 ± 27.09 | 0.565 | 145.12 ± 34.99 | 0.229 |
| Smoking (n, %) | 138 (33.7) | 191 (37.7) | 0.401 | 76 (37.8) | 0.502 | 58 (38.9) | 0.457 | 17 (31.5) | 0.885 | 55 (14.4) | <0.0001 |
| Hypertension (n, %) | 167 (40.8) | 325 (64.1) | 0.0001 | 131 (65.2) | 0.002 | 91 (61.1) | 0.013 | 30 (55.6) | 0.250 | 183 (47.8) | 0.337 |
| Diabetes mellitus (n, %) | 54 (13.2) | 140 (27.6) | <0.0001 | 56 (27.9) | 0.000 | 45 (30.2) | 0.000 | 10 (18.5) | 0.412 | 55 (14.4) | 0.838 |
| Hyperlipidemia (n, %) | 93 (22.7) | 148 (29.2) | 0.094 | 65 (32.3) | 0.061 | 44 (29.5) | <0.0001 | 10 (18.5) | 0.731 | 94 (24.5) | 0.808 |
| PLT (103 cell/μL, mean ± SD) | 242.30 ± 67.58 | 247.37 ± 87.51 | 0.340 | 256.56 ± 88.38 | 0.028 | 236.71 ± 63.26 | 0.553 | 244.93 ± 147.06 | 0.823 | 249.09 ± 71.40 | 0.525 |
| PT (sec, mean ± SD) | 11.77 ± 0.80 | 11.78 ± 1.01 | 0.796 | 11.77 ± 0.76 | 0.979 | 11.64 ± 0.79 | 0.111 | 12.02 ± 1.02 | 0.041 | 12.06 ± 1.11 | <0.0001 |
| aPTT (sec, mean ± SD) | 33.45 ± 18.57 | 30.50 ± 4.49 | 0.001 | 30.42 ± 4.70 | 0.025 | 30.85 ± 4.65 | 0.090 | 30.87 ± 4.25 | 0.320 | 32.17 ± 5.99 | 1.000 |
| Fibrinogen (mg/dL, mean ± SD) | 398.39 ± 120.66 | 425.96 ± 130.43 | 0.024 | 433.11 ± 133.10 | 0.015 | 396.43 ± 111.67 | 0.945 | 450.82 ± 133.66 | 0.010 | 412.72 ± 115.29 | 0.843 |
| Antithrombin (%, mean ± SD) | 94.49 ± 43.73 | 94.29 ± 17.25 | 0.936 | 95.42 ± 15.57 | 0.785 | 95.21 ± 19.77 | 0.321 | 87.25 ± 16.13 | 0.255 | 100.68 ± 12.34 | 0.442 |
| BUN (mg/dL, mean ± SD) | 15.82 ± 5.01 | 15.99 ± 6.29 | 0.667 | 15.35 ± 4.88 | 0.268 | 15.11 ± 5.21 | 0.062 | 18.95 ± 10.97 | 0.000 | 16.22 ± 4.85 | 0.534 |
| Urate (mg/dL, mean ± SD) | 4.65 ± 1.46 | 4.64 ± 1.51 | 0.908 | 4.60 ± 1.42 | 0.646 | 4.56 ± 1.32 | 0.632 | 4.62 ± 1.59 | 0.858 | 4.72 ± 1.75 | 0.810 |
| tHcy (μmol/L, mean ± SD) | 10.06 ± 4.19 | 11.21 ± 6.69 | 0.003 | 11.12 ± 5.80 | 0.011 | 11.12 ± 6.04 | 0.144 | 10.05 ± 4.75 | 0.995 | 12.16 ± 7.71 | 0.001 |
| Folate (ng/mL, mean ± SD) | 8.69 ± 6.26 | 6.91 ± 5.05 | <0.0001 | 6.53 ± 4.32 | <0.0001 | 7.10 ± 5.69 | <0.0001 | 7.65 ± 5.41 | 0.246 | 9.26 ± 7.03 | 0.268 |
| Vit. B12 (pg/mL, mean ± SD) | 746.44 ± 667.38 | 750.07 ± 649.37 | 0.934 | 815.33 ± 910.03 | 0.293 | 658.09 ± 309.93 | 0.082 | 741.30 ± 289.12 | 0.956 | 704.61 ± 719.51 | 0.891 |
| T. chol (mg/dL, mean ± SD) | 192.99 ± 37.56 | 190.84 ± 40.37 | 0.416 | 194.32 ± 45.59 | 0.704 | 189.18 ± 37.19 | 0.277 | 180.02 ± 34.96 | 0.017 | 238.76 ± 44.04 | <0.0001 |
