Developing an Algorithm for Discriminating Oral Cancerous and Normal Tissues Using Raman Spectroscopy
Abstract
:1. Introduction
2. Methods and Materials
2.1. Patients and Samples
2.2. Development of Tissue Classification Models Using the Training Data Set
2.3. Validation of Tissue Classification Models Using a Validation Data Set
2.4. Pre-Processing of Spectra
2.5. Instrumentation
2.6. Univariate and Multivariate Analysis
3. Results and Discussion
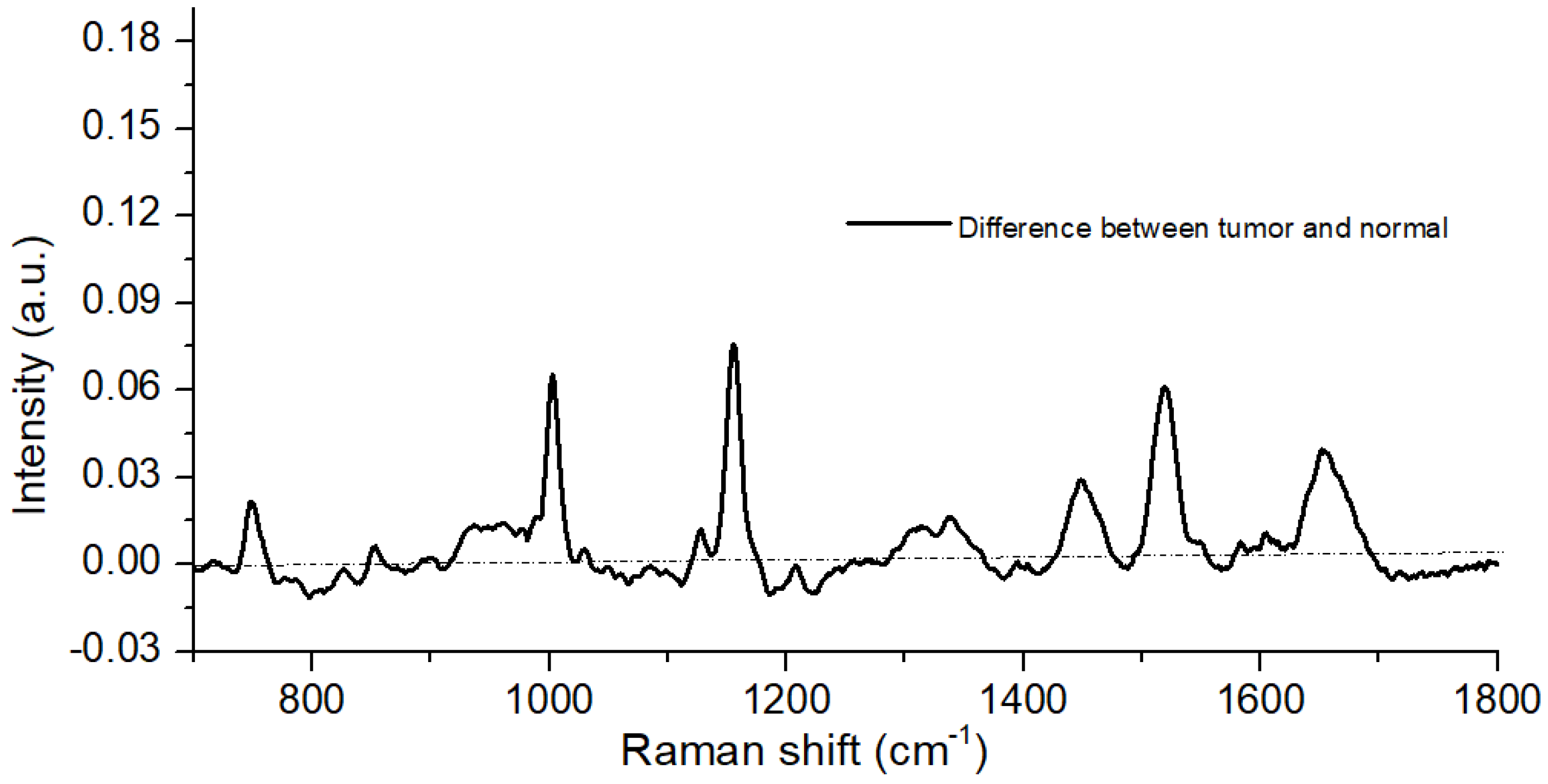
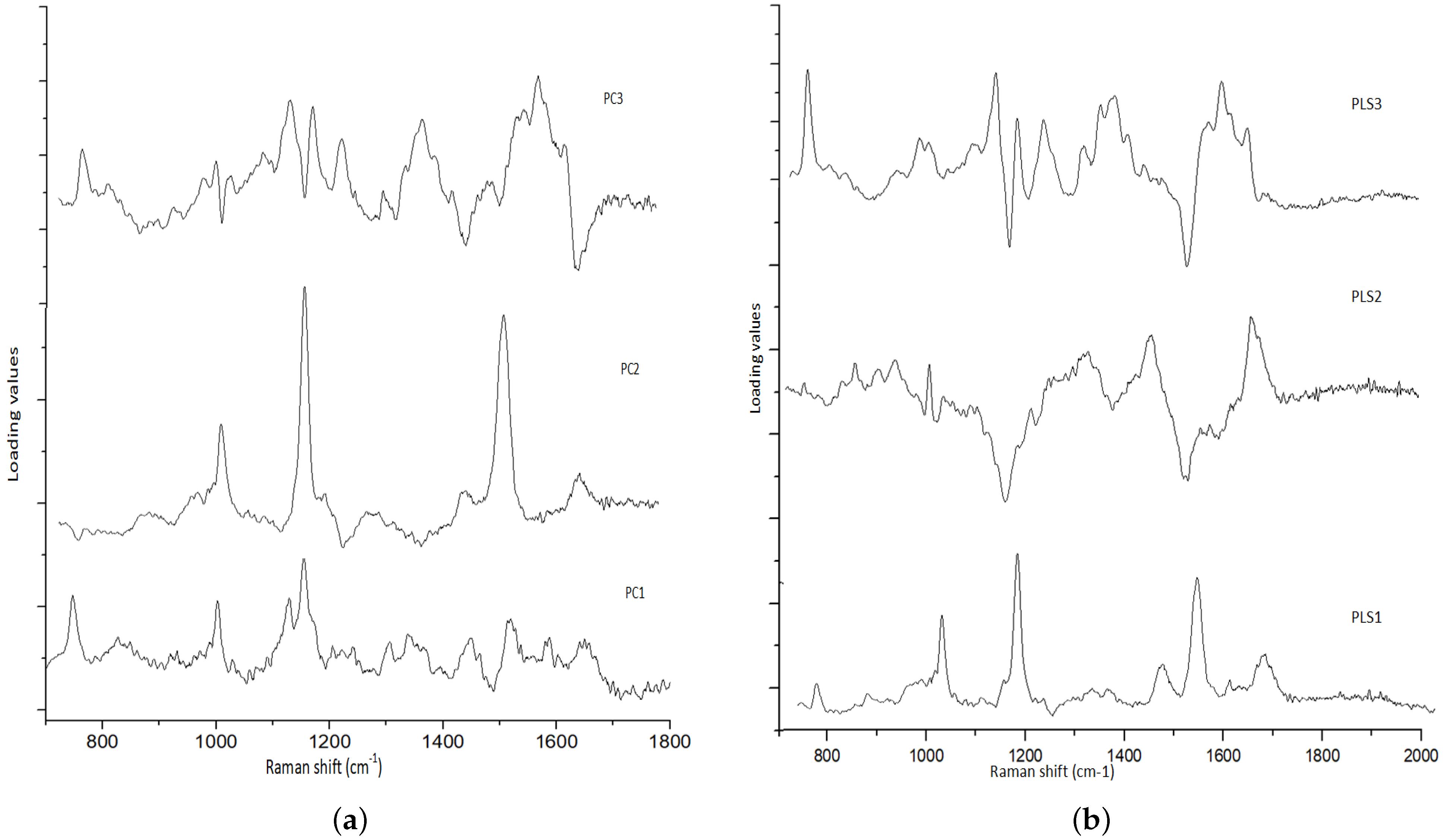
3.1. Spectral Analysis
3.2. Characteristics of the Validation Set
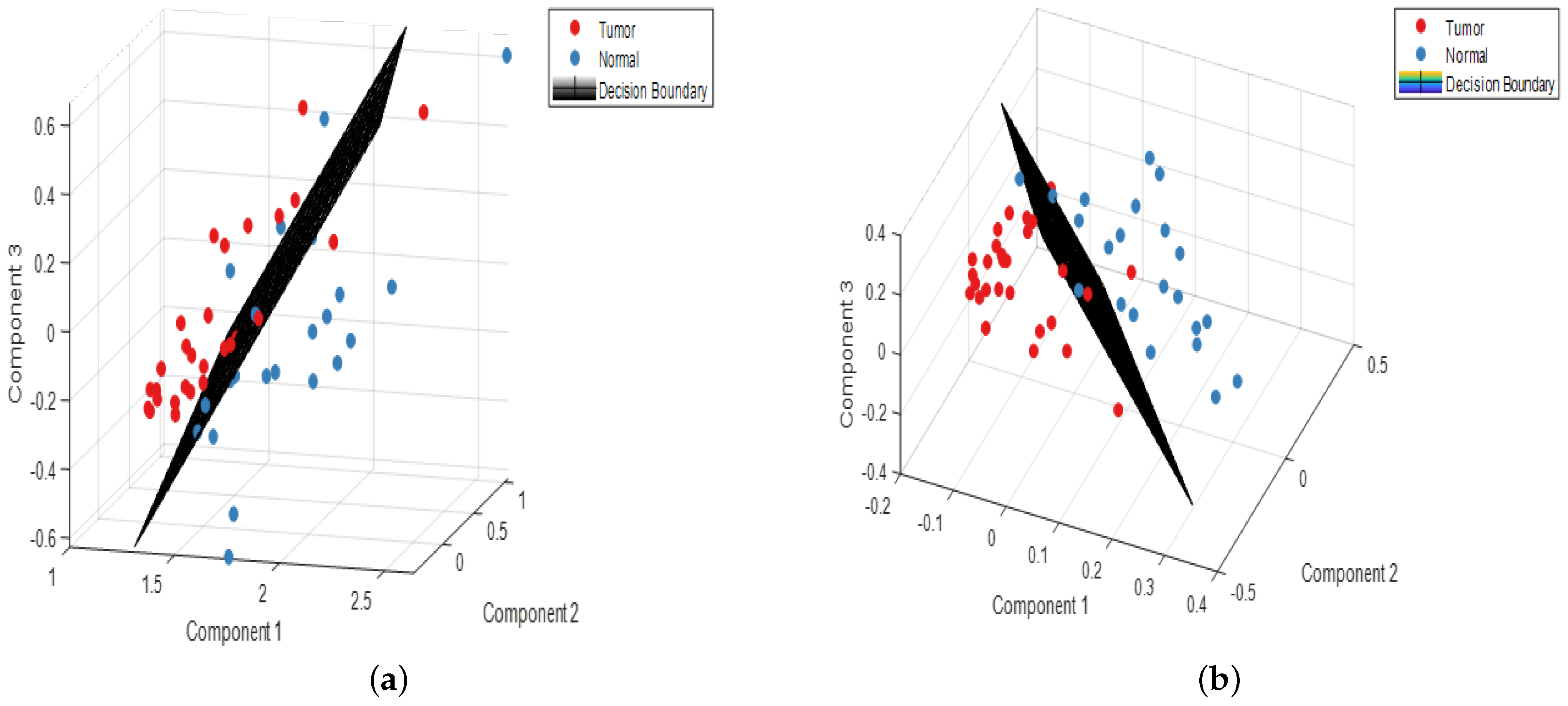
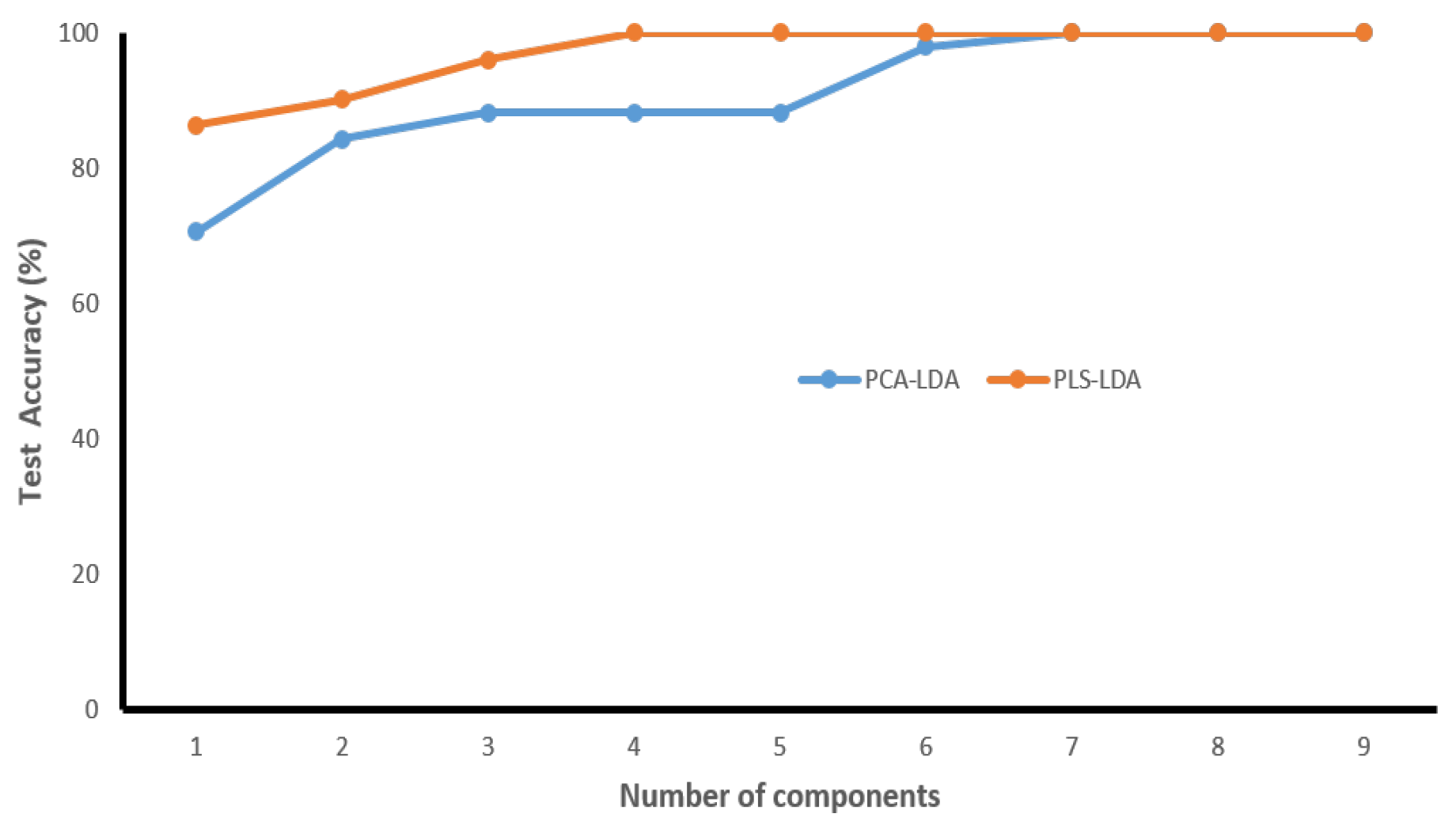
3.3. Validation of the Classification Models
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reddy, S.S.; Sharma, S.; Mysorekar, V. Expression of Epstein–barr virus among oral potentially malignant disorders and oral squamous cell carcinomas in the South Indian tobacco-chewing population. J. Oral Pathol. Med. 2017, 46, 454–459. [Google Scholar] [CrossRef]
- Hung, L.C.; Kung, P.T.; Lung, C.H.; Tsai, M.H.; Liu, S.A.; Chiu, L.T.; Huang, K.H.; Tsai, W.C. Assessment of the risk of oral cancer incidence in a high-risk population and establishment of a predictive model for oral cancer incidence using a population-based cohort in Taiwan. Int. J. Environ. Res. Public Health 2020, 17, 665. [Google Scholar] [CrossRef] [Green Version]
- Chou, H.C.; Lin, H.W.; Yang, J.H.; Lin, P.Y.; Cheng, S.J.; Wu, Y.H.; Kuo, Y.S. Clinical outcomes of oral cancer patients who survive for more than 5 years in Taiwan. J. Formos. Med. Assoc. 2019, 118, 1616–1622. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Tobacco Smoke and Involuntary Smoking; IARC: Lyon, France, 2004; Number 83.
