Expression Analysis and Mutational Status of Histone Methyltransferase KMT2D at Different Upper Tract Urothelial Carcinoma Locations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source and Patient Cohort
2.2. Pathologic Review and Follow-Up
2.3. Immunohistochemistry
2.4. Next-Generation Sequencing
2.5. Statistical Analysis
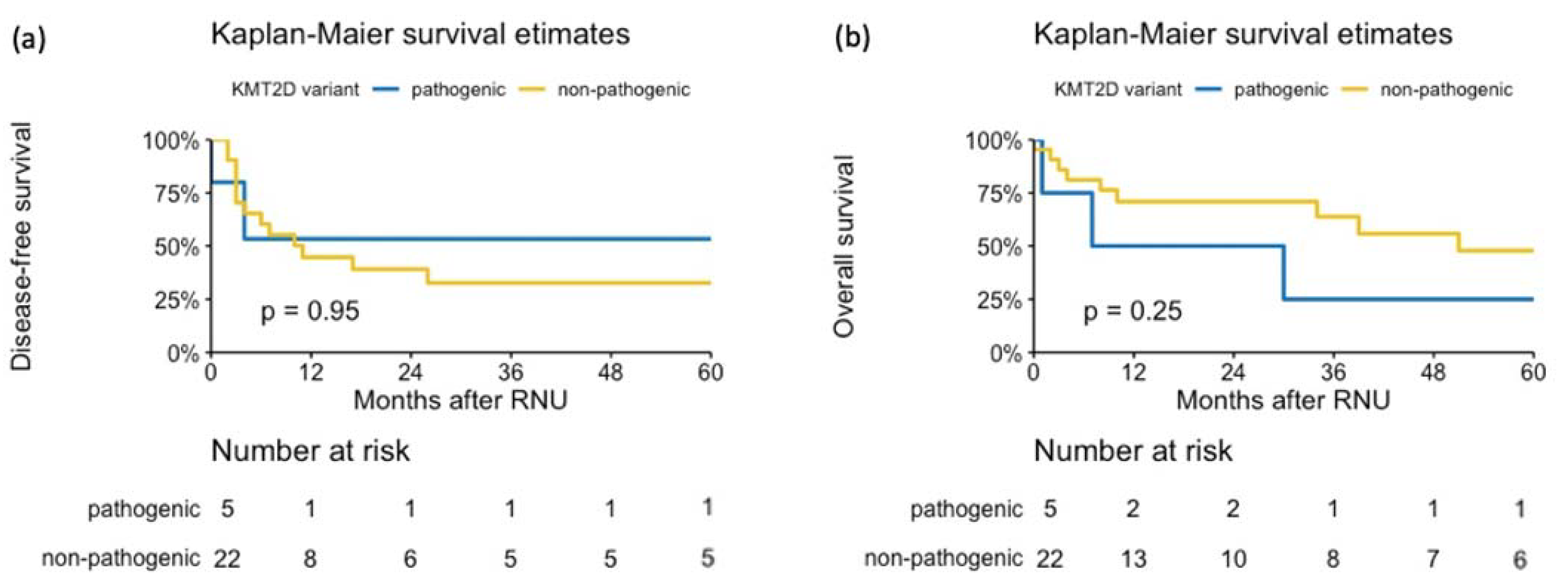
3. Results
3.1. KMT2D Expression
3.2. KMT2D Alterations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lughezzani, G.; Jeldres, C.; Isbarn, H.; Sun, M.; Shariat, S.F.; Alasker, A.; Pharand, D.; Widmer, H.; Arjane, P.; Graefen, M.; et al. Nephroureterectomy and segmental ureterectomy in the treatment of invasive upper tract urothelial carcinoma: A population-based study of 2299 patients. Eur. J. Cancer 2009, 45, 3291–3297. [Google Scholar] [CrossRef]
- Rink, M.; Ehdaie, B.; Cha, E.K.; Green, D.A.; Karakiewicz, P.I.; Babjuk, M.; Margulis, V.; Raman, J.D.; Svatek, R.S.; Fajkovic, H.; et al. Stage-specific impact of tumor location on oncologic outcomes in patients with upper and lower tract urothelial carcinoma following radical surgery. Eur. Urol. 2012, 62, 677–684. [Google Scholar] [CrossRef]
- Rouprêt, M.; Hupertan, V.; Seisen, T.; Colin, P.; Xylinas, E.; Yates, D.R.; Fajkovic, H.; Lotan, Y.; Raman, J.D.; Zigeuner, R.; et al. Prediction of cancer specific survival after radical nephroureterectomy for upper tract urothelial carcinoma: Development of an optimized postoperative nomogram using decision curve analysis. J. Urol. 2013, 189, 1662–1669. [Google Scholar] [CrossRef]
- Rouprêt, M.; Babjuk, M.; Burger, M.; Compérat, E.; Cowan, N.C.; Gontero, P.; Liedberg, F.; Masson-Lecomte, A.; Mostafid, A.H.; Palou, J.; et al. EAU Guidelines on Upper Urinary Tract Urothelial Carcinoma 2021. In European Association of Urology Guidelines; European Association of Urology: Arnhem, The Netherlands, 2021. [Google Scholar]
- Favaretto, R.L.; Shariat, S.F.; Savage, C.; Godoy, G.; Chade, D.C.; Kaag, M.; Bochner, B.H.; Coleman, J.; Dalbagni, G. Combining imaging and ureteroscopy variables in a preoperative multivariable model for prediction of muscle-invasive and non-organ confined disease in patients with upper tract urothelial carcinoma. BJU Int. 2012, 109, 77–82. [Google Scholar] [CrossRef]
- Favaretto, R.L.; Shariat, S.F.; Chade, D.C.; Godoy, G.; Adamy, A.; Kaag, M.; Bochner, B.H.; Coleman, J.; Dalbagni, G. The effect of tumor location on prognosis in patients treated with radical nephroureterectomy at Memorial Sloan-Kettering Cancer Center. Eur. Urol. 2010, 58, 574–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chromecki, T.F.; Cha, E.K.; Fajkovic, H.; Margulis, V.; Novara, G.; Scherr, D.S.; Lotan, Y.; Raman, J.D.; Kassouf, W.; Bensalah, K.; et al. The impact of tumor multifocality on outcomes in patients treated with radical nephroureterectomy. Eur. Urol. 2012, 61, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Janisch, F.; Mostafaei, H.; Lysenko, I.; Kimura, S.; Egawa, S.; Shariat, S.F. Prognostic value of preoperative blood-based biomarkers in upper tract urothelial carcinoma treated with nephroureterectomy: A systematic review and meta-analysis. In Urologic Oncology: Seminars and Original Investigations; 2020; Volume 38, pp. 315–333. [Google Scholar]
