Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression Analysis of the NEK Genes Family
2.2. Survival Analysis of the NEK Genes Family
2.3. DNA Methylation of the NEK Genes Family
2.4. Functional Enrichment and Micro (mi)RNA-Regulated Networks Analysis
2.5. Correlation Analysis between Gene Expressions and Immune Infiltration
3. Results
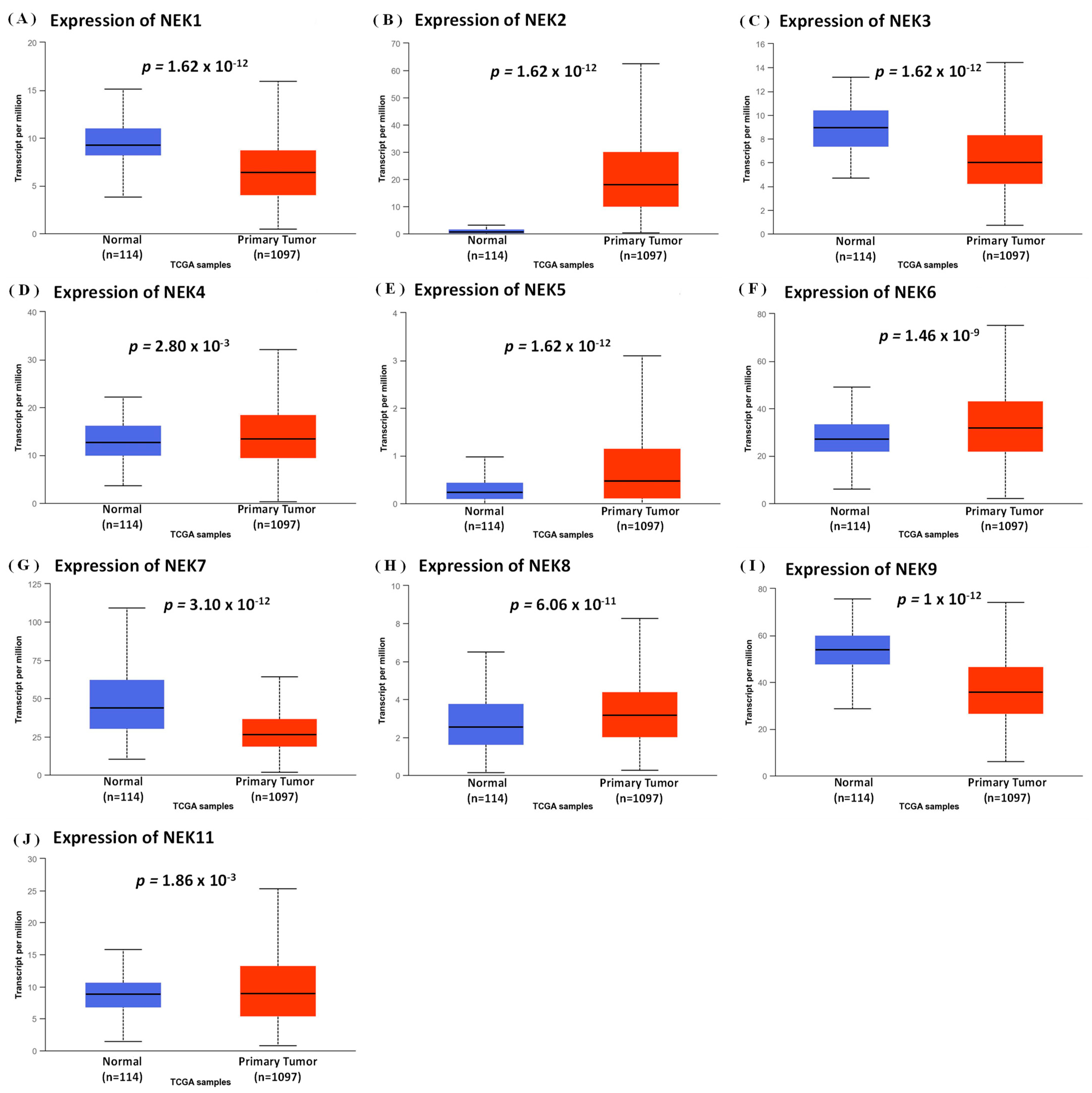
3.1. Expression Analysis of NEK Family Members in Breast Cancer
3.2. Prognostic Value of the NEK Family Members in Breast Cancer
3.3. DNA Methylation Analysis of the NEK Family Members in Breast Cancer
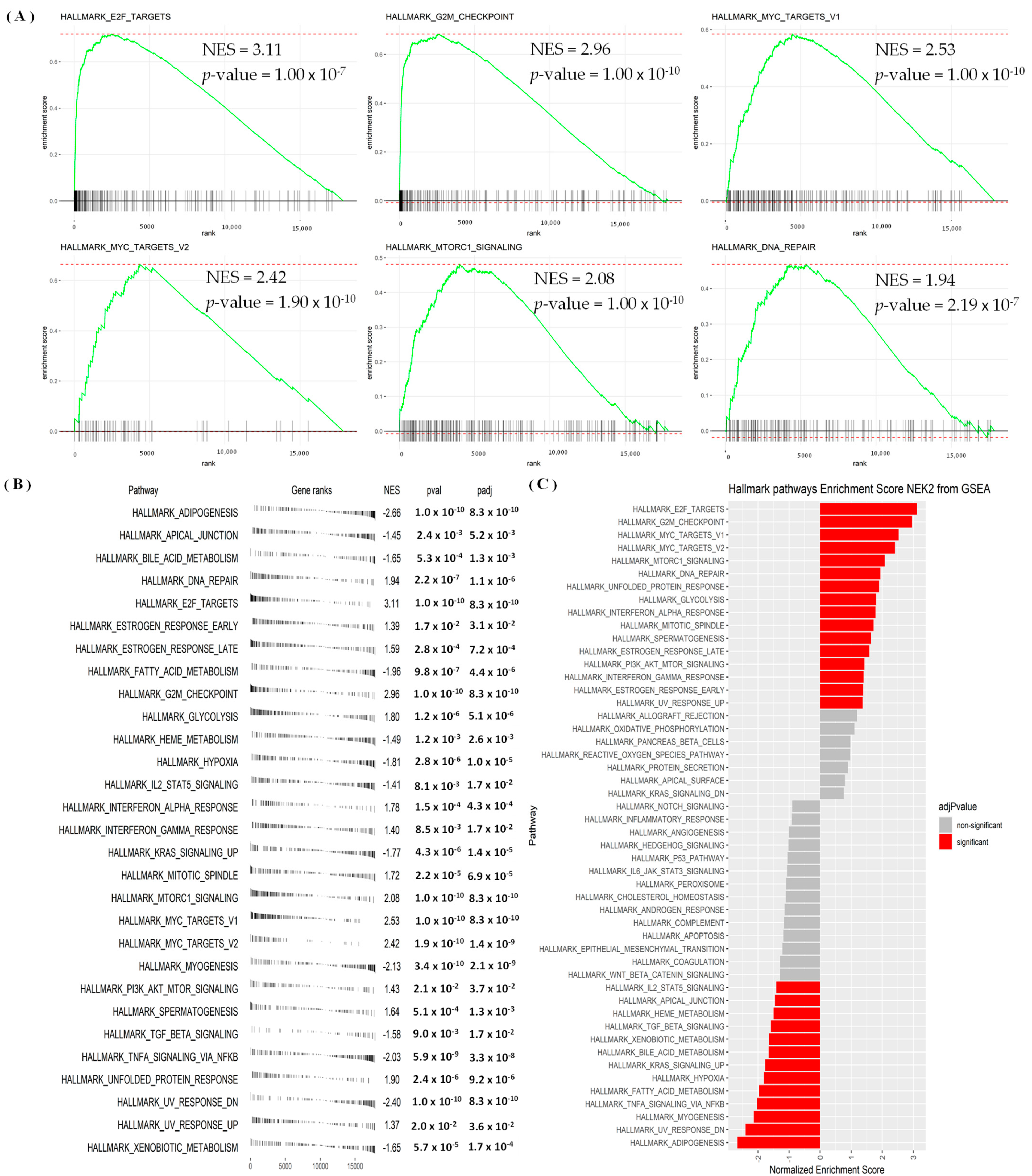
3.4. Regulated Networks of NEK2 Gene Expressions in Breast Cancer
3.5. Levels of Immune Infiltration in Breast Cancer Were Linked to Expression of NEK2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.-Y.; Lee, C.-H.; Chuang, Y.-H.; Lee, J.-Y.; Chiu, Y.-Y.; Wu Lee, Y.-H.; Jong, Y.-J.; Hwang, J.-K.; Huang, S.-H.; Chen, L.-C.; et al. Membrane protein-regulated networks across human cancers. Nat. Commun. 2019, 10, 3131. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.-T.; Huang, C.-S.; Tu, C.-C.; Liu, C.-Y.; Huang, C.-J.; Ho, Y.-S.; Tu, S.-H.; Tseng, L.-M.; Huang, C.-C. Multi-gene signature of microcalcification and risk prediction among Taiwanese breast cancer. Sci. Rep. 2020, 10, 18276. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Liao, Y.C.; Ho, Y.S.; Chen, L.C.; Chang, H.W.; Cheng, T.C.; Liu, D.; Lee, W.R.; Shen, S.C.; Wu, C.H.; et al. The alpha9 Nicotinic Acetylcholine Receptor Mediates Nicotine-Induced PD-L1 Expression and Regulates Melanoma Cell Proliferation and Migration. Cancers 2019, 11, 1991. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.L.; Kuo, Y.C.; Ho, Y.S.; Huang, Y.H. Triple-Negative Breast Cancer: Current Understanding and Future Therapeutic Breakthrough Targeting Cancer Stemness. Cancers 2019, 11, 1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marczyk, M.; Polańska, J.; Wojcik, A.; Lundholm, L. Analysis of the Applicability of microRNAs in Peripheral Blood Leukocytes as Biomarkers of Sensitivity and Exposure to Fractionated Radiotherapy towards Breast Cancer. Int. J. Mol. Sci. 2021, 22, 8705. [Google Scholar] [CrossRef]
- Marczyk, M.; Patwardhan, G.A.; Zhao, J.; Qu, R.; Li, X.; Wali, V.B.; Gupta, A.K.; Pillai, M.M.; Kluger, Y.; Yan, Q.; et al. Multi-Omics Investigation of Innate Navitoclax Resistance in Triple-Negative Breast Cancer Cells. Cancers 2020, 12, 2551. [Google Scholar] [CrossRef] [PubMed]
- Drobin, K.; Marczyk, M.; Halle, M.; Danielsson, D.; Papiez, A.; Sangsuwan, T.; Bendes, A.; Hong, M.G.; Qundos, U.; Harms-Ringdahl, M.; et al. Molecular Profiling for Predictors of Radiosensitivity in Patients with Breast or Head-and-Neck Cancer. Cancers 2020, 12, 753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madsen, R.R.; Longden, J.; Knox, R.G.; Robin, X.; Völlmy, F.; Macleod, K.G.; Moniz, L.S.; Carragher, N.O.; Linding, R.; Vanhaesebroeck, B.; et al. NODAL/TGFβ signalling mediates the self-sustained stemness induced by PIK3CA (H1047R) homozygosity in pluripotent stem cells. Dis. Model. Mech. 2021, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Berenjeno, I.M.; Piñeiro, R.; Castillo, S.D.; Pearce, W.; McGranahan, N.; Dewhurst, S.M.; Meniel, V.; Birkbak, N.J.; Lau, E.; Sansregret, L.; et al. Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling. Nat. Commun. 2017, 8, 1773. [Google Scholar] [CrossRef] [Green Version]
- Alliouachene, S.; Bilanges, B.; Chaussade, C.; Pearce, W.; Foukas, L.C.; Scudamore, C.L.; Moniz, L.S.; Vanhaesebroeck, B. Inactivation of class II PI3K-C2α induces leptin resistance, age-dependent insulin resistance and obesity in male mice. Diabetologia 2016, 59, 1503–1512. [Google Scholar] [CrossRef] [Green Version]
- Moniz, L.; Dutt, P.; Haider, N.; Stambolic, V. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, M.J.; Krien, M.J.; Hunter, T. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 2003, 13, 221–228. [Google Scholar] [CrossRef]
- Pang, K.H.; Esperto, F.; Noon, A.P.; EAU Young Academic Urologists-Urothelial Cancer Working party. Opportunities of next-generation sequencing in non-muscle invasive bladder cancer outcome prediction. Transl. Urol. 2017, 6, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Cabral de Almeida Cardoso, L.; Rodriguez-Laguna, L.; Del Carmen Crespo, M.; Vallespin, E.; Palomares-Bralo, M.; Martin-Arenas, R.; Rueda-Arenas, I.; Silvestre de Faria, P.A.; Group, G.-C.W.; Garcia-Miguel, P.; et al. Array CGH Analysis of Paired Blood and Tumor Samples from Patients with Sporadic Wilms Tumor. PLoS ONE 2015, 10, e0136812. [Google Scholar] [CrossRef]
