Characterization of Biomarkers in Colorectal Cancer Liver Metastases as a Prognostic Tool
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Proteomic Analysis
2.3. Statistical Analysis
3. Results
3.1. Clinical Characteristics
3.2. Proteomics
3.2.1. ECM Pathway
3.2.2. DNA Replication and Repair Pathways
3.2.3. Immune Pathway
4. Discussion
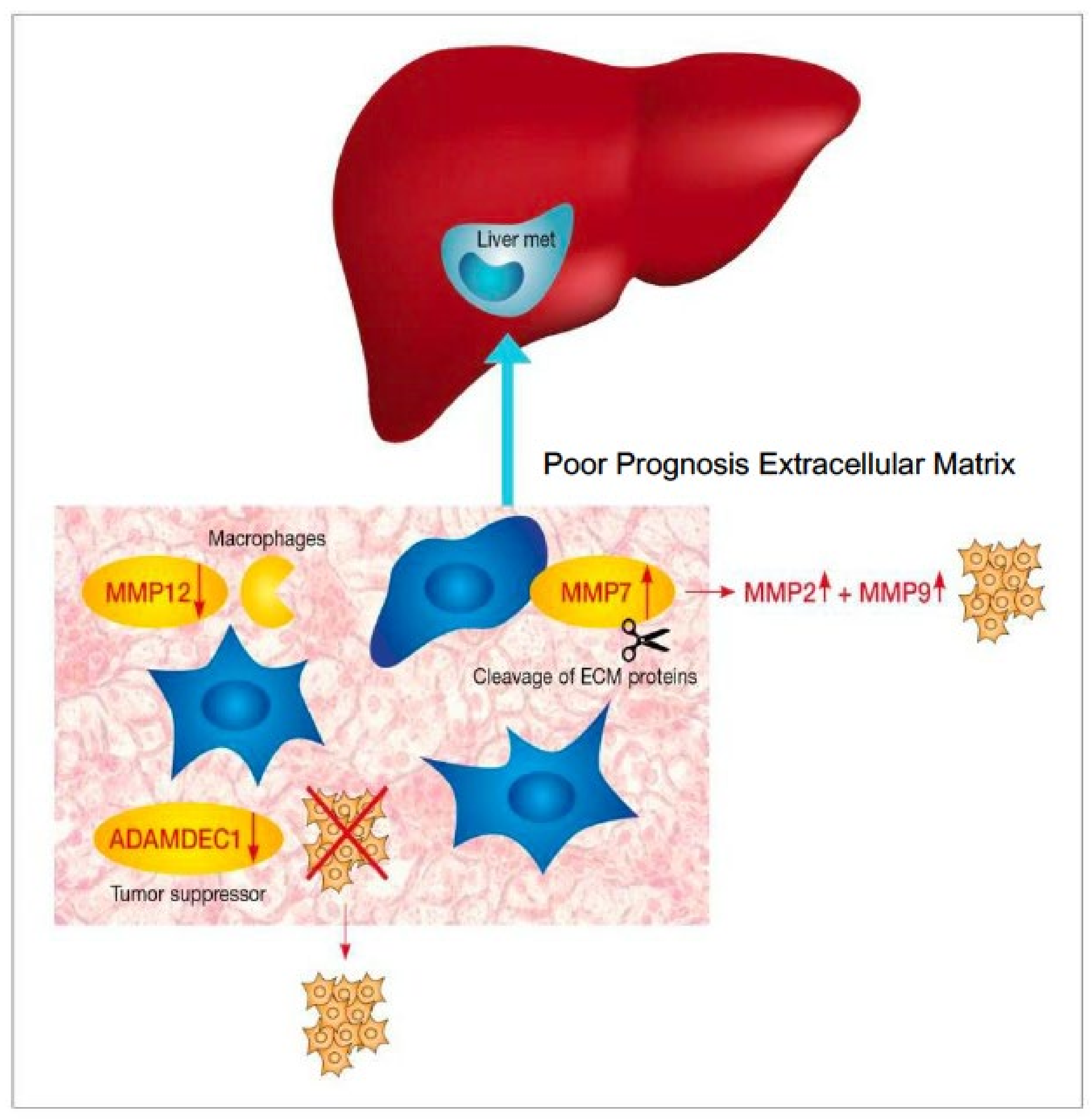
4.1. Matrix Metalloproteinase
4.2. Other ECM Pathways
4.3. Immune Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, R.; de Gramont, A.; Figueras, J.; Kokudo, N.; Kunstlinger, F.; Loyer, E.; Poston, G.; Rougier, P.; Rubbia-Brandt, L.; Sobrero, A.; et al. Managing synchronous liver metastases from colorectal cancer: A multidisciplinary international consensus. Cancer Treat. Rev. 2015, 41, 729–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; van Krieken, J.H.; Aderka, D.; Aguilar, E.A.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Creasy, J.M.; Sadot, E.; Koerkamp, B.G.; Chou, J.F.; Gonen, M.; Kemeny, N.E.; Saltz, L.B.; Balachandran, V.P.; Peter Kingham, T.; DeMatteo, R.P.; et al. The Impact of Primary Tumor Location on Long-Term Survival in Patients Undergoing Hepatic Resection for Metastatic Colon Cancer. Ann. Surg. Oncol. 2018, 25, 431–438. [Google Scholar] [CrossRef]
- Donadon, M.; Lleo, A.; Di Tommaso, L.; Soldani, C.; Franceschini, B.; Roncalli, M.; Torzilli, G. The shifting paradigm of prognostic factors of colorectal liver metastases: From tumor-centered to host immune-centered factors. Front. Oncol. 2018, 8, 181. [Google Scholar] [CrossRef] [Green Version]
- Van Dam, P.J.; Van Der Stok, E.P.; Teuwen, L.A.; Van Den Eynden, G.G.; Illemann, M.; Frentzas, S.; Majeed, A.W.; Eefsen, R.L.; Coebergh Van Den Braak, R.R.J.; Lazaris, A.; et al. International consensus guidelines for scoring the histopathological growth patterns of liver metastasis. Br. J. Cancer 2017, 117, 1427–1441. [Google Scholar] [CrossRef] [Green Version]
- Buisman, F.E.; van der Stok, E.P.; Galjart, B.; Vermeulen, P.B.; Balachandran, V.P.; van den Braak, R.R.J.C.; Creasy, J.M.; Höppener, D.J.; Jarnagin, W.R.; Kingham, T.P.; et al. Histopathological growth patterns as biomarker for adjuvant systemic chemotherapy in patients with resected colorectal liver metastases. Clin. Exp. Metastasis 2020, 37, 593–605. [Google Scholar] [CrossRef]
