New Trends in Precision Medicine: A Pilot Study of Pure Light Scattering Analysis as a Useful Tool for Non-Small Cell Lung Cancer (NSCLC) Diagnosis
Abstract
:Simple Summary
Abstract
1. Introduction
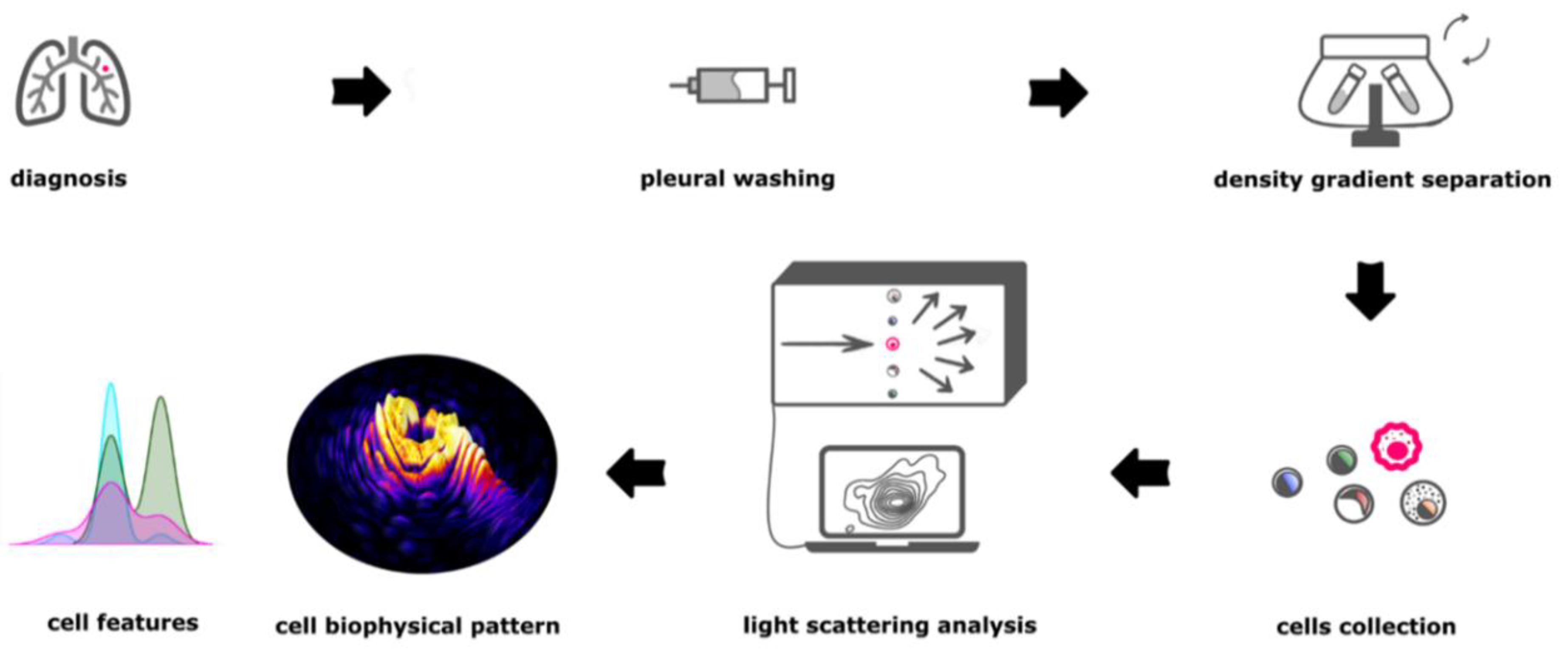
2. Materials and Methods
2.1. Pleural Washing Sample Collection
2.2. Cell Collection
2.3. Bright Field Microscope Evaluation
2.4. Microfluidic Device
2.5. Morphological Single Cell Analysis
2.6. Machine Learning
3. Results
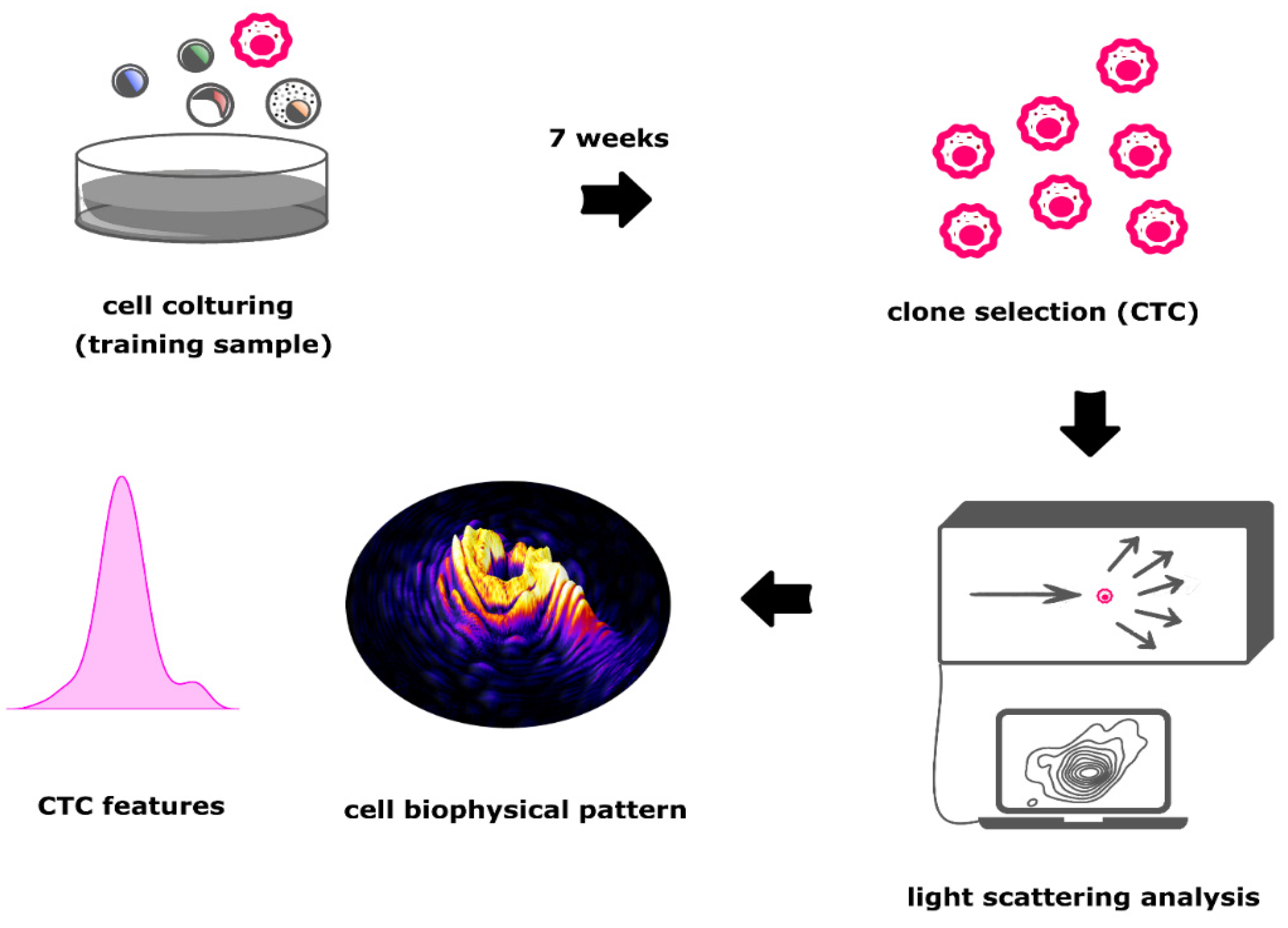
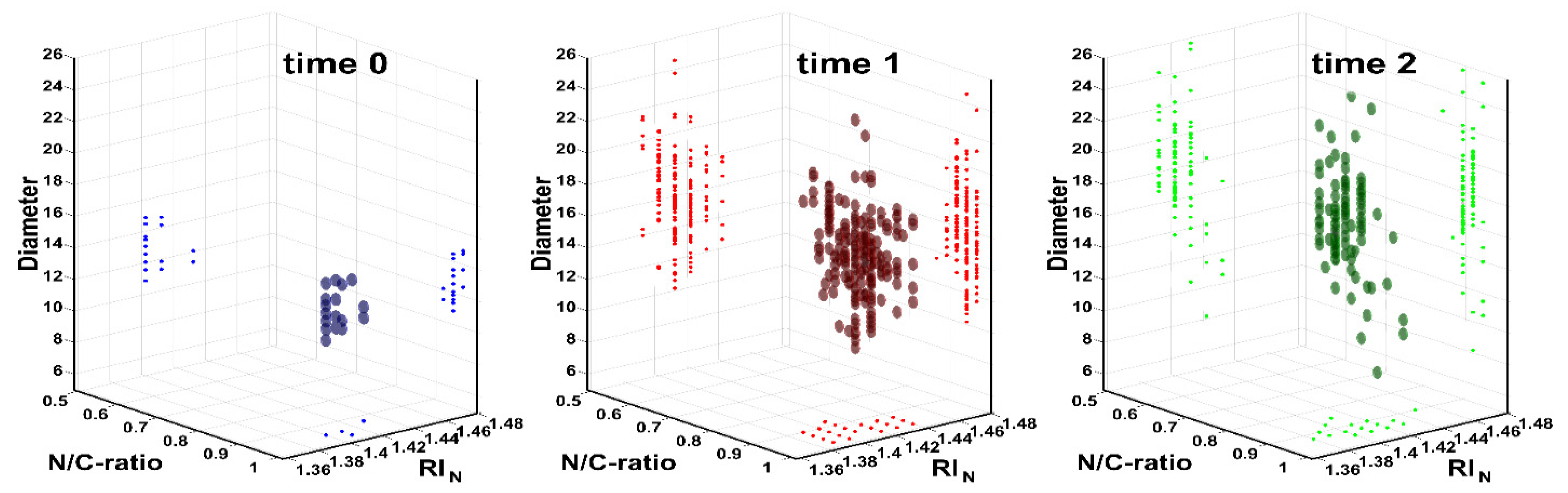
3.1. Training of Morphological Single Cell Analysis
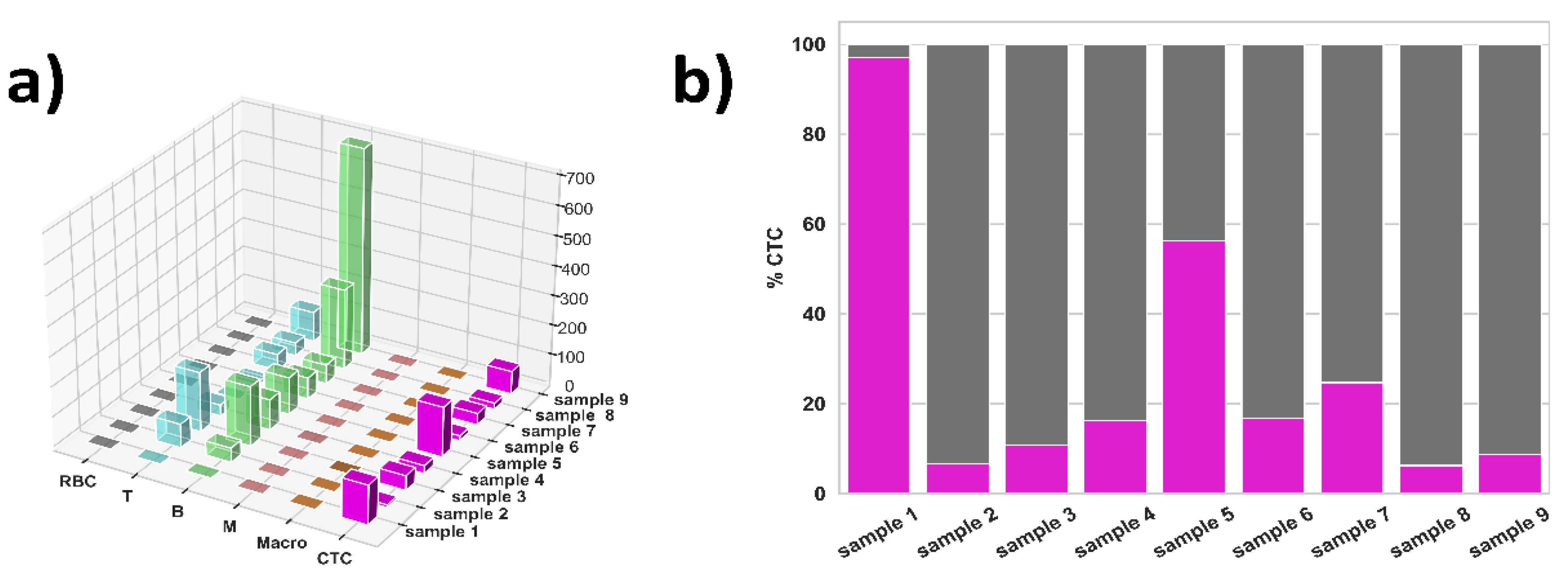
3.2. ML Results for NSCLC Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rodak, O.; Peris-Díaz, M.D.; Olbromski, M.; Podhorska-Okołów, M.; Dzięgiel, P. Current Landscape of Non-Small Cell Lung Cancer: Epidemiology, Histological Classification, Targeted Therapies, and Immunotherapy. Cancers 2021, 13, 4705. [Google Scholar] [CrossRef]
- Boccellino, M.; Pinto, F.; Ieluzzi, V.; Giovane, A.; Quagliuolo, L.; Fariello, C.; Coppola, M.; Carlucci, A.; Santini, M.; Ferati, K.; et al. Proteomics analysis of human serum of patients with non-small-cell lung cancer reveals proteins as diagnostic biomarker candidates. J. Cell. Physiol. 2019, 234, 23798–23806. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Liu, S.V.; Subramaniam, D.S.; Giaccone, G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat. Rev. Clin. Oncol. 