Facial Recognition Intensity in Disease Diagnosis Using Automatic Facial Recognition
Abstract
:1. Introduction
2. Materials and Methods
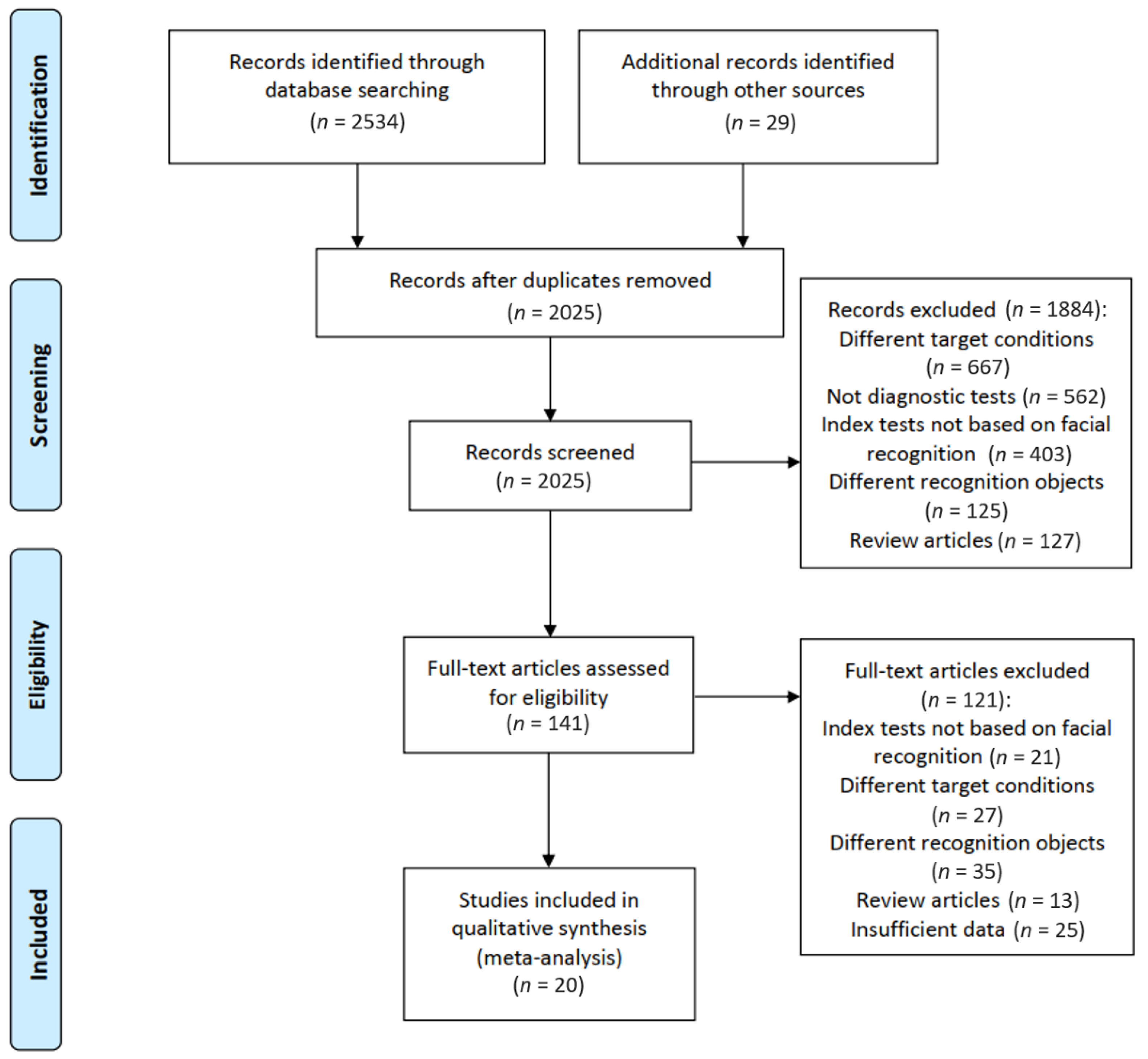
2.1. Study Identification and Selection
2.2. Data Extraction and Quality Assessment
2.3. Definition and Calculation of FRI
2.4. Statistical Methods
3. Results
3.1. Systematic Review
3.2. Risk of Bias Assessment of the Eligible Studies
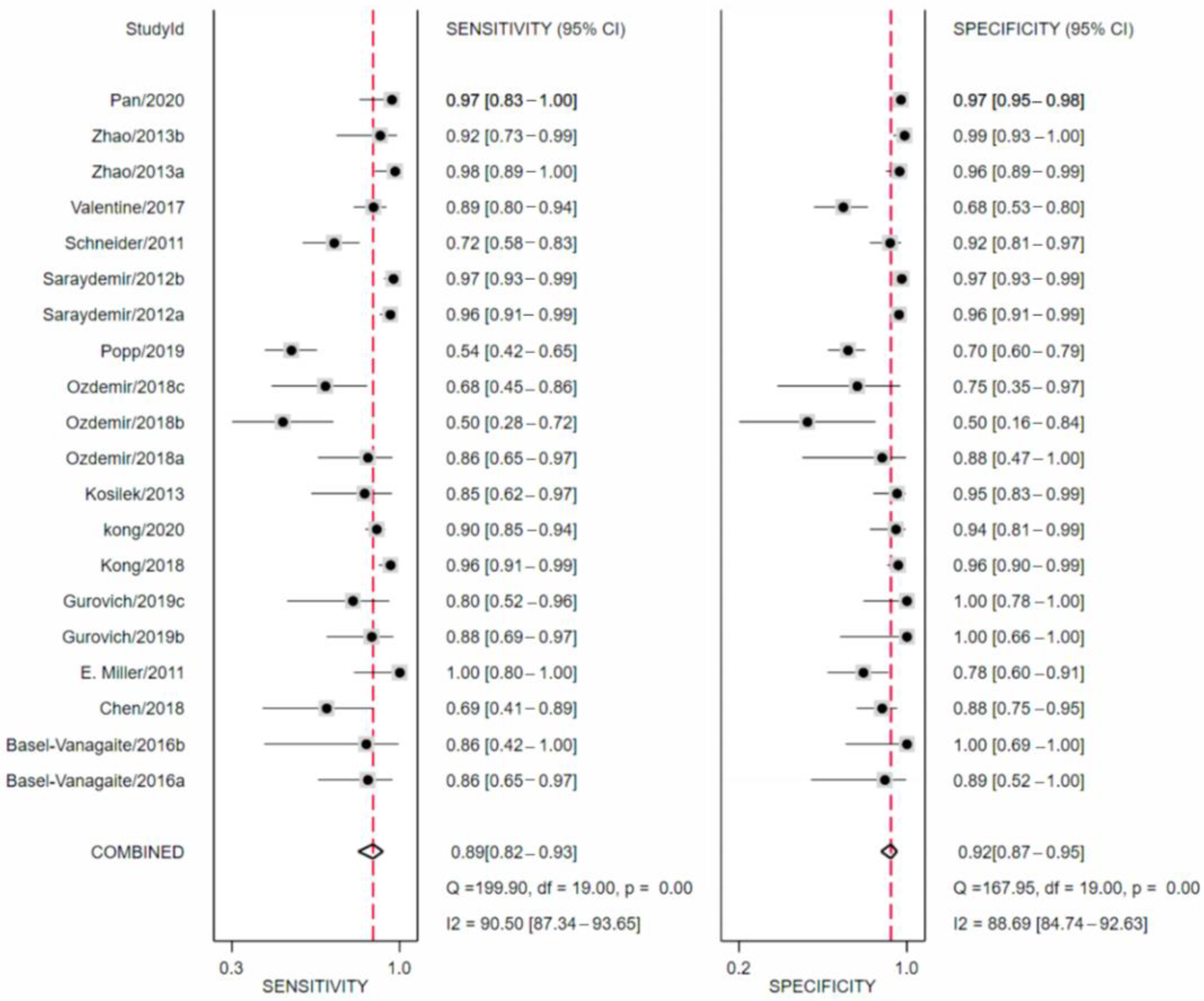
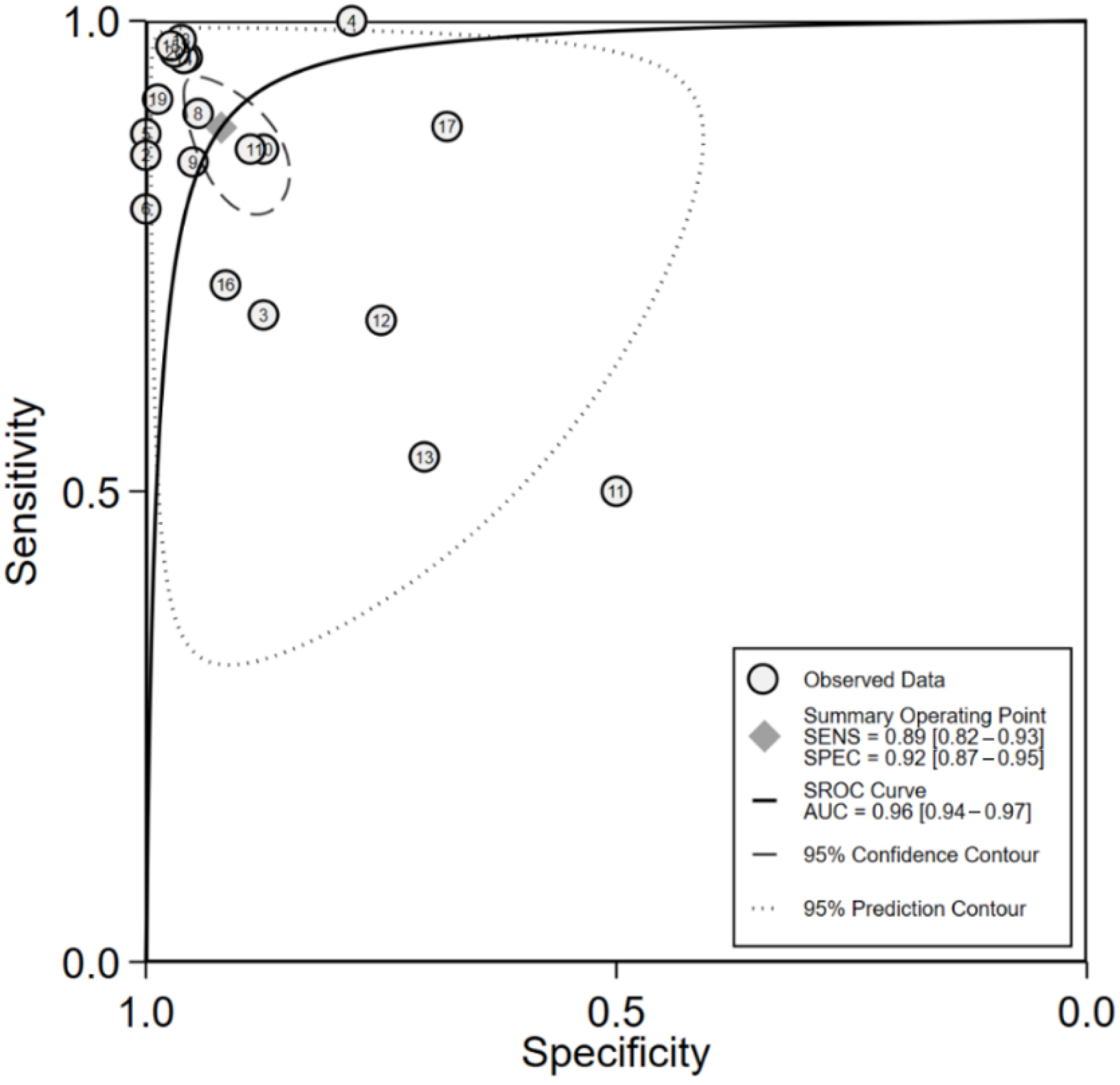
3.3. Meta-Analysis
3.4. Sensitivity Analysis
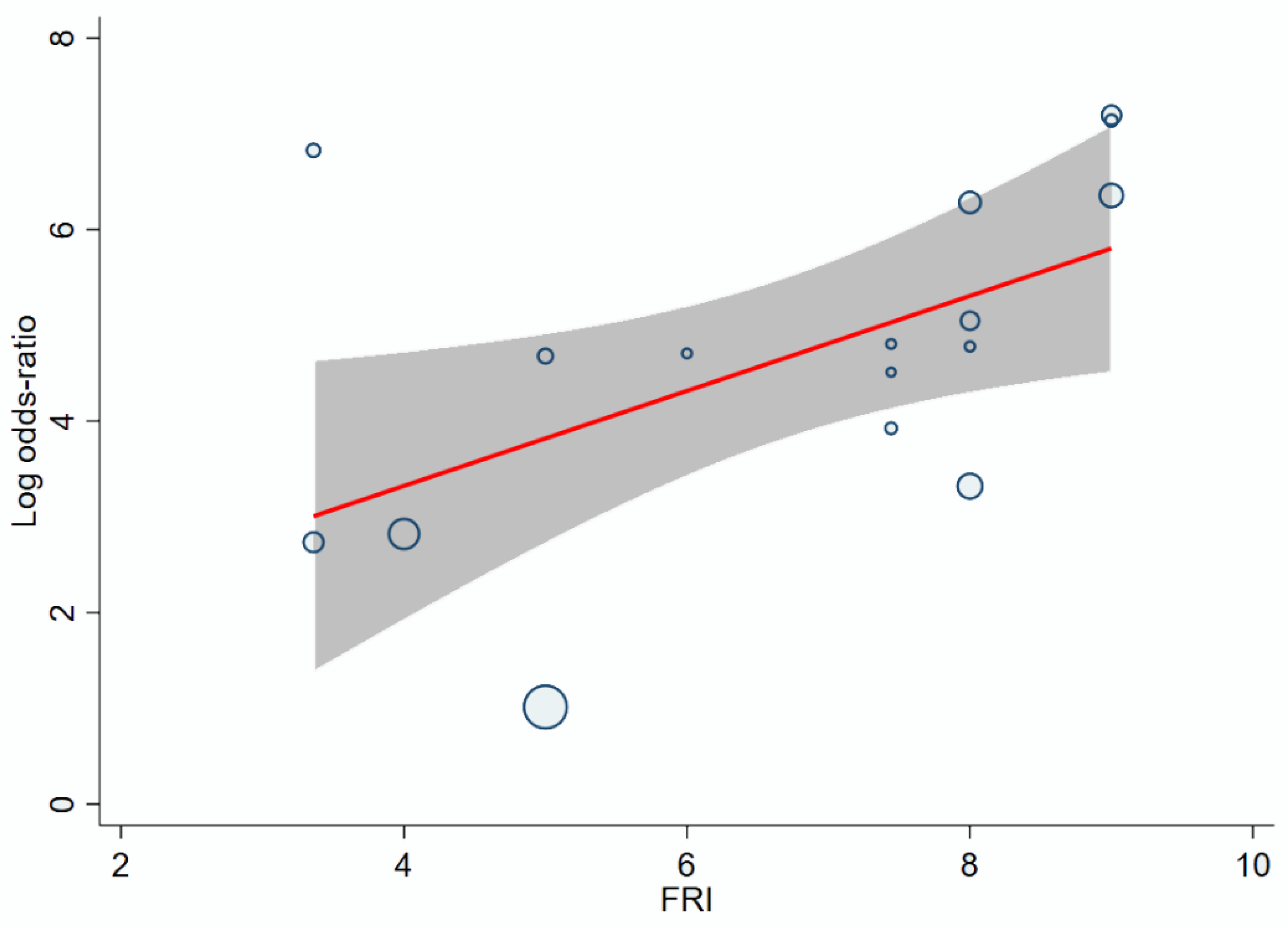
3.5. Evaluation of Facial Recognition Intensity (FRI)
3.6. Effect of FRI on the Accuracy of Facial Recognition
3.7. Effect of Sample Size of the Training Set and AI Model on the Accuracy of Facial Recognition
3.8. Sources of Heterogeneity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hurst, A.C.E. Facial recognition software in clinical dysmorphology. Curr. Opin. Pediatrics 2018, 30, 701–706. [Google Scholar] [CrossRef]
- Gurovich, Y.; Hanani, Y.; Bar, O.; Nadav, G.; Fleischer, N.; Gelbman, D.; Basel-Salmon, L.; Krawitz, P.M.; Kamphausen, S.B.; Zenker, M.; et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019, 25, 60–64. [Google Scholar] [CrossRef]
- Miller, D.D.; Brown, E.W. Artificial Intelligence in Medical Practice: The Question to the Answer? Am. J. Med. 2018, 131, 129–133. [Google Scholar] [CrossRef]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef]
- Loftus, T.J.; Tighe, P.J.; Filiberto, A.C.; Efron, P.A.; Brakenridge, S.C.; Mohr, A.M.; Rashidi, P.; Upchurch, G.R., Jr.; Bihorac, A. Artificial Intelligence and Surgical Decision-making. JAMA Surg. 2020, 155, 148–158. [Google Scholar] [CrossRef]
- Liu, G.; Wei, Y.; Xie, Y.; Li, J.; Qiao, L.; Yang, J.-J. A computer-aided system for ocular myasthenia gravis diagnosis. Tsinghua Sci. Technol. 2021, 26, 749–758. [Google Scholar] [CrossRef]
- Zheng, G.; Han, G.; Soomro, N.Q. An inception module CNN classifiers fusion method on pulmonary nodule diagnosis by signs. Tsinghua Sci. Technol. 2020, 25, 368–383. [Google Scholar] [CrossRef]