- Han, Y.; Chai, F.; Wei, J.; Yue, Y.; Cheng, J.; Gu, D.; Zhang, Y.; Tong, T.; Sheng, W.; Hong, N.; et al. Identification of Predominant Histopathological Growth Patterns of Colorectal Liver Metastasis by Multi-Habitat and Multi-Sequence Based Radiomics Analysis. Front. Oncol. 2020, 10, 1363. [Google Scholar] [CrossRef]
- Kanas, G.P.; Taylor, A.; Primrose, J.N.; Langeberg, W.J.; Kelsh, M.A.; Mowat, F.S.; Alexander, D.D.; Choti, M.A.; Poston, G. Survival after liver resection in metastatic colorectal cancer: Review and meta-analysis of prognostic factors. Clin. Epidemiol. 2012, 4, 283–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasserman, I.; Lee, L.H.; Ogino, S.; Marco, M.R.; Wu, C.; Chen, X.; Datta, J.; Sadot, E.; Szeglin, B.; Guillem, J.G.; et al. Smad4 loss in colorectal cancer patients correlates with recurrence, loss of immune infiltrate, and chemoresistance. Clin. Cancer Res. 2019, 25, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Konishi, M.; Nakagohri, T.; Gotohda, N.; Saito, N.; Kinoshita, T. Short time to recurrence after hepatic resection correlates with poor prognosis in colorectal hepatic metastasis. Jpn. J. Clin. Oncol. 2006, 36, 368–375. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Araujo, R.L.C.; Gönen, M.; Herman, P. Chemotherapy for Patients with Colorectal Liver Metastases Who Underwent Curative Resection Improves Long-Term Outcomes: Systematic Review and Meta-analysis. Ann. Surg. Oncol. 2015, 22, 3070–3078. [Google Scholar] [CrossRef]
- Ciliberto, D.; Prati, U.; Roveda, L.; Barbieri, V.; Staropoli, N.; Abbruzzese, A.; Caraglia, M.; Di Maio, M.; Flotta, D.; Tassone, P.; et al. Role of systemic chemotherapy in the management of resected or resectable colorectal liver metastases: A systematic review and meta-analysis of randomized controlled trials. Oncol. Rep. 2012, 27, 1849–1856. [Google Scholar] [CrossRef] [PubMed]
- Ratti, F.; Fuks, D.; Cipriani, F.; Gayet, B.; Aldrighetti, L. Timing of Perioperative Chemotherapy Does Not Influence Long-Term Outcome of Patients Undergoing Combined Laparoscopic Colorectal and Liver Resection in Selected Upfront Resectable Synchronous Liver Metastases. World J. Surg. 2019, 43, 3110–3119. [Google Scholar] [CrossRef]
- Said, A.H.; Raufman, J.P.; Xie, G. The role of matrix metalloproteinases in colorectal cancer. Cancers 2014, 6, 366–375. [Google Scholar] [CrossRef]
