Age by Single Nucleotide Polymorphism Interactions on Bronchodilator Response in Asthmatics
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Populations
2.2. Outcomes
2.3. Genotyping, Imputation and Quality Control Procedures
2.4. Statistical Analyses
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Global Asthma Report 2018. Available online: http://www.globalasthmareport.org/burden/burden.php (accessed on 11 September 2019).
- Yaghoubi, M.; Adibi, A.; Safari, A.; FitzGerald, J.M.; Sadatsafavi, M. The Projected Economic and Health Burden of Uncontrolled Asthma in the United States. Am. J. Respir. Crit. Care Med. 2019, 200, 1102–1112. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J.; Lee, T.H. Recent advances in asthma. Postgrad. Med. J. 1992, 68, 942–953. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Drazen, J.M.; Silverman, E.K.; Lee, T.H. Heterogeneity of therapeutic responses in asthma. Br. Med. Bull. 2000, 56, 1054–1070. [Google Scholar] [CrossRef]
- Palmer, L.J.; Celedón, J.C.; Chapman, H.A.; Speizer, F.E.; Weiss, S.T.; Silverman, E.K. Genome-wide linkage analysis of bronchodilator responsiveness and post-bronchodilator spirometric phenotypes in chronic obstructive pulmonary disease. Hum. Mol. Genet. 2003, 12, 1199–1210. [Google Scholar] [CrossRef] [PubMed]
- Niu, T.; Rogus, J.J.; Chen, C.; Wang, B.; Yang, J.; Fang, Z.; Weiss, S.T.; Xu, X. Familial aggregation of bronchodilator response: A community-based study. Am. J. Respir. Crit. Care Med. 2000, 162, 1833–1837. [Google Scholar] [CrossRef] [PubMed]