- Kaushik, H.; Singh, D.; Kaur, M.; Alshazly, H.; Zaguia, A.; Hamam, H. Diabetic Retinopathy Diagnosis from Fundus Images Using Stacked Generalization of Deep Models. IEEE Access 2021, 9, 108276–108292. [Google Scholar] [CrossRef]
- Alshazly, H.; Linse, C.; Abdalla, M.; Barth, E.; Martinetz, T. COVID-Nets: Deep CNN architectures for detecting COVID-19 using chest CT scans. PeerJ Comput. Sci. 2021, 7, e655. [Google Scholar] [CrossRef]
- Alshazly, H.; Linse, C.; Barth, E.; Martinetz, T. Explainable COVID-19 Detection Using Chest CT Scans and Deep Learning. Sensors 2021, 21, 455. [Google Scholar] [CrossRef]
- Hong, N.; Park, H.; Rhee, Y. Machine Learning Applications in Endocrinology and Metabolism Research: An Overview. Endocrinol. Metab. 2020, 35, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Marwaha, A.; Chitayat, D.; Meyn, M.S.; Mendoza-Londono, R.; Chad, L. The point-of-care use of a facial phenotyping tool in the genetics clinic: Enhancing diagnosis and education with machine learning. Am. J. Med. Genet. Part A 2021, 185, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Elmas, M.; Gogus, B. Success of Face Analysis Technology in Rare Genetic Diseases Diagnosed by Whole-Exome Sequencing: A Single-Center Experience. Mol. Syndromol. 2020, 11, 4–14. [Google Scholar] [CrossRef]
- Dias, R.; Torkamani, A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019, 11, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraydemir, S.; Taşpınar, N.; Eroğul, O.; Kayserili, H.; Dinçkan, N. Down syndrome diagnosis based on Gabor Wavelet Transform. J. Med. Syst. 2012, 36, 3205–3213. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Z.; Gao, L.; Li, Y.; Wang, S. Incremental face clustering with optimal summary learning via graph convolutional network. Tsinghua Sci. Technol. 2021, 26, 536–547. [Google Scholar] [CrossRef]
- Zhao, Q.; Rosenbaum, K.; Okada, K.; Zand, D.J.; Sze, R.; Summar, M.; Linguraru, M.G. Automated Down syndrome detection using facial photographs. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 3670–3673. [Google Scholar] [CrossRef]