- Koskensalo, S.; Louhimo, J.; Nordling, S.; Hagström, J.; Haglund, C. MMP-7 as a prognostic marker in colorectal cancer. Tumor Biol. 2011, 32, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Vočka, M.; Langer, D.; Fryba, V.; Petrtyl, J.; Hanus, T.; Kalousova, M.; Zima, T.; Petruzelka, L. Serum levels of TIMP-1 and MMP-7 as potential biomarkers in patients with metastatic colorectal cancer. Int. J. Biol. Markers 2019, 34, 292–301. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Hu, Y.; Xiang, W.; Cai, Y.; Wang, Z.; Xiao, Q.; Liu, Y.; Li, Q.; Ding, K. Prognostic significance of matrix metalloproteinase 7 immunohistochemical expression in colorectal cancer: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 3281–3290. [Google Scholar]
- Asano, T.; Tada, M.; Cheng, S.; Takemoto, N.; Kuramae, T.; Abe, M.; Takahashi, O.; Miyamoto, M.; Hamada, J.I.; Moriuchi, T.; et al. Prognostic Values of Matrix Metalloproteinase Family Expression in Human Colorectal Carcinoma. J. Surg. Res. 2008, 146, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Klupp, F.; Neumann, L.; Kahlert, C.; Diers, J.; Halama, N.; Franz, C.; Schmidt, T.; Koch, M.; Weitz, J.; Schneider, M.; et al. Serum MMP7, MMP10 and MMP12 level as negative prognostic markers in colon cancer patients. BMC Cancer 2016, 16, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macartney-Coxson, D.P.; Hood, K.A.; Shi, H.; Ward, T.; Wiles, A.; O’Connor, R.; Hall, D.A.; Lea, R.A.; Royds, J.A.; Stubbs, R.S.; et al. Metastatic susceptibility locus, an 8p hot-spot for tumour progression disrupted in colorectal liver metastases: 13 candidate genes examined at the DNA, mRNA and protein level. BMC Cancer 2008, 8, 187. [Google Scholar] [CrossRef] [Green Version]
- Mochizuki, S.; Okada, Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007, 98, 621–628. [Google Scholar] [CrossRef]
- Van Den Eynden, G.G.; Majeed, A.W.; Illemann, M.; Vermeulen, P.B.; Bird, N.C.; Høyer-Hansen, G.; Eefsen, R.L.; Reynolds, A.R.; Brodt, P. The multifaceted role of the microenvironment in liver metastasis: Biology and clinical implications. Cancer Res. 2013, 73, 2031–2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wang, G.; Liang, Z.; Mei, Z.; Wu, T.; Cui, A.; Liu, C.; Cui, L. Lysyl oxidase: A colorectal cancer biomarker of lung and hepatic metastasis. Thorac. Cancer 2018, 9, 785–793. [Google Scholar] [CrossRef]
- Forsburg, S.L. Eukaryotic MCM Proteins: Beyond Replication Initiation. Microbiol. Mol. Biol. Rev. 2004, 68, 109–131. [Google Scholar] [CrossRef] [Green Version]
- Ahluwalia, P.; Mondal, A.K.; Bloomer, C.; Fulzele, S.; Jones, K.; Ananth, S.; Gahlay, G.K.; Heneidi, S.; Rojiani, A.M.; Kota, V.; et al. Identification and Clinical Validation of a Novel 4 Gene-Signature with Prognostic Utility in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 3818. [Google Scholar] [CrossRef] [Green Version]