- McGeachie, M.J.; Stahl, E.A.; Himes, B.E.; Pendergrass, S.A.; Lima, J.J.; Irvin, C.G.; Peters, S.P.; Ritchie, M.D.; Plenge, R.M.; Tantisira, K.G. Polygenic heritability estimates in pharmacogenetics: Focus on asthma and related phenotypes. Pharmacogenet. Genom. 2013, 23, 324–328. [Google Scholar] [CrossRef]
- Larsen, G.L. Differences between adult and childhood asthma. J. Allergy Clin. Immunol. 2000, 106, S153–S157. [Google Scholar] [CrossRef] [PubMed]
- Litonjua, A.A.; Lasky-Su, J.; Schneiter, K.; Tantisira, K.G.; Lazarus, R.; Klanderman, B.; Lima, J.J.; Irvin, C.G.; Peters, S.P.; Hanrahan, J.P.; et al. ARG1 is a novel bronchodilator response gene: Screening and replication in four asthma cohorts. Am. J. Respir. Crit. Care Med. 2008, 178, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.J.; Walters, E.H.; Perret, J.L.; Lodge, C.J.; Lowe, A.J.; Matheson, M.C.; Dharmage, S.C. Age-of-asthma onset as a determinant of different asthma phenotypes in adults: A systematic review and meta-analysis of the literature. Expert Rev. Respir. Med. 2015, 9, 109–123. [Google Scholar] [CrossRef]
- Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): Design, rationale, and methods. Control. Clin. Trials 1999, 20, 91. [Google Scholar] [CrossRef]
- Sorkness, C.A.; Lemanske, R.F., Jr.; Mauger, D.T.; Boehmer, S.J.; Chinchilli, V.M.; Martinez, F.D.; Strunk, R.C.; Szefler, S.J.; Zeiger, R.S.; Bacharier, L.B.; et al. Long-term comparison of 3 controller regimens for mild-moderate persistent childhood asthma: The Pediatric Asthma Controller Trial. J. Allergy Clin. Immunol. 2007, 119, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Zeiger, R.S.; Szefler, S.J.; Phillips, B.R.; Schatz, M.; Martinez, F.D.; Chinchilli, V.M.; Lemanske, R.F., Jr.; Strunk, R.C.; Larsen, G.; Spahn, J.D.; et al. Response profiles to fluticasone and montelukast in mild-to-moderate persistent childhood asthma. J. Allergy Clin. Immunol. 2006, 117, 45–52. [Google Scholar] [CrossRef] [PubMed]