| Triglyceride (mg/dL, mean ± SD) | 146.73 ± 90.14 | 154.44 ± 114.39 | 0.272 | 152.35 ± 97.94 | 0.486 | 168.18 ± 124.29 | 0.049 | 135.91 ± 179.25 | 0.476 | 239.26 ± 173.87 | 0.064 |
| Genotypes | Controls (n = 409) | Stroke (n = 507) | AOR (95% CI) * | P † | P ‡ | SBI (n = 383) | AOR (95% CI) * | P † | P ‡ |
|---|---|---|---|---|---|---|---|---|---|
| TS 1100 T>C | |||||||||
| TT | 218 (53.3) | 215 (42.4) | 1.000 (reference) | 176 (45.9) | |||||
| TC | 165 (40.3) | 235 (46.4) | 1.486 (1.115–1.980) | 0.007 | 0.011 | 173 (45.2) | 1.397 (0.961–2.031) | 0.080 | 0.120 |
| CC | 26 (6.4) | 57 (11.2) | 2.151 (1.275–3.628) | 0.004 | 0.006 | 34 (8.9) | 1.740 (0.879–3.443) | 0.112 | 0.168 |
| TT vs. TC+CC | 1.576 (1.197–2.074) | 0.001 | 0.002 | 1.443 (1.009–2.063) | 0.045 | 0.068 | |||
| TT+TC vs. CC | 1.758 (1.064–2.905) | 0.028 | 0.042 | 1.489 (0.783–2.833) | 0.225 | 0.338 | |||
| HWE P | 0.480 | 0.547 | 0.354 | ||||||
| TS 1170 A>G | |||||||||
| AA | 190 (46.5) | 320 (63.1) | 1.000 (reference) | 316 (82.5) | |||||
| AG | 184 (45.0) | 170 (33.5) | 0.505 (0.377–0.676) | <0.0001 | 0.0003 | 61 (15.9) | 0.198 (0.127–0.309) | <0.0001 | 0.0003 |
| GG | 35 (8.6) | 17 (3.4) | 0.284 (0.151–0.537) | <0.0001 | 0.0003 | 6 (1.6) | 0.070 (0.016–0.298) | 0.0002 | 0.0006 |
| AA vs. AG+GG | 0.472 (0.357–0.626) | <0.0001 | 0.0003 | 0.179 (0.117–0.276) | <0.0001 | 0.0003 | |||
| AA +AG vs. GG | 0.382 (0.206–0.710) | 0.002 | 0.006 | 0.121 (0.029–0.514) | 0.004 | 0.012 | |||
| HWE P | 0.306 | 0.331 | 0.135 | ||||||
| TS 1494 del>ins | |||||||||
| 0bp0bp | 197 (48.2) | 232 (45.8) | 1.000 (reference) | 184 (48.0) | |||||
| 0bp6bp | 180 (44.0) | 228 (45.0) | 1.127 (0.847–1.500) | 0.411 | 0.411 | 170 (44.4) | 1.121 (0.774–1.623) | 0.546 | 0.546 |
| 6bp6bp | 32 (7.8) | 47 (9.3) | 1.256 (0.754 2.091) | 0.381 | 0.381 | 29 (7.6) | 1.124 (0.570–2.217) | 0.736 | 0.736 |
| 0bp0bp vs. 0bp6bp+6bp6bp | 1.147 (0.872–1.509) | 0.326 | 0.326 | 1.122 (0.786–1.602) | 0.527 | 0.527 | |||
| 0bp0bp +0bp6bp vs. 6bp6bp | 1.302 (0.928–1.825) | 0.506 | 0.506 | 1.040 (0.543–1.994) | 0.905 | 0.905 | |||
| HWE P | 0.300 | 0.398 | 0.228 |
| Haplotypes | Controls (2n = 818) | Stroke (2n = 1014) | OR (95% CI) | P† | P‡ | SBI (2n = 766) | OR (95% CI) | P† | P‡ |
|---|---|---|---|---|---|---|---|---|---|
| TS 1100/1170/1494 | |||||||||
| T-A-0bp | 320 (39.1) | 446 (44.0) | 1.000 (reference) | 379 (49.5) | 1.000 (reference) | ||||
| T-A-6bp | 27 (3.4) | 15 (1.5) | 0.399 (0.209–0.762) | 0.006 | 0.010 | 101 (13.1) | 3.158 (2.014–4.954) | <0.0001 | 0.0001 |