- Piao, C.; Cai, L.; Qiu, S.; Jia, L.; Song, W.; Du, J. Complement 5a enhances hepatic metastases of colon cancer via monocyte chemoattractant protein-1-mediated inflammatory cell infiltration. J. Biol. Chem. 2015, 290, 10667–10676. [Google Scholar] [CrossRef] [Green Version]
- Ding, D.; Song, Y.; Yao, Y.; Zhang, S. Preoperative serum macrophage activated biomarkers soluble mannose receptor (SMR) and soluble haemoglobin scavenger receptor (SCD163), as novel markers for the diagnosis and prognosis of gastric cancer. Oncol. Lett. 2017, 14, 2982–2990. [Google Scholar] [CrossRef] [Green Version]
| GP | PP | p-Value | |||
|---|---|---|---|---|---|
| Median follow up (range) months | 64 (33–149) | 34 (11–153) | NS | ||
| Dem | Gender | Male Female | 14 (48%) 15 (52%) | 18 (62%) 11 (38%) | NS |
| Median age at metastatic disease dg. (Range) years | 62 (45–81) | 64 (34–85) | NS | ||
| Family history of malignancy | 10 (36%) | 16 (56%) | 0.0144 | ||
| Primary tumor | Side of primary tumor | Right-sided tumors Left-sided tumors | 10 (34%) 19 (66%) | 9 (31%) 20 (69%) | NS |
| Primary tumor differentiation | Well Moderate Poor Unknown | 4 (14%) 23 (79%) 1 (3.5%) 1 (3.5%) | 6 (21%) 21 (72%) 1 (3.5%) 1 (3.5%) | NS | |
| Liver mets | Median number of liver mets at dg. (Range) | 1 (1–4) | 1 (1–6) | NS | |
| Median size of largest met. (Range) mm | 23.5 (10–120) | 29 (8–160) | NS | ||
| Bilobar liver mets. Unilobar liver mets | 4 (14%) 25 (86%) | 8 (28%) 21 (72%) | NS | ||
| Metastases appearance | Metachronous disease | 7 (24%) | 14 (48%) | NS | |
| Stage at diagnosis: I II III Unknown | 4/7 (57%) 3/7 (43%) | 1/14 (7%) 8/14 (57%) 4/14 (29%) 1/14 (7%) | NS | ||
| Adjuvant therapy for localized disease Yes No | 6/7 (86%) 1/7 (14%) | 10/14 (71%) 4/14 (29%) | NS | ||
| Synchronous disease | 22 (76%) | 15 (52%) | NS | ||
| Surgery for primary tumor: Prior to CRCLM resection After CRCLM resection Simultaneous procedure | 11/22 (50%) 2/22 (9%) 9/22 (41%) | 10/15 (67%) 2/15 (13%) 3/15 (20%) | NS | ||
| Systemic treatment | Yes No | 26 (90%) 3 (10%) | 24 (83%) 5 (17%) | NS | |
| Neoadjuvant (either alone or perioperative) Adjuvant (either alone or perioperative) Perioperative | 21/26 (81%) 21/26 (81%) 16/26 (62%) | 23/24 (96%) 11/24 (46%) 10/24 (42%) | 0.032 NS NS | ||