- American Lung Association Asthma Clinical Research Centers. Clinical trial of low-dose theophylline and montelukast in patients with poorly controlled asthma. Am. J. Respir. Crit Care Med. 2007, 175, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Drake, K.A.; Torgerson, D.G.; Gignoux, C.R.; Galanter, J.M.; Roth, L.A.; Huntsman, S.; Eng, C.; Oh, S.S.; Yee, S.W.; Lin, L.; et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J. Allergy Clin. Immunol. 2014, 133, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.C.; Bleecker, E.R.; Curran-Everett, D.; Erzurum, S.C.; Ameredes, B.T.; Bacharier, L.; Calhoun, W.J.; Castro, M.; Chung, K.F.; Clark, M.P.; et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J. Allergy Clin. Immunol. 2007, 119, 405–413. [Google Scholar] [CrossRef]
- Himes, B.E.; Jiang, X.; Hu, R.; Wu, A.C.; Lasky-Su, J.A.; Klanderman, B.J.; Ziniti, J.; Senter-Sylvia, J.; Lima, J.J.; Irvin, C.G.; et al. Genome-Wide Association Analysis in Asthma Subjects Identifies SPATS2L as a Novel Bronchodilator Response Gene. PLoS Genet. 2012, 8, e1002824. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- Amare, A.T.; Schubert, K.O.; Tekola-Ayele, F.; Hsu, Y.-H.; Sangkuhl, K.; Jenkins, G.; Whaley, R.M.; Barman, P.; Batzler, A.; Altman, R.B.; et al. Association of the Polygenic Scores for Personality Traits and Response to Selective Serotonin Reuptake Inhibitors in Patients with Major Depressive Disorder. Front. Psychiatry 2018, 9, 65. [Google Scholar] [CrossRef]