| T-G-0bp | 251 (30.7) | 204 (20.1) | 0.583 (0.461–0.737) | <0.0001 | 0.0003 | 45 (5.9) | 0.151 (0.107–0.215) | <0.0001 | 0.0001 |
| T-G-6bp | 3 (0.3) | 0 (0.0) | 0.103 (0.005–1.994) | 0.074 | 0.093 | 0 (0.0) | 0.121 (0.006–2.346) | 0.097 | 0.097 |
| C-A-0bp | 3 (0.4) | 42 (4.1) | 10.04 (3.085–32.70) | <0.0001 | 0.0003 | 96 (12.5) | 27.02 (8.478–86.10) | <0.0001 | 0.0001 |
| C-A-6bp | 214 (26.2) | 307 (30.3) | 1.029 (0.821–1.290) | 0.818 | 0.818 | 118 (15.4) | 0.466 (0.356–0.610) | <0.0001 | 0.0001 |
| TS 1100/1170 | |||||||||
| T-A | 347 (42.4) | 461 (45.5) | 1.000 (reference) | 481 (62.8) | 1.000 (reference) | ||||
| T-G | 254 (31.1) | 204 (20.1) | 0.605 (0.480–0.762) | <0.0001 | 0.0002 | 44 (5.8) | 0.125 (0.088–0.177) | <0.0001 | 0.0002 |
| C-A | 217 (26.5) | 349 (34.4) | 1.211 (0.972–1.508) | 0.095 | 0.095 | 212 (27.7) | 0.705 (0.558–0.891) | 0.003 | 0.003 |
| TS 1100/1494 | |||||||||
| T-0bp | 571 (69.8) | 650 (64.1) | 1.000 (reference) | 424 (55.4) | 1.000 (reference) | ||||
| T-6bp | 30 (3.7) | 15 (1.4) | 0.439 (0.234–0.825) | 0.010 | 0.015 | 101 (13.2) | 4.534 (2.959–6.946) | <0.0001 | 0.0002 |
| C-0bp | 3 (0.4) | 42 (4.1) | 12.30 (3.791–39.90) | <0.0001 | 0.0003 | 114 (14.9) | 51.17 (16.15–162.2) | <0.0001 | 0.0002 |
| C-6bp | 214 (26.2) | 307 (30.3) | 1.260 (1.024–1.551) | 0.031 | 0.031 | 127 (16.6) | 0.799 (0.621–1.029) | 0.082 | 0.082 |
| TS 1170/1494 | |||||||||
| A-0bp | 323 (39.5) | 488 (48.1) | 1.000 (reference) | 475 (62.0) | 1.000 (reference) | ||||
| A-6p | 241 (29.4) | 322 (31.8) | 0.884 (0.711–1.100) | 0.289 | 0.289 | 218 (28.5) | 0.615 (0.488–0.775) | <0.0001 | 0.0002 |
| G-0bp | 251 (30.7) | 204 (20.1) | 0.538 (0.427–0.679) | <0.0001 | 0.0002 | 63 (8.3) | 0.171 (0.125–0.233) | <0.0001 | 0.0002 |
| SNP 1 | SNP 2 | Overall (n = 916) | Controls (n = 409) | Stroke patients (n = 507) | |||
|---|---|---|---|---|---|---|---|
| Mean ± SD (μmol/L) | CV (%) | Mean ± SD (μmol/L) | CV (%) | Mean ± SD (μmol/L) | CV (%) | ||
| MTHFR 677C>T | |||||||
| CC | - | 10.07 ± 6.45 | 64.1 | 9.23 ± 3.19 | 34.6 | 10.92 ± 8.50 | 77.8 |
| CT | - | 9.95 ± 3.63 | 36.5 | 9.84 ± 3.51 | 35.7 | 10.03 ± 3.72 | 37.1 |
| CC+CT | - | 9.99 ± 4.89 | 48.9 | 9.59 ± 3.39 | 35.3 | 10.35 ± 5.88 | 56.8 |
| TT | - | 13.62 ± 7.73 | 56.8 | 12.53 ± 6.54 | 52.2 | 14.25 ± 8.31 | 58.3 |
| P† | <0.001 | <0.001 | <0.001 | ||||
| MTHFR 677C>T | TS 1100T>C | ||||||