- Prasad, G.; McCullough, M. Chemokines and cytokines as salivary biomarkers for the early diagnosis of oral cancer. Int. J. Dent. 2013, 2013, 813756. [Google Scholar] [CrossRef] [Green Version]
- Mortazavi, H.; Baharvand, M.; Mehdipour, M. Oral potentially malignant disorders: An overview of more than 20 entities. J. Dent. Res. Dent. Clin. Dent. Prospect. 2014, 8, 6. [Google Scholar]
- Mascitti, M.; Orsini, G.; Tosco, V.; Monterubbianesi, R.; Balercia, A.; Putignano, A.; Procaccini, M.; Santarelli, A. An overview on current non-invasive diagnostic devices in oral oncology. Front. Physiol. 2018, 9, 1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, E.C.; Tan, M.T.; Schwarz, R.A.; Richards-Kortum, R.R.; Gillenwater, A.M.; Vigneswaran, N. Noninvasive diagnostic adjuncts for the evaluation of potentially premalignant oral epithelial lesions: Current limitations and future directions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 670–681. [Google Scholar] [CrossRef]
- Warnakulasuriya, S. Diagnostic adjuncts on oral cancer and precancer: An update for practitioners. Br. Dent. J. 2017, 223, 663–666. [Google Scholar] [CrossRef]
- Jeng, M.J.; Sharma, M.; Chao, T.Y.; Li, Y.C.; Huang, S.F.; Chang, L.B.; Chow, L. Multiclass classification of autofluorescence images of oral cavity lesions based on quantitative analysis. PLoS ONE 2020, 15, e0228132. [Google Scholar] [CrossRef] [PubMed]
- Cordero, E.; Latka, I.; Matthäus, C.; Schie, I.W.; Popp, J. In-vivo Raman spectroscopy: From basics to applications. J. Biomed. Opt. 2018, 23, 071210. [Google Scholar] [CrossRef]
- Malini, R.; Venkatakrishna, K.; Kurien, J.; Pai, K.M.; Rao, L.; Kartha, V.; Krishna, C.M. Discrimination of normal, inflammatory, premalignant, and malignant oral tissue: A Raman spectroscopy study. Biopolym. Orig. Res. Biomol. 2006, 81, 179–193. [Google Scholar] [CrossRef]
- Chen, P.H.; Shimada, R.; Yabumoto, S.; Okajima, H.; Ando, M.; Chang, C.T.; Lee, L.T.; Wong, Y.K.; Chiou, A.; Hamaguchi, H.O. Automatic and objective oral cancer diagnosis by Raman spectroscopic detection of keratin with multivariate curve resolution analysis. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Czaplicka, M.; Kowalska, A.; Nowicka, A.; Kurzydłowski, D.; Gronkiewicz, Z.; Machulak, A.; Kukwa, W.; Kamińska, A. Raman spectroscopy and surface-enhanced Raman spectroscopy (SERS) spectra of salivary glands carcinoma, tumor and healthy tissues and their homogenates analyzed by chemometry: Towards development of the novel tool for clinical diagnosis. Anal. Chim. Acta 2021, 1177, 338784. [Google Scholar] [CrossRef]
- Zhan, Q.; Li, Y.; Yuan, Y.; Liu, J.; Li, Y. The accuracy of Raman spectroscopy in the detection and diagnosis of oral cancer: A systematic review and meta-analysis. J. Raman Spectrosc. 2020, 51, 2377–2397. [Google Scholar] [CrossRef]
- Barroso, E.; Smits, R.; Bakker Schut, T.; Ten Hove, I.; Hardillo, J.; Wolvius, E.; Baatenburg de Jong, R.J.; Koljenovic, S.; Puppels, G. Discrimination between oral cancer and healthy tissue based on water content determined by Raman spectroscopy. Anal. Chem. 2015, 87, 2419–2426. [Google Scholar] [CrossRef]