- Zhang, Y.-Q.; Zhu, K.-R. The C79G Polymorphism of the β2-Adrenergic Receptor Gene, ADRB2, and Susceptibility to Pediatric Asthma: Meta-Analysis from Review of the Literature. Med. Sci. Monit. 2019, 25, 4005–4013. [Google Scholar] [CrossRef]
- Contopoulos-Ioannidis, D.G.; Alexiou, G.A.; Gouvias, T.C.; Ioannidis, J.P.A. An empirical evaluation of multifarious outcomes in pharmacogenetics: Beta-2 adrenoceptor gene polymorphisms in asthma treatment. Pharm. Genom. 2006, 16, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Verjans, E.; Ohl, K.; Reiss, L.K.; van Wijk, F.; Toncheva, A.A.; Wiener, A.; Yu, Y.; Rieg, A.D.; Gaertner, V.D.; Roth, J.; et al. The cAMP response element modulator (CREM) regulates TH2 mediated inflammation. Oncotarget 2015, 6, 38538–38551. [Google Scholar] [CrossRef]
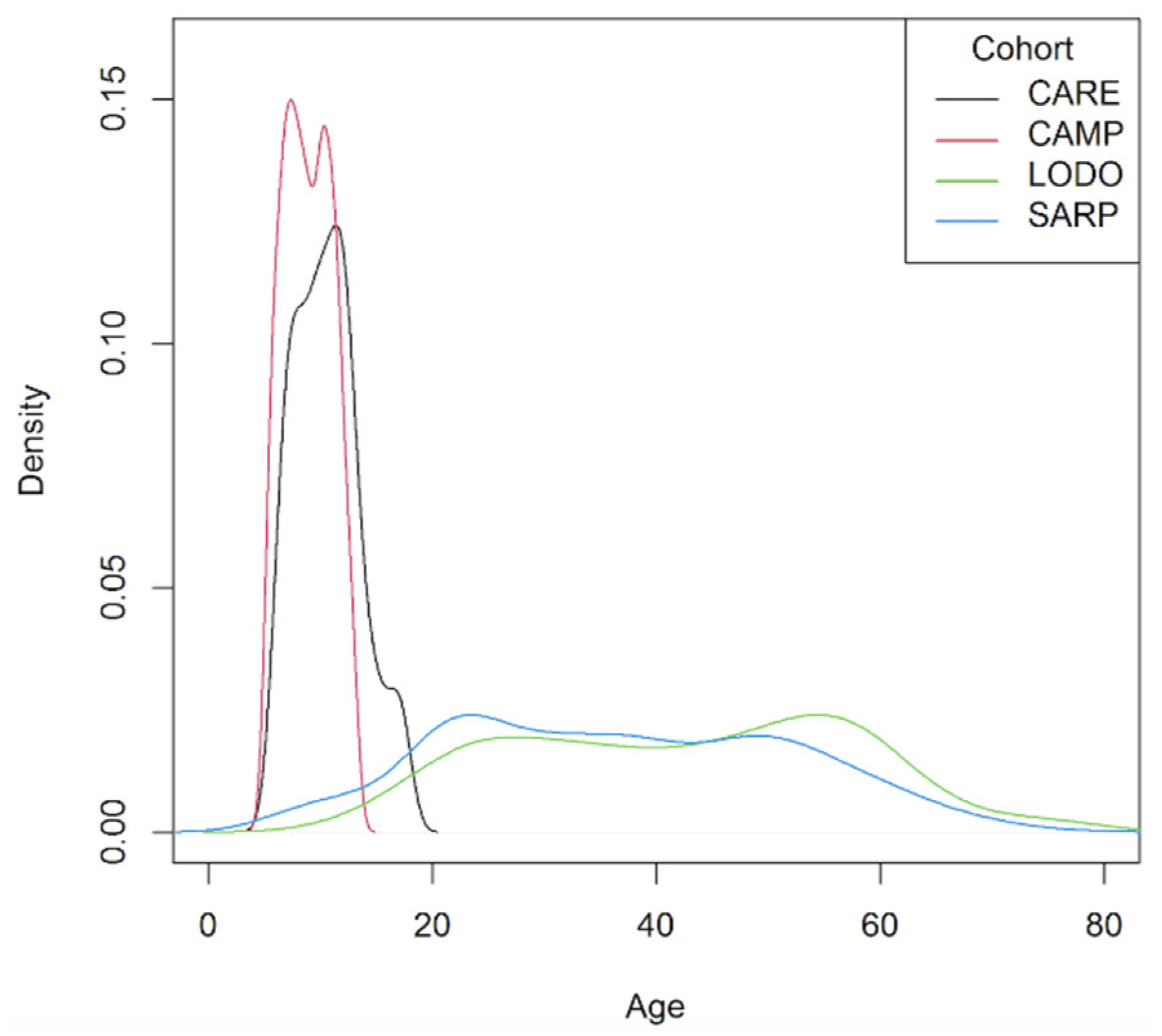
| Meta-Analysis Cohorts | Replication Cohort | |||
|---|---|---|---|---|
| CARE (n = 206) | CAMP (n = 560) | LODO (n = 126) | SARP (n = 559) | |
| Age mean years (SD) | 10.6 (2.9) | 8.9 (2.1) | 42.4 (15.1) | 36.9 (15.3) |
| Age range | 6.0–17.8 | 5.2–13.2 | 15.0–76.0 | 6.4–78.2 |
| Sex (female), n (%) | 78 (37.9) | 225 (40.2) | 94 (74.6) | 366 (65.5) |
| Overweight, n (%) | 37 (18.0) | 87 (15.5) | 32 (25.4) | 160 (28.6) |
| Obese, n (%) | 38 (18.4) | 78 (13.9) | 47 (37.3) | 191(34.2) |
| BDR, mean (SD) | 9.7 (8.4) | 10.9 (10.3) | 8.7 (10.2) | 11.3 (13.4) |
| Meta-Analysis (CAMP, CARE, LODO) | SARP | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Gene | Position/rs# | n | MAF CAMP | MAF CARE | MAF LODO | Beta CAMP | SE CAMP | Beta CARE | SE CARE | Beta LODO | SE LODO | A1 | Z-Score | p | Position/rs# | A1 | Beta | p |