| Type | Chemotherapy- Oxaliplatin based Irinotecan based 5FU only Biological agent- Bevacizumab Cetuximab none | 23/26 (88%) 3/26 (12%) 0 21/26 (81%) 2/26 (8%) 3/26 (11%) | 14/24 (58%) 9/24 (38%) 1/24 (4%) 14/24 (58%) 0 10/24 (42%) | NS NS | |
| Completed six months of therapy | Yes No Unknown | 21/26 (81%) 4/26 (15%) 1/26 (4%) | 19/24 (79%) 3/24 (13%) 2/24 (8%) | NS | |
| Outcome | Median time to recurrence (months) | 23.6 | 5.6 | ||
| Subsequent treatment | Surgery: Yes No unknown Radiotherapy: Yes No unknown Systemic therapy- Yes No Unknown | 14 (48%) 12 (41.5%) 3 (10.5%) 5 (17%) 22 (76%) 2 (7%) 18 (62%) 9 (31%) 2 (7%) | 12 (41.5%) 16 (55%) 1 (3.5%) 6 (21%) 21 (72%) 2 (7%) 24 (83%) 2 (7%) 3 (10%) | NS | |
| Gene Name | Protein Name | Ratio of Protein Expression in Tumor Samples of “Bad Prognosis” vs. “Good Prognosis” | Fold Change (Bad Prognosis vs. Good Prognosis) | FDR Adjusted p-Value | Extracellular Space | Metallo Peptidase Activity | DNA Replication and Repair | Immune System |
|---|---|---|---|---|---|---|---|---|
| AKR1B10 | Aldo-keto reductase family 1 member B10 | Down | 0.95 | 1.2 × 10−4 | V | |||
| APOB | apolipoprotein b | Down | 0.97 | 2.0 × 10−2 | V | |||
| C1RL | Complement C1r subcomponent-like protein | Down | 0.97 | 4.7 × 10−2 | V | V | ||
| C5 | complement component 5 | Down | 0.96 | 1.1 × 10−3 | V | V | ||
| C8A | Complement component C8 alpha chain | Down | 0.95 | 3.2 × 10−6 | V | V | ||
| CD163 | Soluble CD163 | Down | 0.96 | 8.5 × 10−4 | V | V | ||
| CHGA | Chromogranin-A | Up | 1.05 | 5.6 × 10−4 | V | |||
| CMA1 | chymase 1, mast cell | Down | 0.97 | 2.4 × 10−2 | V | V | ||
| DEFA5 | Defensin-5 | Down | 0.94 | 1.9 × 10−6 | V | |||
| GCG | Glucagon | Down | 0.93 | 4.7 × 10−9 | V | |||
| GP2 | Pancreatic secretory granule membrane major glycoprotein GP2 | Down | 0.96 | 7.2 × 10−3 | V | |||
| HLA-B | HLA class I histocompatibility antigen, B-40 alpha chain | Down | 0.94 | 1.3 × 10−6 | V | V | ||
| HSD17B13 | 17-beta-hydroxysteroid dehydrogenase 13 | Down | 0.96 | 4.1 × 10−3 | V | |||
| IGFBP2 | Insulin-like growth factor-binding protein 2 | Up | 1.06 | 7.5 × 10−5 | V | |||
| IGLV3–10 | immunoglobulin lambda variable 3–10 | Up | 1.05 | 2.2 × 10−4 | V | |||
| KRT31 | keratin 31 | Down | 0.96 | 9.9 × 10−3 | V | |||
| KRT85 | keratin 85 | Down | 0.95 | 1.6 × 10−4 | V | |||
| LEFTY1 | Left-right determination factor 1 | Up | 1.07 | 1.7 × 10−7 | V | |||
| LFNG | Beta-1,3-N- acetylglucosaminyltransferase lunatic fringe | Down | 0.93 | 5.2 × 10−10 | V | |||