- Dang, T.T.; Westcott, J.M.; Maine, E.A.; Kanchwala, M.; Xing, C.; Pearson, G.W. DeltaNp63alpha induces the expression of FAT2 and Slug to promote tumor invasion. Oncotarget 2016, 7, 28592–28611. [Google Scholar] [CrossRef] [Green Version]
- Hayward, D.G.; Clarke, R.B.; Faragher, A.J.; Pillai, M.R.; Hagan, I.M.; Fry, A.M. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004, 64, 7370–7376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, S.L.; DeMaria, J.E.; Freier, D.O.; Riegel, A.M.; Clevenger, C.V. Novel association of Vav2 and Nek3 modulates signaling through the human prolactin receptor. Mol. Endocrinol. 2005, 19, 939–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Wu, N.; Liu, L.; Dong, H.; Liu, X. microRNA-128-3p overexpression inhibits breast cancer stem cell characteristics through suppression of Wnt signalling pathway by down-regulating NEK2. J. Cell Mol. Med. 2020, 24, 7353–7369. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Zhang, J.; Yang, X.; Wu, Z.; Sun, C.; Wang, Z.; Wang, B. NEK5 promotes breast cancer cell proliferation through up-regulation of Cyclin A2. Mol. Carcinog. 2019, 58, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Capra, M.; Nuciforo, P.G.; Confalonieri, S.; Quarto, M.; Bianchi, M.; Nebuloni, M.; Boldorini, R.; Pallotti, F.; Viale, G.; Gishizky, M.L.; et al. Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res. 2006, 66, 8147–8154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, M.W.; O’Regan, L.; Mas-Droux, C.; Blot, J.M.; Cheung, J.; Hoelder, S.; Fry, A.M.; Bayliss, R. An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9. Mol. Cell 2009, 36, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Bowers, A.J.; Boylan, J.F. Nek8, a NIMA family kinase member, is overexpressed in primary human breast tumors. Gene 2004, 328, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, A.C.; Beesley, J.; McGuffog, L.; Sinilnikova, O.M.; Healey, S.; Neuhausen, S.L.; Ding, Y.C.; Rebbeck, T.R.; Weitzel, J.N.; Lynch, H.T.; et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: Implications for risk prediction. Cancer Res. 2010, 70, 9742–9754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulligan, A.M.; Couch, F.J.; Barrowdale, D.; Domchek, S.M.; Eccles, D.; Nevanlinna, H.; Ramus, S.J.; Robson, M.; Sherman, M.; Spurdle, A.B.; et al. Common breast cancer susceptibility alleles are associated with tumour subtypes in BRCA1 and BRCA2 mutation carriers: Results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Res. 2011, 13, R110. [Google Scholar] [CrossRef] [Green Version]
- Haider, N.; Dutt, P.; van de Kooij, B.; Ho, J.; Palomero, L.; Pujana, M.A.; Yaffe, M.; Stambolic, V. NEK10 tyrosine phosphorylates p53 and controls its transcriptional activity. Oncogene 2020, 39, 5252–5266. [Google Scholar] [CrossRef] [PubMed]
- Sabir, S.R.; Sahota, N.K.; Jones, G.D.; Fry, A.M. Loss of Nek11 Prevents G2/M Arrest and Promotes Cell Death in HCT116 Colorectal Cancer Cells Exposed to Therapeutic DNA Damaging Agents. PLoS ONE 2015, 10, e0140975. [Google Scholar] [CrossRef] [Green Version]
- Moniz, L.S.; Stambolic, V. Nek10 mediates G2/M cell cycle arrest and MEK autoactivation in response to UV irradiation. Mol. Cell Biol. 2011, 31, 30–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavan, I.C.B.; Peres de Oliveira, A.; Dias, P.R.F.; Basei, F.L.; Issayama, L.K.; Ferezin, C.C.; Silva, F.R.; Rodrigues de Oliveira, A.L.; Alves Dos Reis Moura, L.; Martins, M.B.; et al. On Broken Ne(c)ks and Broken DNA: The Role of Human NEKs in the DNA Damage Response. Cells 2021, 10, 507. [Google Scholar] [CrossRef]
- Schouten, P.C.; Richters, L.K.; Vis, D.J.; Kommoss, S.; van Dyk, E.; Ernst, C.; Kluin, R.J.C.; Marme, F.; Lips, E.H.; Schmidt, S.; et al. Ovarian cancer specific BRCA-like copy number aberration classifiers detect mutations associated with homologous recombination deficiency in the AGO-TR1 trial. Clin. Cancer Res. 2021, 21, 1673. [Google Scholar] [CrossRef]
- Boussios, S.; Karathanasi, A.; Cooke, D.; Neille, C.; Sadauskaite, A.; Moschetta, M.; Zakynthinakis-Kyriakou, N.; Pavlidis, N. PARP Inhibitors in Ovarian Cancer: The Route to “Ithaca”. Diagnostics 2019, 9, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ioannidou, E.; Moschetta, M.; Shah, S.; Parker, J.S.; Ozturk, M.A.; Pappas-Gogos, G.; Sheriff, M.; Rassy, E.; Boussios, S. Angiogenesis and Anti-Angiogenic Treatment in Prostate Cancer: Mechanisms of Action and Molecular Targets. Int. J. Mol. Sci. 2021, 22, 9926. [Google Scholar] [CrossRef] [PubMed]