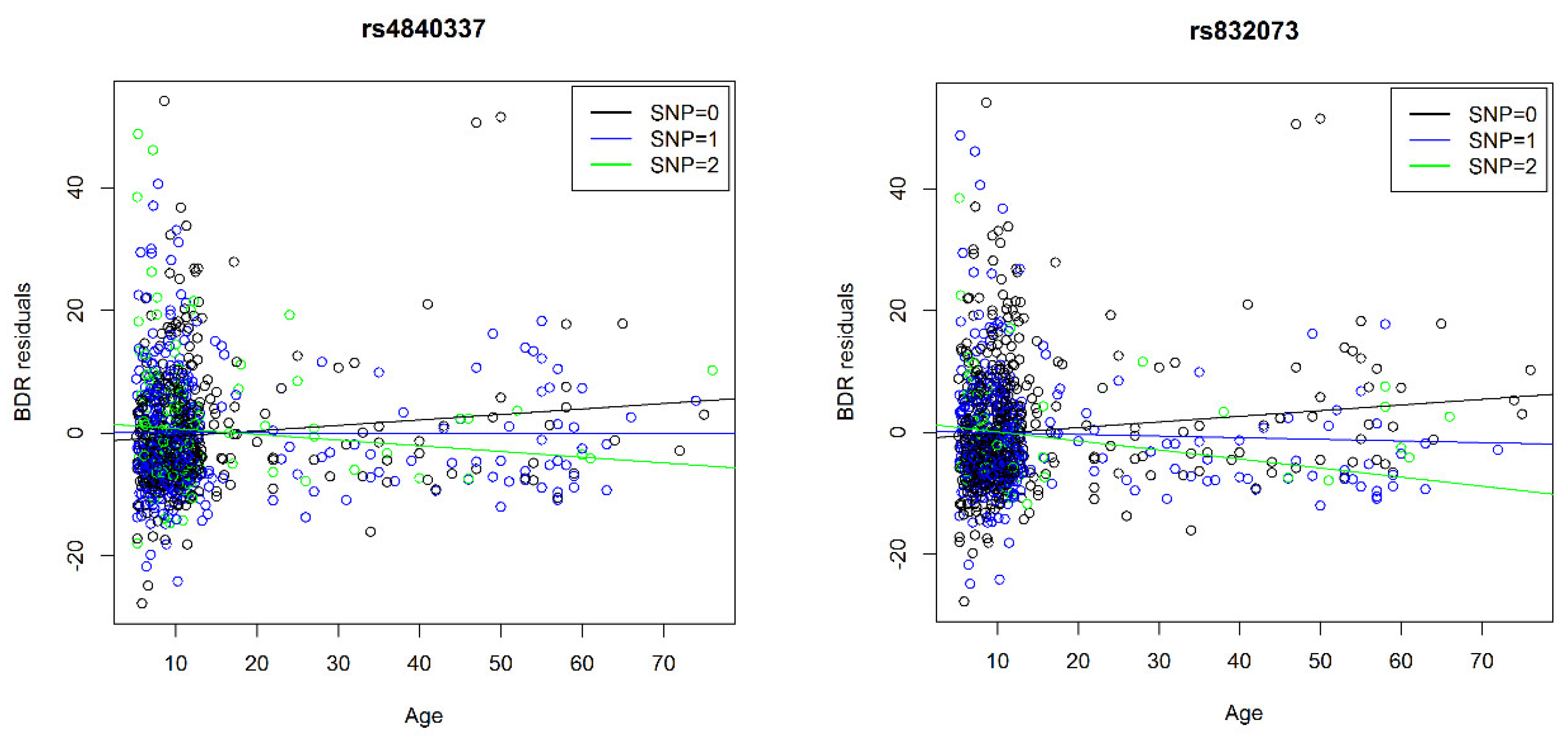
| 3 | 97697002/rs832073 | 892 | 25.3% | 23.3% | 27.8% | −1.3 | 0.3 | −0.7 | 0.3 | −0.1 | 0.1 | T | 4.6 | 3 × 10−6 | 97697002/rs832073 | G | −0.1 | 0.02 | |
| 8 | PRAG1 | 8198306/rs4840337 | 892 | 33.8% | 33.7% | 38.5% | −1.3 | 0.3 | −0.4 | 0.3 | −0.1 | 0.1 | C | 4.8 | 1 × 10−6 | 8198225/rs2945913 | T | −0.2 | 0.04 |
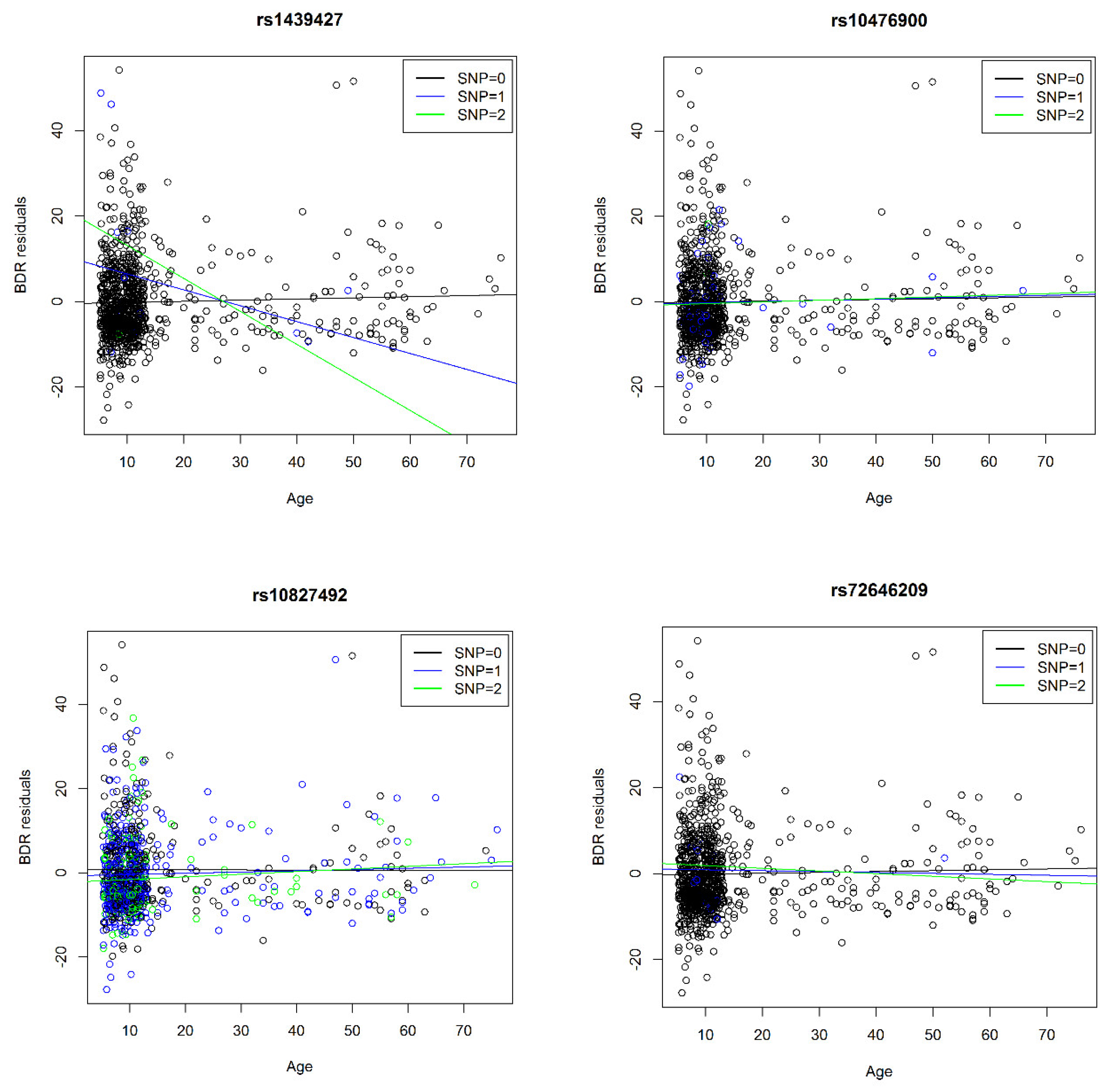
| 18 | 1845637/rs1439427 | 560 | 0.8% | 0.7% | 1.2% | −7.9 | 1.7 | NA | NA | NA | NA | A | −4.6 | 3 × 10−6 | 1846172/rs8091804 | C | −0.2 | 0.006 | |
| Meta-Analysis (CAMP, CARE, LODO) | Candidate GENES | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Gene/Nearest Gene | Position/rs# | n | MAF CAMP | MAF CARE | MAF LODO | Beta CAMP | SE CAMP | Beta CARE | SE CARE | Beta LODO | SE LODO | A1 | Z-Score | p | Position/rs# | A1 | MAF | Beta | p | |
| 1 | PAPPA2 | 176695479/rs77977790 | 892 | 5.9% | 6.8% | 6.0% | −0.3 | 0.6 | 0.7 | 0.6 | 0.1 | 0.2 | T | −0.4 | 0.7 | 176695479/rs77977790 | - | 2.8% | 9.5 | 5 × 10−10 | * |
| 2 | SPATS2L/KCTD18 | 201354935/rs3795969 | 892 | 41.6% | 37.8% | 36.1% | 0.2 | 0.3 | −0.7 | 0.3 | 0.1 | 0.1 | C | 0.2 | 0.9 | 201354866/rs10203042 | - | 2.1% | 4.1 | 2 × 10−3 | * |
| 3 | THRB | 24573150/rs73038406 | 766 | 2.4% | 3.2% | 0.8% | −1.7 | 1.0 | 0.6 | 0.7 | NA | NA | T | −1.0 | 0.3 | 24573150/rs73038406 | - | 1.5% | −4.0 | 0.01 | * |
| 5 | ADRB2 | 148162955/rs10476900 | 892 | 3.0% | 2.9% | 2.4% | 1.6 | 1.0 | 2.7 | 1.1 | 0.1 | 0.3 | A | −2.6 | 9 × 10−3 | 148162955/rs10476900 | - | 10.3% | 1.3 | 0.04 | * |
| 6 | intergenic | 28619532/rs116551936 | 892 | 1.6% | 1.5% | 1.2% | −0.9 | 1.3 | 1.6 | 1.1 | 0.1 | 0.4 | A | 0.3 | 0.8 | 28619532/rs116551936 | - | 0.3% | 27.0 | 6 × 10−9 | * |