- Hassler, M.R.; Bray, F.; Catto, J.W.F.; Grollman, A.P.; Hartmann, A.; Margulis, V.; Matin, S.F.; Roupret, M.; Sfakianos, J.P.; Shariat, S.F.; et al. Molecular Characterization of Upper Tract Urothelial Carcinoma in the Era of Next-generation Sequencing: A Systematic Review of the Current Literature. Eur. Urol. 2020, 78, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Fagan, R.J.; Dingwall, A.K. COMPASS Ascending: Emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer. Cancer Lett. 2019, 458, 56–65. [Google Scholar] [CrossRef]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Du, Y.; Chen, Z.; Xu, D.; Lin, T.; Jin, S.; Wang, G.; Liu, Z.; Lu, M.; Chen, X.; et al. Macroscopic somatic clonal expansion in morphologically normal human urothelium. Science 2020, 370, 82–89. [Google Scholar] [CrossRef]
- Dawkins, J.B.N.; Wang, J.; Maniati, E.; Heward, J.A.; Koniali, L.; Kocher, H.M.; Martin, S.A.; Chelala, C.; Balkwill, F.R.; Fitzgibbon, J.; et al. Reduced expression of histone methyltransferases KMT2C and KMT2D correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Res. 2016, 76, 4861–4871. [Google Scholar] [CrossRef] [Green Version]
- do A. Rabello, D.; da S. Ferreira, V.D.A.; Berzoti-Coelho, M.G.; Burin, S.M.; Magro, C.L.; da C. Cacemiro, M.; Simões, B.P.; Saldanha-Araujo, F.; Castro, F.A.; Pittella-Silva, F. MLL2/KMT2D and MLL3/KMT2C expression correlates with disease progression and response to imatinib mesylate in chronic myeloid leukemia. Cancer Cell Int. 2018, 18, 1–10. [Google Scholar] [CrossRef]
- Bosgana, P.; Nikou, S.; Dimitrakopoulos, F.-I.; Logotheti, S.; Tzelepi, V.; Kalophonos, C.; Bravou, V.; Kourea, E.; Sampsonas, F.; Zolota, V. H3K4 Methylation Status and Lysine Specific Methyltransferase KMT2C Expression Correlate with Prognosis in Lung Adenocarcinoma. Curr. Mol. Pharmacol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Toska, E.; Osmanbeyoglu, H.U.; Castel, P.; Chan, C.; Hendrickson, R.C.; Elkabets, M.; Dickler, M.N.; Scaltriti, M.; Leslie, C.S.; Armstrong, S.A.; et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 2017, 355, 1324–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moss, T.J.; Qi, Y.; Xi, L.; Peng, B.; Kim, T.B.; Ezzedine, N.E.; Mosqueda, M.E.; Guo, C.C.; Czerniak, B.A.; Ittmann, M.; et al. Comprehensive Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur. Urol. 2017, 72, 641–649. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [Green Version]