2015, 12, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Flaherty, L.; Wikman, H.; Pantel, K. Biology and clinical significance of circulating tumor cell subpopulations in lung cancer. Transl. Lung Cancer Res. 2017, 6, 431–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczmarczyk, G.; Lewandowski, R.; Trautsolt, W.; Ziółkowski, A.; Kozielski, J. Cytological examination of pleural cavity lavage accompanied by the study of gene promoter hypermethylation of p16 and O6-methylguanine-DNA-methyltransferase genes in diagnostics of non-small cell lung cancer metastatic changes into pleura. Contemp. Oncol. 2012, 16, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Maly, V.; Maly, O.; Kolostova, K.; Bobek, V. Circulating Tumor Cells in Diagnosis and Treatment of Lung Cancer. Vivo 2019, 33, 1027–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Tian, X.; Gao, L.; Jiang, X.; Fu, R.; Zhang, T.; Ren, T.; Hu, P.; Wu, Y.; Zhao, P.; et al. Clinical significance of circulating tumor cells and tumor markers in the diagnosis of lung cancer. Cancer Med. 2019, 8, 3782–3792. [Google Scholar] [CrossRef] [PubMed]
- Sawabata, N.; Susaki, Y.; Nakamura, T.; Kawaguchi, T.; Yasukawa, M.; Taniguchi, S. Cluster circulating tumor cells in surgical cases of lung cancer. Gen. Thorac. Cardiovasc. Surg. 2020, 68, 975–983. [Google Scholar] [CrossRef]
- Zhong, M.; Zhang, Y.; Pan, Z.; Wang, W.; Zhang, Y.; Weng, Y.; Huang, H.; He, Y.; Liu, O. Clinical Utility of Circulating Tumor Cells in the Early Detection of Lung Cancer in Patients with a Solitary Pulmonary Nodule. Technol. Cancer Res. Treat. 2021, 20, 15330338211041465. [Google Scholar] [CrossRef] [PubMed]