| 6 | ARG1 | 131891820/rs2781659 | 892 | 32.5% | 31.3% | 31.3% | −0.1 | 0.3 | 0.1 | 0.3 | −0.1 | 0.1 | A | 0.4 | 0.7 | 131891820/rs2781659 | A | - | - | 5 × 10−4 | ♦ |
| 6 | IGF2R | 160429357/rs8191725 | 892 | 2.9% | 2.9% | 3.2% | −0.5 | 1.0 | 1.1 | 1.0 | −0.1 | 0.3 | A | 0.0 | 1.0 | 160429357/rs8191725 | - | 0.8% | 10.2 | 3 × 10−9 | * |
| 7 | CRHR2 | 30719049/rs1003929 | 892 | 15.9% | 14.3% | 12.7% | 0.7 | 0.4 | −0.3 | 0.4 | 0.1 | 0.1 | T | 1.2 | 0.2 | 30719049/rs1003929 | C | - | - | 0.01 | ♦ |
| 10 | CREM | 35429825/rs10827492 | 892 | 36.3% | 33.5% | 36.1% | 0.9 | 0.3 | 0.0 | 0.3 | 0.0 | 0.1 | T | 2.3 | 2 × 10−2 | 35429825/rs10827492 | C | - | - | 0.05 | ♦ |
| 11 | SPON1 | 14085131/rs77149876 | 686 | 0.6% | 0.2% | 1.2% | −0.2 | 2.2 | NA | NA | 0.4 | 0.4 | T | −0.3 | 0.7 | 14085131/rs77149876 | - | 0.2% | 32.5 | 1 × 10−8 | * |
| 11 | intergenic | 96962052/rs74973995 | 892 | 1.6% | 1.7% | 0.8% | −0.4 | 1.1 | 0.9 | 1.4 | 0.0 | 0.4 | A | 0.0 | 1.0 | 96962052/rs74973995 | - | 0.2% | 32.6 | 1 × 10−8 | * |
| 12 | CREBL2 | 12796872/rs4555 | 892 | 55.8% | 54.9% | 57.1% | −0.4 | 0.3 | 0.3 | 0.3 | −0.1 | 0.1 | T | 0.9 | 0.4 | 12796872/rs4555 | A | - | - | 0.05 | ♦ |
| 12 | CPM | 69309377/rs1144961 | 892 | 25.4% | 24.8% | 25.8% | −0.3 | 0.4 | −0.3 | 0.4 | 0.1 | 0.1 | A | −0.7 | 0.5 | 69309377/rs1144961 | G | - | - | 0.05 | ♦ |
| 14 | SLC24A4 | 92960148/rs4900131 | 892 | 14.9% | 13.1% | 17.9% | −0.6 | 0.4 | −0.4 | 0.5 | 0.0 | 0.1 | T | −1.7 | 0.1 | 92959857/rs77441273 | - | 0.2% | 23.6 | 4 × 10−10 | * |
| 16 | ADCY9 | 4192082/rs7201216 | 892 | 4.6% | 3.2% | 3.2% | 0.5 | 0.7 | 0.9 | 1.0 | −0.1 | 0.3 | A | −0.9 | 0.4 | 4191522/rs144315541 | - | 0.7% | −6.7 | 4 × 10−4 | * |