- Devlin, L.; Morrison, P.J. Accuracy of the clinical diagnosis of Down syndrome. Ulst. Med. J. 2004, 73, 4–12. [Google Scholar]
- Özdemir, M.E.; Telatar, Z.; Eroğul, O.; Tunca, Y. Classifying dysmorphic syndromes by using artificial neural network based hierarchical decision tree. Australas. Phys. Eng. Sci. Med. 2018, 41, 451–461. [Google Scholar] [CrossRef]
- Basel-Vanagaite, L.; Wolf, L.; Orin, M.; Larizza, L.; Gervasini, C.; Krantz, I.D.; Deardoff, M.A. Recognition of the Cornelia de Lange syndrome phenotype with facial dysmorphology novel analysis. Clin. Genet. 2016, 89, 557–563. [Google Scholar] [CrossRef]
- Chen, S.; Pan, Z.X.; Zhu, H.J.; Wang, Q.; Yang, J.J.; Lei, Y.; Li, J.Q.; Pan, H. Development of a computer-aided tool for the pattern recognition of facial features in diagnosing Turner syndrome: Comparison of diagnostic accuracy with clinical workers. Sci. Rep. 2018, 8, 9317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Z.; Shen, Z.; Zhu, H.; Bao, Y.; Liang, S.; Wang, S.; Li, X.; Niu, L.; Dong, X.; Shang, X.; et al. Clinical application of an automatic facial recognition system based on deep learning for diagnosis of Turner syndrome. Endocrine 2020, 72, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.E.; Learned-Miller, E.G.; Trainer, P.; Paisley, A.; Blanz, V. Early diagnosis of acromegaly: Computers vs clinicians. Clin. Endocrinol. 2011, 75, 226–231. [Google Scholar] [CrossRef]
- Kong, X.; Gong, S.; Su, L.; Howard, N.; Kong, Y. Automatic Detection of Acromegaly from Facial Photographs Using Machine Learning Methods. EBioMedicine 2018, 27, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Kong, Y.; Kong, X.; He, C.; Liu, C.; Wang, L.; Su, L.; Gao, J.; Guo, Q.; Cheng, R. Constructing an automatic diagnosis and severity-classification model for acromegaly using facial photographs by deep learning. J. Hematol. Oncol. 2020, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.J.; Kosilek, R.P.; Günther, M.; Roemmler, J.; Stalla, G.K.; Sievers, C.; Reincke, M.; Schopohl, J.; Würtz, R.P. A novel approach to the detection of acromegaly: Accuracy of diagnosis by automatic face classification. J. Clin. Endocrinol. Metab. 2011, 96, 2074–2080. [Google Scholar] [CrossRef] [Green Version]