- Revythis, A.; Shah, S.; Kutka, M.; Moschetta, M.; Ozturk, M.A.; Pappas-Gogos, G.; Ioannidou, E.; Sheriff, M.; Rassy, E.; Boussios, S. Unraveling the Wide Spectrum of Melanoma Biomarkers. Diagnostics 2021, 11, 1341. [Google Scholar] [CrossRef] [PubMed]
- Saxby, H.; Mikropoulos, C.; Boussios, S. An Update on the Prognostic and Predictive Serum Biomarkers in Metastatic Prostate Cancer. Diagnostics 2020, 10, 549. [Google Scholar] [CrossRef]
- Malumbres, M.; Barbacid, M. Cell cycle kinases in cancer. Curr. Opin. Genet. Dev. 2007, 17, 60–65. [Google Scholar] [CrossRef]
- de Carcer, G.; Perez de Castro, I.; Malumbres, M. Targeting cell cycle kinases for cancer therapy. Curr. Med. Chem. 2007, 14, 969–985. [Google Scholar] [CrossRef]
- Lin, J.C.; Liu, T.P.; Yang, P.M. CDKN2A-Inactivated Pancreatic Ductal Adenocarcinoma Exhibits Therapeutic Sensitivity to Paclitaxel: A Bioinformatics Study. J. Clin. Med. 2020, 9, 4019. [Google Scholar] [CrossRef]
- Lin, T.Y.; Wang, P.W.; Huang, C.H.; Yang, P.M.; Pan, T.L. Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics. Int. J. Mol. Sci. 2020, 21, 6077. [Google Scholar] [CrossRef]
- Liu, L.W.; Hsieh, Y.Y.; Yang, P.M. Bioinformatics Data Mining Repurposes the JAK2 (Janus Kinase 2) Inhibitor Fedratinib for Treating Pancreatic Ductal Adenocarcinoma by Reversing the KRAS (Kirsten Rat Sarcoma 2 Viral Oncogene Homolog)-Driven Gene Signature. J. Pers. Med. 2020, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.M.; Hsieh, Y.Y.; Du, J.L.; Yen, S.C.; Hung, C.F. Sequential Interferon β-Cisplatin Treatment Enhances the Surface Exposure of Calreticulin in Cancer Cells via an Interferon Regulatory Factor 1-Dependent Manner. Biomolecules 2020, 10, 643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.M.; Lin, L.S.; Liu, T.P. Sorafenib Inhibits Ribonucleotide Reductase Regulatory Subunit M2 (RRM2) in Hepatocellular Carcinoma Cells. Biomolecules 2020, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Cooke, D.L.; McCoy, D.B.; Halbach, V.V.; Hetts, S.W.; Amans, M.R.; Dowd, C.F.; Higashida, R.T.; Lawson, D.; Nelson, J.; Wang, C.Y.; et al. Endovascular Biopsy: In Vivo Cerebral Aneurysm Endothelial Cell Sampling and Gene Expression Analysis. Transl. Stroke Res. 2018, 9, 20–33. [Google Scholar] [CrossRef]
- Weng, T.Y.; Wu, H.F.; Li, C.Y.; Hung, Y.H.; Chang, Y.W.; Chen, Y.L.; Hsu, H.P.; Chen, Y.H.; Wang, C.Y.; Chang, J.Y.; et al. Homoharringtonine induced immune alteration for an Efficient Anti-tumor Response in Mouse Models of Non-small Cell Lung Adenocarcinoma Expressing Kras Mutation. Sci. Rep. 2018, 8, 8216. [Google Scholar] [CrossRef]
- Wang, C.Y.; Li, C.Y.; Hsu, H.P.; Cho, C.Y.; Yen, M.C.; Weng, T.Y.; Chen, W.C.; Hung, Y.H.; Lee, K.T.; Hung, J.H.; et al. PSMB5 plays a dual role in cancer development and immunosuppression. Am. J. Cancer Res. 2017, 7, 2103–2120. [Google Scholar] [PubMed]
- Lanczky, A.; Nagy, A.; Bottai, G.; Munkacsy, G.; Szabo, A.; Santarpia, L.; Gyorffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016, 160, 439–446. [Google Scholar] [CrossRef]
- Nagy, A.; Munkacsy, G.; Gyorffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Wang, C.Y.; Chang, Y.C.; Kuo, Y.L.; Lee, K.T.; Chen, P.S.; Cheung, C.H.A.; Chang, C.P.; Phan, N.N.; Shen, M.R.; Hsu, H.P. Mutation of the PTCH1 gene predicts recurrence of breast cancer. Sci. Rep. 2019, 9, 16359. [Google Scholar] [CrossRef]
- Cho, C.Y.; Lee, K.T.; Chen, W.C.; Wang, C.Y.; Chang, Y.S.; Huang, H.L.; Hsu, H.P.; Yen, M.C.; Lai, M.Z.; Lai, M.D. MST3 promotes proliferation and tumorigenicity through the VAV2/Rac1 signal axis in breast cancer. Oncotarget 2016, 7, 14586–14604. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.L.; Chen, W.C.; Hsu, H.P.; Cho, C.Y.; Hung, Y.H.; Wang, C.Y.; Lai, M.D. Argininosuccinate lyase is a potential therapeutic target in breast cancer. Oncol. Rep. 2015, 34, 3131–3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martens, J.W.; Margossian, A.L.; Schmitt, M.; Foekens, J.; Harbeck, N. DNA methylation as a biomarker in breast cancer. Future Oncol. 2009, 5, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Modhukur, V.; Iljasenko, T.; Metsalu, T.; Lokk, K.; Laisk-Podar, T.; Vilo, J. MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018, 10, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Walter, W.; Sanchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO:: TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergushichev, A.A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2021. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Damrauer, J.S.; Hoadley, K.A.; Chism, D.D.; Fan, C.; Tiganelli, C.J.; Wobker, S.E.; Yeh, J.J.; Milowsky, M.I.; Iyer, G.; Parker, J.S.; et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. USA 2014, 111, 3110. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Xiang, L.; Li, T.; Bai, Z. Cancer Hallmarks, Biomarkers and Breast Cancer Molecular Subtypes. J. Cancer 2016, 7, 1281–1294. [Google Scholar] [CrossRef] [Green Version]