| 20 | NCOA3 | 46282645/rs72646209 | 766 | 0.5% | 1.7% | 0.4% | −3.9 | 2.1 | −2.9 | 1.9 | NA | NA | A | −2.4 | 2 × 10−2 | 46282708/rs115501901 | - | 0.2% | 31.5 | 4 × 10−8 | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voorhies, K.; Sordillo, J.E.; McGeachie, M.; Ampleford, E.; Wang, A.L.; Lasky-Su, J.; Tantisira, K.; Dahlin, A.; Kelly, R.S.; Ortega, V.E.; et al. Age by Single Nucleotide Polymorphism Interactions on Bronchodilator Response in Asthmatics. J. Pers. Med. 2021, 11, 59. https://doi.org/10.3390/jpm11010059
Voorhies K, Sordillo JE, McGeachie M, Ampleford E, Wang AL, Lasky-Su J, Tantisira K, Dahlin A, Kelly RS, Ortega VE, et al. Age by Single Nucleotide Polymorphism Interactions on Bronchodilator Response in Asthmatics. Journal of Personalized Medicine. 2021; 11(1):59. https://doi.org/10.3390/jpm11010059
Chicago/Turabian StyleVoorhies, Kirsten, Joanne E. Sordillo, Michael McGeachie, Elizabeth Ampleford, Alberta L. Wang, Jessica Lasky-Su, Kelan Tantisira, Amber Dahlin, Rachel S. Kelly, Victor E. Ortega, and et al. 2021. "Age by Single Nucleotide Polymorphism Interactions on Bronchodilator Response in Asthmatics" Journal of Personalized Medicine 11, no. 1: 59. https://doi.org/10.3390/jpm11010059
APA StyleVoorhies, K., Sordillo, J. E., McGeachie, M., Ampleford, E., Wang, A. L., Lasky-Su, J., Tantisira, K., Dahlin, A., Kelly, R. S., Ortega, V. E., Lutz, S. M., & Wu, A. C. (2021). Age by Single Nucleotide Polymorphism Interactions on Bronchodilator Response in Asthmatics. Journal of Personalized Medicine, 11(1), 59. https://doi.org/10.3390/jpm11010059