- Hoseok, I.; Cho, J.-Y. Lung Cancer Biomarkers. Adv. Appl. Microbiol. 2015, 72, 107–170. [Google Scholar] [CrossRef]
- Nappi, A.; Gallicchio, R.; Simeon, V.; Nardelli, A.; Pelagalli, A.; Zupa, A.; Vita, G.; Venetucci, A.; Di Cosola, M.; Barbato, F.; et al. [F-18] FDG-PET/CT parameters as predictors of outcome in inoperable NSCLC patients. Radiol. Oncol. 2015, 49, 320–326. [Google Scholar] [CrossRef] [Green Version]
- Łaczmańska, I.; Dębicka, I.; Gil, J.; Michałowska, D.; Pawlak, I.; Sąsiadek, M.M. Personalised medicine in lung cancer. Genet. Oncol. Nowotw. J. Oncol. 2021, 71, 122–128. [Google Scholar] [CrossRef]
- Berdon, A.L.B.; Pozzi, D.; Caracciolo, G.; Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Riccioli, A.; Palchetti, S.; Laganà, A. Time Evolution of Nanoparticle–Protein Corona in Human Plasma: Relevance for Targeted Drug Delivery. Langmuir 2013, 29, 6485–6494. [Google Scholar] [CrossRef]
- Caputo, D.; Pozzi, D.; Farolfi, T.; Passa, R.; Coppola, R.; Caracciolo, G. Nanotechnology and pancreatic cancer management: State of the art and further perspectives. World J. Gastrointest. Oncol. 2021, 13, 231–237. [Google Scholar] [CrossRef]
- Digiacomo, L.; Caputo, D.; Coppola, R.; Cascone, C.; Giulimondi, F.; Palchetti, S.; Pozzi, D.; Caracciolo, G. Efficient pancreatic cancer detection through personalized protein corona of gold nanoparticles. Biointerphases 2021, 16, 011010. [Google Scholar] [CrossRef] [PubMed]
- La Barbera, G.; Capriotti, A.L.; Caracciolo, G.; Cavaliere, C.; Cerrato, A.; Montone, C.M.; Piovesana, S.; Pozzi, D.; Quagliarini, E.; Laganà, A. A comprehensive analysis of liposomal biomolecular corona upon human plasma incubation: The evolution towards the lipid corona. Talanta 2020, 209, 120487. [Google Scholar] [CrossRef]