- Cirillo, E.; Parnell, L.D.; Evelo, C.T. A Review of Pathway-Based Analysis Tools That Visualize Genetic Variants. Front. Genet. 2017, 8, 174. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, W.; Wang, X.; Xu, M.; Zhang, L.; Ding, L.; Guo, R.; Shi, Y. Network pharmacology-based strategy to investigate pharmacological mechanisms of Zuojinwan for treatment of gastritis. BMC Complement. Altern Med. 2018, 18, 292. [Google Scholar] [CrossRef] [PubMed]
- Shahi, P.; Wang, C.Y.; Lawson, D.A.; Slorach, E.M.; Lu, A.; Yu, Y.; Lai, M.D.; Gonzalez Velozo, H.; Werb, Z. ZNF503/Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression. Proc. Natl. Acad. Sci. USA 2017, 114, 3169–3174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, N.N.; Wang, C.Y.; Lin, Y.C. The novel regulations of MEF2A, CAMKK2, CALM3, and TNNI3 in ventricular hypertrophy induced by arsenic exposure in rats. Toxicology 2014, 324, 123–135. [Google Scholar] [CrossRef]
- Choy, T.K.; Wang, C.Y.; Phan, N.N.; Khoa Ta, H.D.; Anuraga, G.; Liu, Y.H.; Wu, Y.F.; Lee, K.H.; Chuang, J.Y.; Kao, T.J. Identification of Dipeptidyl Peptidase (DPP) Family Genes in Clinical Breast Cancer Patients via an Integrated Bioinformatics Approach. Diagnostics 2021, 11, 1204. [Google Scholar] [CrossRef]
- Chen, P.S.; Hsu, H.P.; Phan, N.N.; Yen, M.C.; Chen, F.W.; Liu, Y.W.; Lin, F.P.; Feng, S.Y.; Cheng, T.L.; Yeh, P.H.; et al. CCDC167 as a potential therapeutic target and regulator of cell cycle-related networks in breast cancer. Aging (Albany NY) 2021, 13, 4157–4181. [Google Scholar] [CrossRef]
- Bakre, A.; Andersen, L.E.; Meliopoulos, V.; Coleman, K.; Yan, X.; Brooks, P.; Crabtree, J.; Tompkins, S.M.; Tripp, R.A. Identification of Host Kinase Genes Required for Influenza Virus Replication and the Regulatory Role of MicroRNAs. PLoS ONE 2013, 8, e66796. [Google Scholar] [CrossRef] [Green Version]
- Ting, G.; Li, X.; Kwon, H.Y.; Ding, T.; Zhang, Z.; Chen, Z.; Li, C.; Liu, Y.; Yang, Y. microRNA-219-5p targets NEK6 to inhibit hepatocellular carcinoma progression. Am. J. Transl. Res. 2020, 12, 7528–7541. [Google Scholar]
- Falzone, L.; Grimaldi, M.; Celentano, E.; Augustin, L.S.A.; Libra, M. Identification of Modulated MicroRNAs Associated with Breast Cancer, Diet, and Physical Activity. Cancers 2020, 12, 2555. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Saito, M.; Robles, A.I.; Nishida, M.; Takeshita, F.; Watanabe, M.; Ochiya, T.; Yokota, J.; Kohno, T.; Harris, C.C.; et al. A Nucleolar Stress-Specific p53-miR-101 Molecular Circuit Functions as an Intrinsic Tumor-Suppressor Network. EBioMedicine 2018, 33, 33–48. [Google Scholar] [CrossRef] [Green Version]
- Frontiers Editorial, O. Retraction: Exosome-Derived miR-486-5p Regulates Cell Cycle, Proliferation and Metastasis in Lung Adenocarcinoma via Targeting NEK2. Front. Bioeng. Biotechnol. 2021, 9, 681918. [Google Scholar] [CrossRef]
- Luo, M.; Zeng, H.; Ma, X.Y.; Ma, X.L. Identification of Hub Genes for Ovarian Cancer Stem Cell Properties with Weighted Gene Co-expression Network Analysis. Sichuan Da Xue Xue Bao Yi Xue Ban 2021, 52, 248–258. [Google Scholar] [CrossRef]
- de Almeida, B.P.; Apolónio, J.D.; Binnie, A.; Castelo-Branco, P. Roadmap of DNA methylation in breast cancer identifies novel prognostic biomarkers. BMC Cancer 2019, 19, 219. [Google Scholar] [CrossRef] [Green Version]
- Pouliot, M.C.; Labrie, Y.; Diorio, C.; Durocher, F. The Role of Methylation in Breast Cancer Susceptibility and Treatment. Anticancer Res. 2015, 35, 4569–4574. [Google Scholar] [PubMed]
- Garaud, S.; Buisseret, L.; Solinas, C.; Gu-Trantien, C.; de Wind, A.; Van den Eynden, G.; Naveaux, C.; Lodewyckx, J.N.; Boisson, A.; Duvillier, H.; et al. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight 2019, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Disis, M.L.; Stanton, S.E. Triple-Negative Breast Cancer: Immune Modulation as the New Treatment Paradigm. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e25–e30. [Google Scholar] [CrossRef]
- Kuroda, H.; Jamiyan, T.; Yamaguchi, R.; Kakumoto, A.; Abe, A.; Harada, O.; Masunaga, A. Tumor-infiltrating B cells and T cells correlate with postoperative prognosis in triple-negative carcinoma of the breast. BMC Cancer 2021, 21, 286. [Google Scholar] [CrossRef]