| LOXL1 | Lysyl oxidase homolog 1 | Down | 0.96 | 8.5 × 10−4 | V | |||
| MXRA5 | Matrix-remodeling-associated protein 5 | Down | 0.96 | 7.4 × 10−3 | V | |||
| OLFM4 | Olfactomedin-4 | Down | 0.93 | 4.5 × 10−11 | V | |||
| OLFML1 | Olfactomedin-like protein 1 | Up | 1.05 | 2.5 × 10−4 | V | |||
| OSCAR | Osteoclast-associated immunoglobulin-like receptor | Down | 0.97 | 3.8 × 10−2 | V | |||
| PROM1 | Prominin-1 | Down | 0.94 | 1.7 × 10−6 | V | |||
| PXDN | Peroxidasin homolog | Down | 0.95 | 1.8 × 10−4 | V | |||
| TNFSF13 | Tumor necrosis factor ligand superfamily member 13 | Down | 0.97 | 3.0 × 10−2 | V | |||
| DPEP1 | Dipeptidase 1 | Up | 1.04 | 9.1 × 10−3 | V | V | ||
| MEP1A | Metalloendopeptidase; Meprin A subunit alpha | Down | 0.93 | 5.2 × 10−10 | V | V | ||
| MMP12 | Macrophage metalloelastase | Down | 0.95 | 2.7 × 10−4 | V | V | ||
| MMP7 | matrix metallopeptidase 7 | Up | 1.04 | 6.4 × 10−3 | V | V | ||
| ADAMDEC1 | adam-like, decysin 1 | Down | 0.96 | 4.1 × 10−3 | V | |||
| ENPEP | Glutamyl aminopeptidase | Down | 0.97 | 2.1 × 10−2 | V | |||
| METAP1 | Methionine aminopeptidase 1 | Down | 0.96 | 3.2 × 10−4 | V | |||
| MSH2 | MutS homolog 2 | Up | 1.05 | 1.0 × 10−4 | V | |||
| MCM4 | Minichromosome maintenance 4 | Up | 1.05 | 3.4 × 10−4 | V |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michal, S.; Tal, G.-L.; Gali, P.; Miki, G.; Elana, B.; Baroch, B.; Hanoch, K.; Irit, B.A.; Riad, H. Characterization of Biomarkers in Colorectal Cancer Liver Metastases as a Prognostic Tool. J. Pers. Med. 2021, 11, 1059. https://doi.org/10.3390/jpm11111059
Michal S, Tal G-L, Gali P, Miki G, Elana B, Baroch B, Hanoch K, Irit BA, Riad H. Characterization of Biomarkers in Colorectal Cancer Liver Metastases as a Prognostic Tool. Journal of Personalized Medicine. 2021; 11(11):1059. https://doi.org/10.3390/jpm11111059
Chicago/Turabian StyleMichal, Sternschuss, Goshen-Lago Tal, Perl Gali, Goldenfeld Miki, Brook Elana, Brenner Baroch, Kashtan Hanoch, Ben Aharon Irit, and Haddad Riad. 2021. "Characterization of Biomarkers in Colorectal Cancer Liver Metastases as a Prognostic Tool" Journal of Personalized Medicine 11, no. 11: 1059. https://doi.org/10.3390/jpm11111059
APA StyleMichal, S., Tal, G.-L., Gali, P., Miki, G., Elana, B., Baroch, B., Hanoch, K., Irit, B. A., & Riad, H. (2021). Characterization of Biomarkers in Colorectal Cancer Liver Metastases as a Prognostic Tool. Journal of Personalized Medicine, 11(11), 1059. https://doi.org/10.3390/jpm11111059