| CC+CT | TT | 9.84 ± 3.96 | 40.2 | 9.46 ± 3.34 | 35.3 | 10.24 ± 4.51 | 44.0 |
| CC+CT | TC+CC | 10.13 ± 5.60 | 55.3 | 9.72 ± 3.45 | 35.5 | 10.42 ± 6.73 | 64.6 |
| TT | TT | 12.85 ± 7.54 | 58.7 | 11.61 ± 5.44 | 46.9 | 13.85 ± 8.82 | 63.7 |
| TT | TC+CC | 14.32 ± 7.87 | 55.0 | 13.83 ± 7.76 | 56.1 | 14.53 ± 7.97 | 54.9 |
| P† | <0.001 | <0.001 | <0.001 | ||||
| MTHFR 677C>T | TS 1170A>G | ||||||
| CC+CT | AA | 9.98 ± 3.96 | 39.7 | 9.78 ± 3.67 | 37.5 | 10.10 ± 4.13 | 40.9 |
| CC+CT | AG+GG | 10.01 ± 5.84 | 58.3 | 9.43 ± 3.14 | 33.3 | 10.77 ± 8.05 | 74.7 |
| TT | AA | 13.92 ± 7.80 | 56.0 | 12.25 ± 6.59 | 53.8 | 14.68 ± 8.22 | 56.0 |
| TT | AG+GG | 13.20 ± 7.67 | 58.1 | 12.80 ± 6.58 | 51.4 | 13.53 ± 8.50 | 62.8 |
| P† | <0.001 | <0.001 | <0.001 | ||||
| MTHFR 677C>T | TS 1494 ins/del | ||||||
| CC+CT | 0bp0bp | 9.72 ± 3.44 | 35.4 | 9.37 ± 2.90 | 30.9 | 10.03 ± 3.83 | 38.2 |
| CC+CT | 0bp6bp+6bp6bp | 10.23 ± 5.86 | 57.3 | 9.78 ± 3.76 | 38.4 | 10.61 ± 7.16 | 67.5 |
| TT | 0bp0bp | 12.99 ± 7.80 | 60.0 | 11.57 ± 5.52 | 47.7 | 13.97 ± 8.97 | 64.2 |
| TT | 0bp6bp+6bp6bp | 14.24±7.65 | 53.7 | 13.71±7.55 | 55.1 | 14.49±7.75 | 53.5 |
| P† | <0.001 | <0.001 | <0.001 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.O.; Park, H.S.; Ko, E.J.; Sung, J.H.; Kim, J.; Oh, S.H.; Kim, O.J.; Kim, N.K. The 3′-UTR Polymorphisms in the Thymidylate Synthase (TS) Gene Associated with the Risk of Ischemic Stroke and Silent Brain Infarction. J. Pers. Med. 2021, 11, 200. https://doi.org/10.3390/jpm11030200
Kim JO, Park HS, Ko EJ, Sung JH, Kim J, Oh SH, Kim OJ, Kim NK. The 3′-UTR Polymorphisms in the Thymidylate Synthase (TS) Gene Associated with the Risk of Ischemic Stroke and Silent Brain Infarction. Journal of Personalized Medicine. 2021; 11(3):200. https://doi.org/10.3390/jpm11030200
Chicago/Turabian StyleKim, Jung Oh, Han Sung Park, Eun Ju Ko, Jung Hoon Sung, Jinkwon Kim, Seung Hun Oh, Ok Joon Kim, and Nam Keun Kim. 2021. "The 3′-UTR Polymorphisms in the Thymidylate Synthase (TS) Gene Associated with the Risk of Ischemic Stroke and Silent Brain Infarction" Journal of Personalized Medicine 11, no. 3: 200. https://doi.org/10.3390/jpm11030200
APA StyleKim, J. O., Park, H. S., Ko, E. J., Sung, J. H., Kim, J., Oh, S. H., Kim, O. J., & Kim, N. K. (2021). The 3′-UTR Polymorphisms in the Thymidylate Synthase (TS) Gene Associated with the Risk of Ischemic Stroke and Silent Brain Infarction. Journal of Personalized Medicine, 11(3), 200. https://doi.org/10.3390/jpm11030200