- Guze, K.; Pawluk, H.C.; Short, M.; Zeng, H.; Lorch, J.; Norris, C.; Sonis, S. Pilot study: Raman spectroscopy in differentiating premalignant and malignant oral lesions from normal mucosa and benign lesions in humans. Head Neck 2015, 37, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Knipfer, C.; Motz, J.; Adler, W.; Brunner, K.; Gebrekidan, M.T.; Hankel, R.; Agaimy, A.; Will, S.; Braeuer, A.; Neukam, F.W.; et al. Raman difference spectroscopy: A non-invasive method for identification of oral squamous cell carcinoma. Biomed. Opt. Express 2014, 5, 3252–3265. [Google Scholar] [CrossRef] [Green Version]
- Jeng, M.J.; Sharma, M.; Sharma, L.; Chao, T.Y.; Huang, S.F.; Chang, L.B.; Wu, S.L.; Chow, L. Raman spectroscopy analysis for optical diagnosis of oral cancer detection. J. Clin. Med. 2019, 8, 1313. [Google Scholar] [CrossRef]
- Jeng, M.J.; Sharma, M.; Sharma, L.; Huang, S.F.; Chang, L.B.; Wu, S.L.; Chow, L. Novel Quantitative Analysis Using Optical Imaging (VELscope) and Spectroscopy (Raman) Techniques for Oral Cancer Detection. Cancers 2020, 12, 3364. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Casp. J. Int. Med. 2013, 4, 627. [Google Scholar]
- Fan, J.; Upadhye, S.; Worster, A. Understanding receiver operating characteristic (ROC) curves. Can. J. Emerg. Med. 2006, 8, 19–20. [Google Scholar] [CrossRef]
- Maitra, S.; Yan, J. Principle component analysis and partial least squares: Two dimension reduction techniques for regression. Appl. Multivar. Stat. Model. 2008, 79, 79–90. [Google Scholar]
- Liu, W.; Sun, Z.; Chen, J.; Jing, C. Raman spectroscopy in colorectal cancer diagnostics: Comparison of PCA-LDA and PLS-DA models. J. Spectrosc. 2016, 2016, 1603609. [Google Scholar] [CrossRef]
- Abramczyk, H.; Brozek-Pluska, B.; Surmacki, J.; Jablonska, J.; Kordek, R. The label-free Raman imaging of human breast cancer. J. Mol. Liq. 2011, 164, 123–131. [Google Scholar] [CrossRef]
- Rau, J.V.; Fosca, M.; Graziani, V.; Taffon, C.; Rocchia, M.; Caricato, M.; Pozzilli, P.; Muda, A.O.; Crescenzi, A. Proof-of-concept Raman spectroscopy study aimed to differentiate thyroid follicular patterned lesions. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rimai, L.; Kilponen, R.; Gill, D. Excitation profiles of laser Raman spectra in the resonance region of two carotenoid pigments in solution. J. Am. Chem. Soc. 1970, 92, 3824–3825. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Sahu, A.; Tawde, S.; Pai, V.; Gera, P.; Chaturvedi, P.; Nair, S.; Krishna, C.M. Raman spectroscopy and cytopathology of oral exfoliated cells for oral cancer diagnosis. Anal. Methods 2015, 7, 7548–7559. [Google Scholar] [CrossRef]
- Lloyd, G.R.; Orr, L.E.; Christie-Brown, J.; McCarthy, K.; Rose, S.; Thomas, M.; Stone, N. Discrimination between benign, primary and secondary malignancies in lymph nodes from the head and neck utilising Raman spectroscopy and multivariate analysis. Analyst 2013, 138, 3900–3908. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Yang, J.; Yang, J.Y. Why can LDA be performed in PCA transformed space? Pattern Recognit. 2003, 36, 563–566. [Google Scholar] [CrossRef]
- Nocairi, H.; Qannari, E.M.; Vigneau, E.; Bertrand, D. Discrimination on latent components with respect to patterns. Application to multicollinear data. Comput. Stat. Data Anal. 2005, 48, 139–147. [Google Scholar] [CrossRef]