- Kosilek, R.P.; Schopohl, J.; Grunke, M.; Reincke, M.; Dimopoulou, C.; Stalla, G.K.; Würtz, R.P.; Lammert, A.; Günther, M.; Schneider, H.J. Automatic face classification of Cushing’s syndrome in women—A novel screening approach. Exp. Clin. Endocrinol. Diabetes 2013, 121, 561–564. [Google Scholar] [CrossRef] [Green Version]
- Popp, K.H.; Kosilek, R.P.; Frohner, R.; Stalla, G.K.; Athanasoulia-Kaspar, A.; Berr, C.; Zopp, S.; Reincke, M.; Witt, M.; Würtz, R.P.; et al. Computer Vision Technology in the Differential Diagnosis of Cushing’s Syndrome. Exp. Clin. Endocrinol. Diabetes 2019, 127, 685–690. [Google Scholar] [CrossRef] [Green Version]
- Valentine, M.; Bihm, D.C.J.; Wolf, L.; Hoyme, H.E.; May, P.A.; Buckley, D.; Kalberg, W.; Abdul-Rahman, O.A. Computer-Aided Recognition of Facial Attributes for Fetal Alcohol Spectrum Disorders. Pediatrics 2017, 140, e20162028. [Google Scholar] [CrossRef]
- Kline, A.D.; Moss, J.F.; Selicorni, A.; Bisgaard, A.M.; Deardorff, M.A.; Gillett, P.M.; Ishman, S.L.; Kerr, L.M.; Levin, A.V.; Mulder, P.A.; et al. Diagnosis and management of Cornelia de Lange syndrome: First international consensus statement. Nat. Rev. Genet. 2018, 19, 649–666. [Google Scholar] [CrossRef] [Green Version]
- Kruszka, P.; Addissie, Y.A.; Tekendo-Ngongang, C.; Jones, K.L.; Savage, S.K.; Gupta, N.; Sirisena, N.D.; Dissanayake, V.H.W.; Paththinige, C.S.; Aravena, T.; et al. Turner syndrome in diverse populations. Am. J. Med. Genet. Part A 2020, 182, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, V.A. Cochrane Handbook for Systematic Reviews of Interventions, 2nd ed.; John Wiley & Sons: Chichester, UK, 2019. [Google Scholar]
- Wu, H.Q.; Shan, Y.X.; Wu, H.; Zhu, D.R.; Tao, H.M.; Wei, H.G.; Shen, X.Y.; Sang, A.M.; Dong, J.C. Computer aided diabetic retinopathy detection based on ophthalmic photography: A systematic review and Meta-analysis. Int. J. Ophthal. 2019, 12, 1908–1916. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, Y.; Lei, S.; Zhu, H.; Li, J.; Wang, Q.; Yang, J.; Chen, S.; Pan, H. Performance of deep neural network-based artificial intelligence method in diabetic retinopathy screening: A systematic review and meta-analysis of diagnostic test accuracy. Eur. J. Endocrinol. 2020, 183, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Posso, M.; Puig, T.; Carles, M.; Rué, M.; Canelo-Aybar, C.; Bonfill, X. Effectiveness and cost-effectiveness of double reading in digital mammography screening: A systematic review and meta-analysis. Eur. J. Radiol. 2017, 96, 40–49. [Google Scholar] [CrossRef] [Green Version]
- Dorrius, M.D.; Jansen-van der Weide, M.C.; van Ooijen, P.M.; Pijnappel, R.M.; Oudkerk, M. Computer-aided detection in breast MRI: A systematic review and meta-analysis. Eur. Radiol. 2011, 21, 1600–1608. [Google Scholar] [CrossRef] [Green Version]
- Hassan, C.; Spadaccini, M.; Iannone, A.; Maselli, R.; Jovani, M.; Chandrasekar, V.T.; Antonelli, G.; Yu, H.; Areia, M.; Dinis-Ribeiro, M.; et al. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: A systematic review and meta-analysis. Gastrointest. Endosc. 2021, 93, 77–85.e76. [Google Scholar] [CrossRef]
- Hughes, K.S.; Zhou, J.; Bao, Y.; Singh, P.; Wang, J.; Yin, K. Natural language processing to facilitate breast cancer research and management. Breast J. 2020, 26, 92–99. [Google Scholar] [CrossRef]
- Hartmanis, J.; Stearns, R.E. On the computational complexity of algorithms. Trans. Am. Math. Soc. 1965, 117, 285–306. [Google Scholar] [CrossRef]
- Wolpert, D.H. The Lack of a Priori Distinctions between Learning Algorithms. Neural Comput. 1996, 8, 1341–1390. [Google Scholar] [CrossRef]
- Wolpert, D.H.; Macready, W.G. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef] [Green Version]
- Ho, Y.C.; Pepyne, D.L. Simple explanation of the no-free-lunch theorem and its implications. J. Optim. Theory Appl. 2002, 115, 549–570. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, J.; Liu, J.; Hao, W.; Nie, S.; Wang, S.; Peng, W. Computer-aided diagnosis of ground-glass opacity pulmonary nodules using radiomic features analysis. Phys. Med. Biol. 2019, 64, 135015. [Google Scholar] [CrossRef]