- Rink, M.; Fajkovic, H.; Cha, E.K.; Gupta, A.; Karakiewicz, P.I.; Chun, F.K.; Lotan, Y.; Shariat, S.F. Death certificates are valid for the determination of cause of death in patients with upper and lower tract urothelial carcinoma. Eur. Urol. 2012, 61, 854–855. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, R.C.; Dou, Y. Hijacked in cancer: The KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 2015, 15, 334–346. [Google Scholar] [CrossRef] [Green Version]
- Milne, T.A.; Briggs, S.D.; Brock, H.W.; Martin, M.E.; Gibbs, D.; Allis, C.D.; Hess, J.L. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 2002, 10, 1107–1117. [Google Scholar] [CrossRef]
- Herz, H.-M.; Hu, D.; Shilatifard, A. Enhancer malfunction in cancer. Mol. Cell 2014, 53, 859–866. [Google Scholar] [CrossRef] [Green Version]
- Audenet, F.; Isharwal, S.; Cha, E.K.; Donoghue, M.T.A.; Drill, E.N.; Ostrovnaya, I.; Pietzak, E.J.; Sfakianos, J.P.; Bagrodia, A.; Murugan, P.; et al. Clonal relatedness and mutational differences between upper tract and bladder urothelial carcinoma. Clin. Cancer Res. 2019, 25, 967–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sfakianos, J.P.; Cha, E.K.; Iyer, G.; Scott, S.N.; Zabor, E.C.; Shah, R.H.; Ren, Q.; Bagrodia, A.; Kim, P.H.; Hakimi, A.A.; et al. Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur. Urol. 2015, 68, 970–977. [Google Scholar] [CrossRef] [Green Version]
- De Lorenzis, E.; Albo, G.; Longo, F.; Bebi, C.; Boeri, L.; Montanari, E. Current knowledge on genomic profiling of upper tract urothelial carcinoma. Genes 2021, 12, 333. [Google Scholar] [CrossRef]
- Yafi, F.A.; Novara, G.; Shariat, S.F.; Gupta, A.; Matsumoto, K.; Walton, T.J.; Fritsche, H.-M.; El-Hakim, A.; Trischler, S.; Martínez-Salamanca, J.I.; et al. Impact of tumour location versus multifocality in patients with upper tract urothelial carcinoma treated with nephroureterectomy and bladder cuff excision: A homogeneous series without perioperative chemotherapy. BJU Int. 2012, 110, E7–E13. [Google Scholar] [CrossRef] [PubMed]
- Ouzzane, A.; Colin, P.; Xylinas, E.; Pignot, G.; Ariane, M.M.; Saint, F.; Hoarau, N.; Adam, E.; Azemar, M.D.; Bensadoun, H.; et al. Ureteral and multifocal tumours have worse prognosis than renal pelvic tumours in urothelial carcinoma of the upper urinary tract treated by nephroureterectomy. Eur. Urol. 2011, 60, 1258–1265. [Google Scholar] [CrossRef]
- Zhang, X.; Bu, R.; Liu, Z.; Wu, B.; Bai, S. Development and Validation of a Model for Predicting Intravesical Recurrence in Organ-confined Upper Urinary Tract Urothelial Carcinoma Patients after Radical Nephroureterectomy: A Retrospective Study in One Center with Long-term Follow-up. Pathol. Oncol. Res. 2020, 26, 1741–1748. [Google Scholar] [CrossRef]
- Zhang, P.; Huang, Y. Genomic alterations in KMT2 family predict outcome of immune checkpoint therapy in multiple cancers. J. Hematol. Oncol. 2021, 14, 39. [Google Scholar] [CrossRef]