- Rossi, D.; Dannhauser, D.; Telesco, M.; Netti, P.A.; Causa, F. CD4+versusCD8+ T-lymphocyte identification in an integrated microfluidic chip using light scattering and machine learning. Lab Chip 2019, 19, 3888–3898. [Google Scholar] [CrossRef] [PubMed]
- Dannhauser, D.; Rossi, D.; Memmolo, P.; Finizio, A.; Ferraro, P.; Netti, P.; Causa, F. Biophysical investigation of living monocytes in flow by collaborative coherent imaging techniques. Biomed. Opt. Express 2018, 9, 5194–5204. [Google Scholar] [CrossRef]
- Dannhauser, D.; Rossi, D.; Ripaldi, M.; Netti, P.; Causa, F. Single-cell screening of multiple biophysical properties in leukemia diagnosis from peripheral blood by pure light scattering. Sci. Rep. 2017, 7, 12666. [Google Scholar] [CrossRef] [Green Version]
- Dannhauser, D.; Rossi, D.; Memmolo, P.; Causa, F.; Finizio, A.; Ferraro, P.; Netti, P. Label-free analysis of mononuclear human blood cells in microfluidic flow by coherent imaging tools. J. Biophotonics 2016, 10, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Dannhauser, D.; Rossi, D.; Causa, F.; Memmolo, P.; Finizio, A.; Wriedt, T.; Hellmers, J.; Eremin, Y.; Ferraro, P.; Netti, P.A. Optical signature of erythrocytes by light scattering in microfluidic flows. Lab Chip 2015, 15, 3278–3285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Domenico, M.; Pozzi, D.; Palchetti, S.; Digiacomo, L.; Iorio, R.; Astarita, C.; Fiorelli, A.; Pierdiluca, M.; Santini, M.; Barbarino, M.; et al. Nanoparticle-biomolecular corona: A new approach for the early detection of non-small-cell lung cancer. J. Cell. Physiol. 2019, 234, 9378–9386. [Google Scholar] [CrossRef] [PubMed]