- Beig, N.; Khorrami, M.; Alilou, M.; Prasanna, P.; Braman, N.; Orooji, M.; Rakshit, S.; Bera, K.; Rajiah, P.; Ginsberg, J.; et al. Perinodular and Intranodular Radiomic Features on Lung CT Images Distinguish Adenocarcinomas from Granulomas. Radiology 2019, 290, 783–792. [Google Scholar] [CrossRef]
- Phene, S.; Dunn, R.C.; Hammel, N.; Liu, Y.; Krause, J.; Kitade, N.; Schaekermann, M.; Sayres, R.; Wu, D.J.; Bora, A.; et al. Deep Learning and Glaucoma Specialists: The Relative Importance of Optic Disc Features to Predict Glaucoma Referral in Fundus Photographs. Ophthalmology 2019, 126, 1627–1639. [Google Scholar] [CrossRef] [Green Version]
- Aziz, M.; Fatima, R.; Dong, C.; Lee-Smith, W.; Nawras, A. The impact of deep convolutional neural network-based artificial intelligence on colonoscopy outcomes: A systematic review with meta-analysis. J. Gastroenterol. Hepatol. 2020, 35, 1676–1683. [Google Scholar] [CrossRef]
- Wang, P.; Berzin, T.M.; Glissen Brown, J.R.; Bharadwaj, S.; Becq, A.; Xiao, X.; Liu, P.; Li, L.; Song, Y.; Zhang, D.; et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: A prospective randomised controlled study. Gut 2019, 68, 1813–1819. [Google Scholar] [CrossRef] [Green Version]
| Disease | Prevalence | Maximum Penetrance (Pmax) | Facial Phenotypes | Facial Recognition Intensity (FRI) | |
|---|---|---|---|---|---|
| Independent Facial Phenotypes | Number of Facial Phenotypes (Nf) | ||||
| Down syndrome [16,17] | 1/300~1000 | 100% | Short face Upward slanting eyes Epicanthus Brushfield spots (white spots on the colored part of the eyes) Low-set ears Small ears Flattened nose Small mouth Protruding tongue | 9 | 9 |
| Acromegaly [23,24,25,26] | 7/1000 | 100% | Forehead bulge Prominent jaw Prominent zygomatic arch Deep nasolabial folds Enlarged nose Enlarged brow Enlarged ear Enlarged lip | 8 | 8 |
| Cornelia de Lange Syndrome [2,20] | 1/10,000~1/30,000 | 82.7% | Short face Small jaw Arched eyebrows Joined eyebrows Short nose Forward nostril Long philtrum Thin upper lip Upturned corners of the mouth | 9 | 7.443 |