- Lilo, T.; Morais, C.L.; Ashton, K.M.; Pardilho, A.; Davis, C.; Dawson, T.P.; Gurusinghe, N.; Martin, F.L. Spectrochemical differentiation of meningioma tumours based on attenuated total reflection Fourier-transform infrared (ATR-FTIR) spectroscopy. Anal. Bioanal. Chem. 2020, 412, 1077–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cals, F.L.; Koljenović, S.; Hardillo, J.A.; de Jong, R.J.B.; Schut, T.C.B.; Puppels, G.J. Development and validation of Raman spectroscopic classification models to discriminate tongue squamous cell carcinoma from non-tumorous tissue. Oral Oncol. 2016, 60, 41–47. [Google Scholar] [CrossRef] [Green Version]
- Cals, F.L.; Schut, T.B.; Caspers, P.; De Jong, R.B.; Koljenović, S.; Puppels, G.J. Raman spectroscopic analysis of the molecular composition of oral cavity squamous cell carcinoma and healthy tongue tissue. Analyst 2018, 143, 4090–4102. [Google Scholar] [CrossRef] [PubMed]

| Subsites | Tongue | Buccal Mucosa | Gingiva | Total |
|---|---|---|---|---|
| Tumor | 20 | 30 | 17 | 67 |
| Normal | 18 | 29 | 17 | 64 |
| Raman Bands (cm) | Compound/Assignments |
|---|---|
| 749 | Symmetric Breathing of Tryptophan (Protein Assignment) |
| 848 | Tyrosine (protein assignment) |
| 1004 | Phenylalanine (ring breathing mode) |
| 1064 | Skeletal C-C stretch of lipids |
| 1123 | (C-N), proteins (protein assignment) |
| 1156 | C-C(and C-N) stretching of proteins/ carotenoid |
| 1168 | Lipids |
| 1220 | =CH bending (lipids) |
| 1302 | Amide III (protein) |
| 1450 | CH2 bending in proteins and lipids |
| 1517 | beta -carotene or porphyrin |
| 1650 | In normal tissue due to C=C of lipids and in tumor tissue due to |
| Amide-I-proteins |
| Dataset | Confusion Table | Performance Parameters | ||||
|---|---|---|---|---|---|---|
| PCA-LDA | Normal | Tumor | Total | Accuracy (%) | Sensitivity (%) | Specificity (%) |
| Normal | 28 | 0 | 28 | 90.2 | 78.3 | 100 |
| Tumor | 5 | 18 | 23 | |||
| PLS-LDA | Normal | Tumor | Total | Accuracy (%) | Sensitivity (%) | Specificity (%) |
| Normal | 28 | 0 | 28 | 100 | 100 | 100 |
| Tumor | 0 | 23 | 23 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, M.; Jeng, M.-J.; Young, C.-K.; Huang, S.-F.; Chang, L.-B. Developing an Algorithm for Discriminating Oral Cancerous and Normal Tissues Using Raman Spectroscopy. J. Pers. Med. 2021, 11, 1165. https://doi.org/10.3390/jpm11111165
Sharma M, Jeng M-J, Young C-K, Huang S-F, Chang L-B. Developing an Algorithm for Discriminating Oral Cancerous and Normal Tissues Using Raman Spectroscopy. Journal of Personalized Medicine. 2021; 11(11):1165. https://doi.org/10.3390/jpm11111165
Chicago/Turabian StyleSharma, Mukta, Ming-Jer Jeng, Chi-Kuang Young, Shiang-Fu Huang, and Liann-Be Chang. 2021. "Developing an Algorithm for Discriminating Oral Cancerous and Normal Tissues Using Raman Spectroscopy" Journal of Personalized Medicine 11, no. 11: 1165. https://doi.org/10.3390/jpm11111165
APA StyleSharma, M., Jeng, M.-J., Young, C.-K., Huang, S.-F., & Chang, L.-B. (2021). Developing an Algorithm for Discriminating Oral Cancerous and Normal Tissues Using Raman Spectroscopy. Journal of Personalized Medicine, 11(11), 1165. https://doi.org/10.3390/jpm11111165