- Melo-Hanchuk, T.D.; Martins, M.B.; Cunha, L.L.; Soares, F.A.; Ward, L.S.; Vassallo, J.; Kobarg, J. Expression of the NEK family in normal and cancer tissue: An immunohistochemical study. BMC Cancer 2020, 20, 23. [Google Scholar] [CrossRef]
- Peres de Oliveira, A.; Kazuo Issayama, L.; Betim Pavan, I.C.; Riback Silva, F.; Diniz Melo-Hanchuk, T.; Moreira Simabuco, F.; Kobarg, J. Checking NEKs: Overcoming a Bottleneck in Human Diseases. Molecules 2020, 25, 1778. [Google Scholar] [CrossRef]
- Basei, F.L.; Meirelles, G.V.; Righetto, G.L.; Dos Santos Migueleti, D.L.; Smetana, J.H.; Kobarg, J. New interaction partners for Nek4.1 and Nek4.2 isoforms: From the DNA damage response to RNA splicing. Proteome Sci. 2015, 13, 11. [Google Scholar] [CrossRef] [Green Version]
- Melo Hanchuk, T.D.; Papa, P.F.; La Guardia, P.G.; Vercesi, A.E.; Kobarg, J. Nek5 interacts with mitochondrial proteins and interferes negatively in mitochondrial mediated cell death and respiration. Cell Signal. 2015, 27, 1168–1177. [Google Scholar] [CrossRef]
- Ino, Y.; Yamazaki-Itoh, R.; Shimada, K.; Iwasaki, M.; Kosuge, T.; Kanai, Y.; Hiraoka, N. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer 2013, 108, 914–923. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Q.; Yu, C.; Cao, Y.; Zuo, Y.; Yang, L. Immune cell infiltration-based signature for prognosis and immunogenomic analysis in breast cancer. Brief. Bioinform. 2021, 22, 2020–2031. [Google Scholar] [CrossRef]
- Lv, Y.; Lv, D.; Lv, X.; Xing, P.; Zhang, J.; Zhang, Y. Immune Cell Infiltration-Based Characterization of Triple-Negative Breast Cancer Predicts Prognosis and Chemotherapy Response Markers. Front. Genet. 2021, 12, 354. [Google Scholar] [CrossRef]
- Longo, V.; Longo, A.; Adamo, G.; Fiannaca, A.; Picciotto, S.; La Paglia, L.; Romancino, D.; La Rosa, M.; Urso, A.; Cibella, F.; et al. 2,2′4,4′-Tetrabromodiphenyl Ether (PBDE-47) Modulates the Intracellular miRNA Profile, sEV Biogenesis and Their miRNA Cargo Exacerbating the LPS-Induced Pro-Inflammatory Response in THP-1 Macrophages. Front. Immunol. 2021, 12, 664534. [Google Scholar] [CrossRef] [PubMed]
- Fiannaca, A.; La Rosa, M.; Urso, A.; Rizzo, R.; Gaglio, S. A knowledge-based decision support system in bioinformatics: An application to protein complex extraction. BMC Bioinform. 2013, 14, S5. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Graveel, C.R.; Calderone, H.M.; Westerhuis, J.J.; Winn, M.E.; Sempere, L.F. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer 2015, 7, 59–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, N.; Zhang, X.; Qin, D.; Yang, J.; Wu, A.; Wang, L.; Sun, Y.; Li, H.; Shen, X.; Lin, J.; et al. Identification of Core Genes Related to Progression and Prognosis of Hepatocellular Carcinoma and Small-Molecule Drug Predication. Front. Genet. 2021, 12, 608017. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Rivera, Y.; Marina, M.; Jusino, S.; Lee, M.; Velazquez, J.V.; Chardon-Colon, C.; Vargas, G.; Padmanabhan, J.; Chellappan, S.P.; Saavedra, H.I. The Nek2 centrosome-mitotic kinase contributes to the mesenchymal state, cell invasion, and migration of triple-negative breast cancer cells. Sci. Rep. 2021, 11, 9016. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.; Moschetta, M.; Pappas-Gogos, G.; Sheriff, M.; Boussios, S. Genetic Aberrations of DNA Repair Pathways in Prostate Cancer: Translation to the Clinic. Int. J. Mol. Sci. 2021, 22, 9783. [Google Scholar] [CrossRef]
- Chen, F.; Feng, Z.; Zhu, J.; Liu, P.; Yang, C.; Huang, R.; Deng, Z. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol. 2018, 19, 1139–1152. [Google Scholar] [CrossRef] [Green Version]
- Orenay-Boyacioglu, S.; Kasap, E.; Gerceker, E.; Yuceyar, H.; Demirci, U.; Bilgic, F.; Korkmaz, M. Expression profiles of histone modification genes in gastric cancer progression. Mol. Biol. Rep. 2018, 45, 2275–2282. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Xia, Y.; Yang, J.; Jiang, J.; Chen, L.; Ni, R.; Li, L.; Gu, Z. Clinical and biological significance of never in mitosis gene A-related kinase 6 (NEK6) expression in hepatic cell cancer. Pathol. Oncol. Res. 2012, 18, 201–207. [Google Scholar] [CrossRef]
- He, Z.; Ni, X.; Xia, L.; Shao, Z. Overexpression of NIMA-related kinase 6 (NEK6) contributes to malignant growth and dismal prognosis in Human Breast Cancer. Pathol. Res. Pract. 2018, 214, 1648–1654. [Google Scholar] [CrossRef]