| Angelman syndrome [2] | 1/20,000~1/12,000 | 100% | Narrow bifrontal diameter Huge jaw Almond-shaped palpebral fissures Narrow nasal bridge Thin upper lip Protruding tongue | 6 | 6 |
| Cushing’s syndrome [27,28] | 4/100,000 | 100% | Red face Full moon face Acne Excessive hair Chemosis conjunctiva | 5 | 5 |
| Fetal alcohol spectrum disorders (FASDs) [29] | 7.7/1000 | 100% | Small head Short palpebral fissures Smooth philtrum Thin vermilion border of the upper lip | 4 | 4 |
| Turner syndrome [21,22] | 1/2500 | 56% | Small jaw Epicanthus Ptosis Ocular hypertelorism Low-set ears Multiple facial nevi | 6 | 3.36 |
| Covariate | Coefficient [95 Cl] | p Value |
|---|---|---|
| Facial recognition intensity (FRI) | 0.4939 [0.0710,0.9169] | 0.022 |
| Sample size of the training set | 0.0004 [−0.0006,0.0014] | 0.467 |
| Sensitivity | Specificity | OR | ln(OR) | FRI |
|---|---|---|---|---|
| 85% | 85% | 32.11 | 3.47 | 4.05 |
| 90% | 85% | 51.00 | 3.93 | 4.98 |
| 90% | 90% | 81.00 | 4.39 | 5.92 |
| 95% | 90% | 171.00 | 5.14 | 7.42 |
| 95% | 95% | 361.00 | 5.89 | 8.93 |
| FRI | Minimum Sample Size of Training Set | Range of Sample Size of Training Set | Range of Accuracies | AI Models | Range of Accuracies | ||
|---|---|---|---|---|---|---|---|
| Sensitivities | Specificities | Sensitivities | Specificities | ||||
| >8 | 30 | <100 | 0.967 (0.960~0.973) | 0.967 (0.960~0.973) | Non-DL | 0.973 (0.960~0.977) | 0.962 (0.960~0.973) |
| 100~200 | 0.977 | 0.962 | |||||
| 6~8 | 49 | <100 | 0.710 | 1.000 | Non-DL DL | 0.810 (0.719~0.901) 0.860 (0.800~0.960) | 0.972 (0.944~1.000) 1.000 (0.890~1.000) |
| 100~1000 | 0.790 (0.719~0.860) | 0.903 (0.890~0.915) | |||||
| >1000 | 0.901 (0.800~0.960) | 1.000 (0.944~1.000) | |||||
| <6 | 60 | <100 | 0.769 (0.688~0.850) | 0.913 (0.875~0.950) | Non-DL | 0.688 | 0.875 |
| 100~1000 | 0.714 (0.537~0.890) | 0.697 (0.690~0.704) | DL | 0.929 (0.890~0.967) | 0.830 (0.690~0.970) | ||
| >1000 | 0.967 | 0.970 | |||||
| Subgroup Variables | Numbers of Eligible Studies | Sensitivity, % [95 Cl] | Specificity, % [95 Cl] | p for Interaction |
|---|---|---|---|---|
| Image resolution | 0.415 | |||