- Di Capua, D.; Bracken-Clarke, D.; Ronan, K.; Baird, A.M.; Finn, S. The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments. Cancers 2021, 13, 3923. [Google Scholar] [CrossRef]

| Reference | Cell Type | D [µm] | RIC | RIN | N/C-Ratio |
|---|---|---|---|---|---|
| CTC | 18.44 ± 3.49 | 1.36 | 1.40 | 0.920 | |
| [23] | RBC | 7.64 ± 0.91 | 1.40 | - | 1.00 |
| [21] | T | 6.60 ± 0.36 | 1.36 | 1.40 | 0.950 |
| [20] | B | 7.42 ± 0.51 | 1.36 | 1.42 | 0.975 |
| [20] | M | 9.57 ± 1.02 | 1.36 | 1.38 | 0.784 |
| RBC (%) | T (%) | B (%) | M (%) | Macro (%) | CTC (%) | |
|---|---|---|---|---|---|---|
| Time step_1 | 0.00 | 25.71 | 47.14 | 1.43 | 0.00 | 25.71 |
| Time step_2 | 0.00 | 0.46 | 11.06 | 0.00 | 0.46 | 88.02 |
| Time step_3 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 100.00 |
 |  |  |  |  |  |
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Sample 7 | Sample 8 | Sample 9 | |
|---|---|---|---|---|---|---|---|---|---|
| RBC | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T | 0 | 80 | 201 | 37 | 4 | 13 | 55 | 44 | 101 |
| B | 3 | 48 | 201 | 103 | 125 | 72 | 64 | 282 | 700 |