- Wang, J.; Xiu, J.; Baca, Y.; Battaglin, F.; Arai, H.; Kawanishi, N.; Soni, S.; Zhang, W.; Millstein, J.; Salhia, B.; et al. Large-scale analysis of KMT2 mutations defines a distinctive molecular subset with treatment implication in gastric cancer. Oncogene 2021, 40, 4894–4905. [Google Scholar] [CrossRef]
- Wang, G.; Chow, R.D.; Zhu, L.; Bai, Z.; Ye, L.; Zhang, F.; Renauer, P.A.; Dong, M.B.; Dai, X.; Zhang, X.; et al. CRISPR-GEMM Pooled Mutagenic Screening Identifies KMT2D as a Major Modulator of Immune Checkpoint Blockade. Cancer Discov. 2020, 10, 1912–1933. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Brownell, J.E. Biochemical perspectives on targeting KMT2 methyltransferases in cancer. Trends Pharmacol. Sci. 2021, 42, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.S.; Lee, M.G. Cancer-epigenetic function of the histone methyltransferase KMT2D and therapeutic opportunities for the treatment of KMT2D-deficient tumors. Oncotarget 2021, 12, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef] [PubMed]



| IHC | NGS | |||||||
|---|---|---|---|---|---|---|---|---|
| Overall | Stratified by KMT2D Staining | Overall | Stratified by KMT2D Variant | |||||
| N = 51 | Negative, N = 19 | Positive, N = 32 | p-Value | N = 27 | Pathogenic, N = 5 | Non-Pathogenic, N = 22 | p-Value | |
| Age | 73 (60, 79) | 73 (54, 78) | 73 (63, 78) | 0.7 | 71 (61, 80) | 61 (45, 71) | 73 (64, 80) | 0.2 |
| Sex | 0.5 | 0.6 | ||||||
| female | 24 (47%) | 10 (53%) | 14 (44%) | 12 (44%) | 3 (60%) | 9 (41%) | ||
| male | 27 (53%) | 9 (47%) | 18 (56%) | 15 (56%) | 2 (40%) | 13 (59%) | ||
| Pathological tumor stage | 0.8 | >0.9 | ||||||
| pT1 | 14 (27%) | 6 (32%) | 8 (25%) | 8 (30%) | 2 (40%) | 6 (27%) | ||
| pT2 | 6 (12%) | 1 (5.3%) | 5 (16%) | 1 (3.7%) | 0 (0%) | 1 (4.5%) | ||
| pT3 | 20 (39%) | 8 (42%) | 12 (38%) | 11 (41%) | 2 (40%) | 9 (41%) | ||
| pT4 | 7 (14%) | 2 (11%) | 5 (16%) | 5 (19%) | 1 (20%) | 4 (18%) | ||
| pTa | 4 (7.8%) | 2 (11%) | 2 (6.2%) | 2 (7.4%) | 0 (0%) | 2 (9.1%) | ||
| N stage | 0.3 | 0.5 | ||||||
| pN0 | 8 (16%) | 2 (11%) | 6 (19%) | 4 (15%) | 0 (0%) | 4 (18%) | ||
| pN1 | 6 (12%) | 2 (11%) | 4 (12%) | 5 (19%) | 2 (40%) | 3 (14%) | ||
| pN2 | 4 (7.8%) | 0 (0%) | 4 (12%) | 3 (11%) | 0 (0%) | 3 (14%) | ||
| pNx | 33 (65%) | 15 (79%) | 18 (56%) | 15 (56%) | 3 (60%) | 12 (55%) | ||
| M stage | 0.8 | >0.9 | ||||||
| M0 | 45 (88%) | 16 (84%) | 29 (91%) | 23 (85%) | 5 (100%) | 18 (82%) | ||
| M1 | 3 (5.9%) | 2 (11%) | 1 (3.1%) | 2 (7.4%) | 0 (0%) | 2 (9.1%) | ||
| Mx | 3 (5.9%) | 1 (5.3%) | 2 (6.2%) | 2 (7.4%) | 0 (0%) | 2 (9.1%) | ||
| Grade | 0.7 | >0.9 | ||||||
| HG | 39 (76%) | 14 (74%) | 25 (78%) | 19 (70%) | 4 (80%) | 15 (68%) | ||
| LG | 12 (24%) | 5 (26%) | 7 (22%) | 8 (30%) | 1 (20%) | 7 (32%) | ||
| Location | 0.8 | 0.02 | ||||||
| multifocal | 9 (18%) | 3 (16%) | 6 (19%) | 6 (22%) | 3 (60%) | 3 (14%) | ||
| pelvis | 34 (67%) | 13 (68%) | 21 (66%) | 19 (70%) | 1 (20%) | 18 (82%) | ||
| proximal | 6 (12%) | 3 (16%) | 3 (9.4%) | 2 (7.4%) | 1 (20%) | 1 (4.5%) | ||
| distal | 2 (3.9%) | 0 (0%) | 2 (6.2%) | |||||