| <30,000 pixels | 7 | 0.85 [0.73–0.97] | 0.90 [0.82–0.98] | |
| ≥30,000 pixels | 7 | 0.90 [0.82–0.98] | 0.94 [0.89–0.98] | |
| Sample size of training set | 0.145 | |||
| <1000 | 14 | 0.87 [0.80–0.93] | 0.89 [0.84–0.95] | |
| ≥1000 | 6 | 0.92 [0.86–0.99] | 0.97 [0.93–1.00] | |
| Model/system of AI | 0.802 | |||
| Neural network | 7 | 0.91 [0.83–0.99] | 0.93 [0.85–1.00] | |
| Non-neural network | 8 | 0.92 [0.86–0.97] | 0.92 [0.86–0.98] | |
| Number of diseases | 0.930 | |||
| 1 | 16 | 0.90 [0.86–0.95] | 0.78 [0.60–0.97] | |
| >1 | 4 | 0.93 [0.89–0.97] | 0.88 [0.74–1.00] | |
| Selection of control group | 0.573 | |||
| Healthy | 9 | 0.85 [0.75–0.95] | 0.94 [0.89–0.99] | |
| Other diseases | 11 | 0.90 [0.84–0.96] | 0.91 [0.86–0.97] | |
| Facial recognition intensity (FRI) | 0.003 | |||
| ≤6 | 7 | 0.81 [0.71–0.90] | 0.90 [0.83–0.96] | |
| >6 | 9 | 0.95 [0.92–0.98] | 0.95 [0.91–0.98] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, D.; Chen, S.; Zhang, Y.; Zhang, H.; Wang, Q.; Li, J.; Fu, Y.; Wang, S.; Yang, H.; Du, H.; et al. Facial Recognition Intensity in Disease Diagnosis Using Automatic Facial Recognition. J. Pers. Med. 2021, 11, 1172. https://doi.org/10.3390/jpm11111172
Wu D, Chen S, Zhang Y, Zhang H, Wang Q, Li J, Fu Y, Wang S, Yang H, Du H, et al. Facial Recognition Intensity in Disease Diagnosis Using Automatic Facial Recognition. Journal of Personalized Medicine. 2021; 11(11):1172. https://doi.org/10.3390/jpm11111172
Chicago/Turabian StyleWu, Danning, Shi Chen, Yuelun Zhang, Huabing Zhang, Qing Wang, Jianqiang Li, Yibo Fu, Shirui Wang, Hongbo Yang, Hanze Du, and et al. 2021. "Facial Recognition Intensity in Disease Diagnosis Using Automatic Facial Recognition" Journal of Personalized Medicine 11, no. 11: 1172. https://doi.org/10.3390/jpm11111172
APA StyleWu, D., Chen, S., Zhang, Y., Zhang, H., Wang, Q., Li, J., Fu, Y., Wang, S., Yang, H., Du, H., Zhu, H., Pan, H., & Shen, Z. (2021). Facial Recognition Intensity in Disease Diagnosis Using Automatic Facial Recognition. Journal of Personalized Medicine, 11(11), 1172. https://doi.org/10.3390/jpm11111172