| M | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| Macro | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| CTC | 132 | 9 | 49 | 27 | 168 | 17 | 39 | 21 | 77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, D.; Dannhauser, D.; Nastri, B.M.; Ballini, A.; Fiorelli, A.; Santini, M.; Netti, P.A.; Scacco, S.; Marino, M.M.; Causa, F.; et al. New Trends in Precision Medicine: A Pilot Study of Pure Light Scattering Analysis as a Useful Tool for Non-Small Cell Lung Cancer (NSCLC) Diagnosis. J. Pers. Med. 2021, 11, 1023. https://doi.org/10.3390/jpm11101023
Rossi D, Dannhauser D, Nastri BM, Ballini A, Fiorelli A, Santini M, Netti PA, Scacco S, Marino MM, Causa F, et al. New Trends in Precision Medicine: A Pilot Study of Pure Light Scattering Analysis as a Useful Tool for Non-Small Cell Lung Cancer (NSCLC) Diagnosis. Journal of Personalized Medicine. 2021; 11(10):1023. https://doi.org/10.3390/jpm11101023
Chicago/Turabian StyleRossi, Domenico, David Dannhauser, Bianca Maria Nastri, Andrea Ballini, Alfonso Fiorelli, Mario Santini, Paolo Antonio Netti, Salvatore Scacco, Maria Michela Marino, Filippo Causa, and et al. 2021. "New Trends in Precision Medicine: A Pilot Study of Pure Light Scattering Analysis as a Useful Tool for Non-Small Cell Lung Cancer (NSCLC) Diagnosis" Journal of Personalized Medicine 11, no. 10: 1023. https://doi.org/10.3390/jpm11101023
APA StyleRossi, D., Dannhauser, D., Nastri, B. M., Ballini, A., Fiorelli, A., Santini, M., Netti, P. A., Scacco, S., Marino, M. M., Causa, F., Boccellino, M., & Di Domenico, M. (2021). New Trends in Precision Medicine: A Pilot Study of Pure Light Scattering Analysis as a Useful Tool for Non-Small Cell Lung Cancer (NSCLC) Diagnosis. Journal of Personalized Medicine, 11(10), 1023. https://doi.org/10.3390/jpm11101023