| Tumor side | 0.5 | 0.6 | ||||||
| left | 24 (47%) | 10 (53%) | 14 (44%) | 14 (52%) | 2 (40%) | 12 (55%) | ||
| right | 27 (53%) | 9 (47%) | 18 (56%) | 13 (48%) | 3 (60%) | 10 (45%) | ||
| Number of positive lymph nodes | >0.9 | 0.3 | ||||||
| 0 | 8 (53%) | 2 (67%) | 6 (50%) | 4 (44%) | 0 (0%) | 4 (57%) | ||
| 1 | 4 (27%) | 1 (33%) | 3 (25%) | 3 (33%) | 2 (100%) | 1 (14%) | ||
| 3 | 2 (13%) | 0 (0%) | 2 (17%) | 1 (11%) | 0 (0%) | 1 (14%) | ||
| 5 | 1 (6.7%) | 0 (0%) | 1 (8.3%) | 1 (11%) | 0 (0%) | 1 (14%) | ||
| Chemotherapy | 13 (26%) | 3 (17%) | 10 (31%) | 0.3 | 9 (35%) | 1 (25%) | 8 (36%) | >0.9 |
| Metastasis | 19 (37%) | 6 (32%) | 13 (41%) | 0.5 | 13 (48%) | 2 (40%) | 11 (50%) | >0.9 |
| History of bladder cancer | 8 (16%) | 0 (0%) | 8 (25%) | 0.02 | 2 (7.4%) | 0 (0%) | 2 (9.1%) | >0.9 |
| Recurrence | 22 (43%) | 7 (37%) | 15 (47%) | 0.5 | 11 (41%) | 1 (20%) | 10 (45%) | 0.6 |
| Site of recurrence | >0.9 | 0.6 | ||||||
| bladder | 19 (37%) | 7 (37%) | 12 (38%) | 9 (33%) | 1 (20%) | 8 (36%) | ||
| no | 32 (63%) | 12 (63%) | 20 (62%) | 18 (67%) | 4 (80%) | 14 (64%) | ||
| Patient died | 23 (45%) | 7 (37%) | 16 (50%) | 0.4 | 14 (52%) | 3 (60%) | 11 (50%) | >0.9 |
| Months to Death/Censor | 29 (8, 80) | 10 (4, 26) | 36 (16, 102) | 0.036 | 18 (6, 60) | 7 (1, 30) | 20 (8, 64) | 0.2 |
| Median (IQR); n (%) | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laukhtina, E.; Lemberger, U.; Bruchbacher, A.; Ilijazi, D.; Korn, S.; Berndl, F.; D’Andrea, D.; Susani, M.; Enikeev, D.; Compérat, E.; et al. Expression Analysis and Mutational Status of Histone Methyltransferase KMT2D at Different Upper Tract Urothelial Carcinoma Locations. J. Pers. Med. 2021, 11, 1147. https://doi.org/10.3390/jpm11111147
Laukhtina E, Lemberger U, Bruchbacher A, Ilijazi D, Korn S, Berndl F, D’Andrea D, Susani M, Enikeev D, Compérat E, et al. Expression Analysis and Mutational Status of Histone Methyltransferase KMT2D at Different Upper Tract Urothelial Carcinoma Locations. Journal of Personalized Medicine. 2021; 11(11):1147. https://doi.org/10.3390/jpm11111147
Chicago/Turabian StyleLaukhtina, Ekaterina, Ursula Lemberger, Andreas Bruchbacher, Dafina Ilijazi, Stephan Korn, Florian Berndl, David D’Andrea, Martin Susani, Dmitry Enikeev, Eva Compérat, and et al. 2021. "Expression Analysis and Mutational Status of Histone Methyltransferase KMT2D at Different Upper Tract Urothelial Carcinoma Locations" Journal of Personalized Medicine 11, no. 11: 1147. https://doi.org/10.3390/jpm11111147
APA StyleLaukhtina, E., Lemberger, U., Bruchbacher, A., Ilijazi, D., Korn, S., Berndl, F., D’Andrea, D., Susani, M., Enikeev, D., Compérat, E., Shariat, S. F., & Hassler, M. R. (2021). Expression Analysis and Mutational Status of Histone Methyltransferase KMT2D at Different Upper Tract Urothelial Carcinoma Locations. Journal of Personalized Medicine, 11(11), 1147. https://doi.org/10.3390/jpm11111147