- Ding, X.F.; Chen, J.; Zhou, J.; Chen, G.; Wu, Y.L. Never-in-mitosis A-related kinase 8, a novel target of von-Hippel-Lindau tumor suppressor protein, promotes gastric cancer cell proliferation. Oncol. Lett. 2018, 16, 5900–5906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Gao, Y.; Lu, Y.; Zhang, J.; Li, L.; Yin, F. Downregulation of NEK11 is associated with drug resistance in ovarian cancer. Int. J. Oncol. 2014, 45, 1266–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vassilev, A.; DePamphilis, M.L. Links between DNA Replication, Stem Cells and Cancer. Genes 2017, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Liu, Y.; Ma, X.; Hu, H. The Influence of Cell Cycle Regulation on Chemotherapy. Int. J. Mol. Sci. 2021, 22, 6923. [Google Scholar] [CrossRef]
- Pirali, M.; Taheri, M.; Zarei, S.; Majidi, M.; Ghafouri, H. Artesunate, as a HSP70 ATPase activity inhibitor, induces apoptosis in breast cancer cells. Int. J. Biol. Macromol. 2020, 164, 3369–3375. [Google Scholar] [CrossRef]
- Khajah, M.A.; Mathew, P.M.; Luqmani, Y.A. Na+/K+ ATPase activity promotes invasion of endocrine resistant breast cancer cells. PLoS ONE 2018, 13, e0193779. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, A.H.; Kim, S.-H.; Bertos, N.; El-Assaad, W.; Nandi, I.; Smith, H.; Yang, J.; Chen, O.J.; Gamache, I.; Rao, T.; et al. Elevated V–ATPase Activity Following PTEN Loss Is Required for Enhanced Oncogenic Signaling in Breast Cancer. Mol. Cancer Res. 2020, 18, 1477. [Google Scholar] [CrossRef] [PubMed]
- De Luca, M.; Romano, R.; Bucci, C. Role of the V1G1 subunit of V-ATPase in breast cancer cell migration. Sci. Rep. 2021, 11, 4615. [Google Scholar] [CrossRef]
- Yu, W.; Kanaan, Y.; Bae, Y.K.; Gabrielson, E. Chromosomal changes in aggressive breast cancers with basal-like features. Cancer Genet. Cytogenet. 2009, 193, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Fry, A.M.; O’Regan, L.; Sabir, S.R.; Bayliss, R. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 2012, 125, 4423–4433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Han, B.; Guo, L.; Fan, Z. Molecular mechanisms associated with breast cancer based on integrated gene expression profiling by bioinformatics analysis. J. Obs. Gynaecol. 2016, 36, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ye, H. Screening of the prognostic targets for breast cancer based co-expression modules analysis. Mol. Med. Rep. 2017, 16, 4038–4044. [Google Scholar] [CrossRef] [Green Version]
- Xue, D.; Cheng, P.; Han, M.; Liu, X.; Xue, L.; Ye, C.; Wang, K.; Huang, J. An integrated bioinformatical analysis to evaluate the role of KIF4A as a prognostic biomarker for breast cancer. OncoTargets Ther. 2018, 11, 4755–4768. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Qi, J.; Dou, Z.; Hu, J.; Lu, L.; Dai, H.; Wang, H.; Yang, W. Systematic expression analysis of WEE family kinases reveals the importance of PKMYT1 in breast carcinogenesis. Cell Prolif. 2020, 53, e12741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Lu, C.; Li, Q.; Xie, J.; Chen, T.; Tan, Y.; Wu, C.; Jiang, J. The role of Kif4A in doxorubicin-induced apoptosis in breast cancer cells. Mol. Cells 2014, 37, 812–818. [Google Scholar] [CrossRef] [Green Version]
- Bayley, R.; Ward, C.; Garcia, P. MYBL2 amplification in breast cancer: Molecular mechanisms and therapeutic potential. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188407. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X. MYBL2 Is Targeted by miR-143-3p and Regulates Breast Cancer Cell Proliferation and Apoptosis. Oncol. Res. 2018, 26, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Wan, L.; Zhou, X.; Wang, Z.; Wei, W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharm. Ther. 2015, 151, 141–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visconti, R.; Della Monica, R.; Grieco, D. Cell cycle checkpoint in cancer: A therapeutically targetable double-edged sword. J. Exp. Clin. Cancer Res. 2016, 35, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bièche, I.; Vacher, S.; Lallemand, F.; Tozlu-Kara, S.; Bennani, H.; Beuzelin, M.; Driouch, K.; Rouleau, E.; Lerebours, F.; Ripoche, H.; et al. Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: Role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy. Mol. Cancer 2011, 10, 23. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Trainer, A.H.; Friedman, L.S.; Thistlethwaite, F.C.; Evans, M.J.; Ponder, B.A.J.; Venkitaraman, A.R. Mitotic Checkpoint Inactivation Fosters Transformation in Cells Lacking the Breast Cancer Susceptibility Gene, Brca2. Mol. Cell 1999, 4, 1–10. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, H.; Shen, J.; Yang, S.; Deng, K.; Li, Q.; Cui, W. MiR-1236-3p Inhibits the Proliferation, Invasion, and Migration of Colon Cancer Cells and Hinders Epithelial-Mesenchymal Transition by Targeting DCLK3. Front. Oncol. 2021, 11, 688882. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Wang, J.; Ouyang, T.; Lu, N.; Lu, J.; Shen, Y.; Bai, Y.; Xie, X.; Ge, Q. MiR-486-5p Serves as a Good Biomarker in Nonsmall Cell Lung Cancer and Suppresses Cell Growth with the Involvement of a Target PIK3R1. Front. Genet. 2019, 10, 688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Chen, T.; Yan, L.; Xu, H.; Wang, Y.; Li, Y.; Wang, H.; Chen, S.; Wang, W.; Chen, C.; et al. MiR-155-3p acts as a tumor suppressor and reverses paclitaxel resistance via negative regulation of MYD88 in human breast cancer. Gene 2019, 700, 85–95. [Google Scholar] [CrossRef]
- Bower, J.J.; Vance, L.D.; Psioda, M.; Smith-Roe, S.L.; Simpson, D.A.; Ibrahim, J.G.; Hoadley, K.A.; Perou, C.M.; Kaufmann, W.K. Patterns of cell cycle checkpoint deregulation associated with intrinsic molecular subtypes of human breast cancer cells. NPJ Breast Cancer 2017, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Thu, K.L.; Soria-Bretones, I.; Mak, T.W.; Cescon, D.W. Targeting the cell cycle in breast cancer: Towards the next phase. Cell Cycle 2018, 17, 1871–1885. [Google Scholar] [CrossRef] [Green Version]
- Tenga, M.J.; Lazar, I.M. Proteomic snapshot of breast cancer cell cycle: G1/S transition point. Proteomics 2013, 13, 48–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simizu, S.; Osada, H. Mutations in the Plk gene lead to instability of Plk protein in human tumour cell lines. Nat. Cell Biol. 2000, 2, 852–854. [Google Scholar] [CrossRef] [PubMed]
- Kops, G.J.P.L.; Weaver, B.A.A.; Cleveland, D.W. On the road to cancer: Aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 2005, 5, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, S.; Matsumoto, Y.; Morishima, K.; Izumi, H.; Matsumoto, H.; Ito, E.; Tsutsui, K.; Kobayashi, J.; Tauchi, H.; Kajiwara, Y.; et al. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am. J. Med. Genet. A 2006, 140, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Lapenna, S.; Giordano, A. Cell cycle kinases as therapeutic targets for cancer. Nat. Rev. Drug Discov. 2009, 8, 547–566. [Google Scholar] [CrossRef]
- Barr, A.R.; Gergely, F. Aurora-A: The maker and breaker of spindle poles. J. Cell Sci. 2007, 120, 2987–2996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez de Castro, I.; de Cárcer, G.; Malumbres, M. A census of mitotic cancer genes: New insights into tumor cell biology and cancer therapy. Carcinogenesis 2007, 28, 899–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kastan, M.B.; Bartek, J. Cell-cycle checkpoints and cancer. Nature 2004, 432, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Abrams, S.L.; Akula, S.M.; Meher, A.K.; Steelman, L.S.; Gizak, A.; Duda, P.; Rakus, D.; Martelli, A.M.; Ratti, S.; Cocco, L.; et al. GSK-3β Can Regulate the Sensitivity of MIA-PaCa-2 Pancreatic and MCF-7 Breast Cancer Cells to Chemotherapeutic Drugs, Targeted Therapeutics and Nutraceuticals. Cells 2021, 10, 816. [Google Scholar] [CrossRef]
- Candido, S.; Tomasello, B.M.R.; Lavoro, A.; Falzone, L.; Gattuso, G.; Libra, M. Novel Insights into Epigenetic Regulation of IL6 Pathway: In Silico Perspective on Inflammation and Cancer Relationship. Int. J. Mol. Sci. 2021, 22, 10172. [Google Scholar] [CrossRef]
- Vivarelli, S.; Falzone, L.; Candido, S.; Bonavida, B.; Libra, M. YY1 Silencing Induces 5-Fluorouracil-Resistance and BCL2L15 Downregulation in Colorectal Cancer Cells: Diagnostic and Prognostic Relevance. Int. J. Mol. Sci. 2021, 22, 8481. [Google Scholar] [CrossRef] [PubMed]
- Vivarelli, S.; Candido, S.; Caruso, G.; Falzone, L.; Libra, M. Patient-Derived Tumor Organoids for Drug Repositioning in Cancer Care: A Promising Approach in the Era of Tailored Treatment. Cancers 2020, 12, 3636. [Google Scholar] [CrossRef] [PubMed]
- Crimi, S.; Falzone, L.; Gattuso, G.; Grillo, C.M.; Candido, S.; Bianchi, A.; Libra, M. Droplet Digital PCR Analysis of Liquid Biopsy Samples Unveils the Diagnostic Role of hsa-miR-133a-3p and hsa-miR-375-3p in Oral Cancer. Biology 2020, 9, 379. [Google Scholar] [CrossRef]
- Gu, Y.; Liu, Y.; Fu, L.; Zhai, L.; Zhu, J.; Han, Y.; Jiang, Y.; Zhang, Y.; Zhang, P.; Jiang, Z.; et al. Tumor-educated B cells selectively promote breast cancer lymph node metastasis by HSPA4-targeting IgG. Nat. Med. 2019, 25, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Hong, Y.; Qi, P.; Lu, G.; Mai, X.; Xu, S.; He, X.; Guo, Y.; Gao, L.; Jing, Z.; et al. Atlas of breast cancer infiltrated B-lymphocytes revealed by paired single-cell RNA-sequencing and antigen receptor profiling. Nat. Commun. 2021, 12, 2186. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Liao, J.; Liu, J.; Huang, D.; He, C.; Chen, F.; Yang, L.; Wu, W.; Chen, J.; Lin, L.; et al. Blocking the recruitment of naive CD4(+) T cells reverses immunosuppression in breast cancer. Cell Res. 2017, 27, 461–482. [Google Scholar] [CrossRef]
- Wu, L.; Saxena, S.; Goel, P.; Prajapati, D.R.; Wang, C.; Singh, R.K. Breast Cancer Cell-Neutrophil Interactions Enhance Neutrophil Survival and Pro-Tumorigenic Activities. Cancers 2020, 12, 2884. [Google Scholar] [CrossRef] [PubMed]
- Soto-Perez-de-Celis, E.; Chavarri-Guerra, Y.; Leon-Rodriguez, E.; Gamboa-Dominguez, A. Tumor-Associated Neutrophils in Breast Cancer Subtypes. Asian Pac. J. Cancer Prev. 2017, 18, 2689–2693. [Google Scholar] [CrossRef] [PubMed]
- Gago-Dominguez, M.; Matabuena, M.; Redondo, C.M.; Patel, S.P.; Carracedo, A.; Ponte, S.M.; Martínez, M.E.; Castelao, J.E. Neutrophil to lymphocyte ratio and breast cancer risk: Analysis by subtype and potential interactions. Sci. Rep. 2020, 10, 13203. [Google Scholar] [CrossRef] [PubMed]
- Palucka, K.; Coussens, L.M.; O’Shaughnessy, J. Dendritic cells, inflammation, and breast cancer. Cancer J. 2013, 19, 511–516. [Google Scholar] [CrossRef] [Green Version]
- da Cunha, A.; Michelin, M.A.; Murta, E.F. Pattern response of dendritic cells in the tumor microenvironment and breast cancer. World J. Clin. Oncol. 2014, 5, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Michea, P.; Noël, F.; Zakine, E.; Czerwinska, U.; Sirven, P.; Abouzid, O.; Goudot, C.; Scholer-Dahirel, A.; Vincent-Salomon, A.; Reyal, F.; et al. Adjustment of dendritic cells to the breast-cancer microenvironment is subset specific. Nat. Immunol. 2018, 19, 885–897. [Google Scholar] [CrossRef]
- Lenahan, C.; Avigan, D. Dendritic cell defects in patients with cancer: Mechanisms and significance. Breast Cancer Res. 2006, 8, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brockwell, N.K.; Rautela, J.; Owen, K.L.; Gearing, L.J.; Deb, S.; Harvey, K.; Spurling, A.; Zanker, D.; Chan, C.L.; Cumming, H.E.; et al. Tumor inherent interferon regulators as biomarkers of long-term chemotherapeutic response in TNBC. NPJ Precis. Oncol. 2019, 3, 21. [Google Scholar] [CrossRef] [Green Version]
- Savas, P.; Virassamy, B.; Ye, C.; Salim, A.; Mintoff, C.P.; Caramia, F.; Salgado, R.; Byrne, D.J.; Teo, Z.L.; Dushyanthen, S.; et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018, 24, 986–993. [Google Scholar] [CrossRef]
- Fuertes, M.B.; Kacha, A.K.; Kline, J.; Woo, S.R.; Kranz, D.M.; Murphy, K.M.; Gajewski, T.F. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J. Exp. Med. 2011, 208, 2005–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Approved Symbol | HGNC ID | Gene ID | Aliases | Location on Chromosome |
|---|---|---|---|---|
| NEK1 | 7744 | 4750 | NY-REN-55; KIAA1901 | 4q33 |
| NEK2 | 7745 | 4751 | NLK1; PPP1R111; RP67 | 1q32.3 |
| NEK3 | 7746 | 4752 | HSPK36 | 13q14.3 |
| NEK4 | 11399 | 6787 | Pp12301; NRK2; STK2 | 3p21.1 |
| NEK5 | 7748 | 341676 | EC 2.7.11.1 | 13q14.3 |
| NEK6 | 7749 | 10783 | SID6-1512 | 9q33.3 |
| NEK7 | 13386 | 140609 | EC 2.7.11.1 | 1q31.3 |
| NEK8 | 13387 | 284086 | NPHP9 | 17q11.2 |
| NEK9 | 18591 | 91754 | NERCC; DKFZp434D0935 | 14q24.3 |
| NEK10 | 18592 | 152110 | FLJ32685; CILD44 | 3p24.1 |
| NEK11 | 18593 | 79858 | FLJ23495; EC 2.7.11.1 | 3q22.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anuraga, G.; Wang, W.-J.; Phan, N.N.; An Ton, N.T.; Ta, H.D.K.; Berenice Prayugo, F.; Minh Xuan, D.T.; Ku, S.-C.; Wu, Y.-F.; Andriani, V.; et al. Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer. J. Pers. Med. 2021, 11, 1089. https://doi.org/10.3390/jpm11111089
Anuraga G, Wang W-J, Phan NN, An Ton NT, Ta HDK, Berenice Prayugo F, Minh Xuan DT, Ku S-C, Wu Y-F, Andriani V, et al. Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer. Journal of Personalized Medicine. 2021; 11(11):1089. https://doi.org/10.3390/jpm11111089
Chicago/Turabian StyleAnuraga, Gangga, Wei-Jan Wang, Nam Nhut Phan, Nu Thuy An Ton, Hoang Dang Khoa Ta, Fidelia Berenice Prayugo, Do Thi Minh Xuan, Su-Chi Ku, Yung-Fu Wu, Vivin Andriani, and et al. 2021. "Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer" Journal of Personalized Medicine 11, no. 11: 1089. https://doi.org/10.3390/jpm11111089
APA StyleAnuraga, G., Wang, W.-J., Phan, N. N., An Ton, N. T., Ta, H. D. K., Berenice Prayugo, F., Minh Xuan, D. T., Ku, S.-C., Wu, Y.-F., Andriani, V., Athoillah, M., Lee, K.-H., & Wang, C.-Y. (2021). Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer. Journal of Personalized Medicine, 11(11), 1089. https://doi.org/10.3390/jpm11111089