Pharmacogenomics at the Point of Care: A Community Pharmacy Project in British Columbia
Abstract
:1. Introduction
2. Materials and Methods
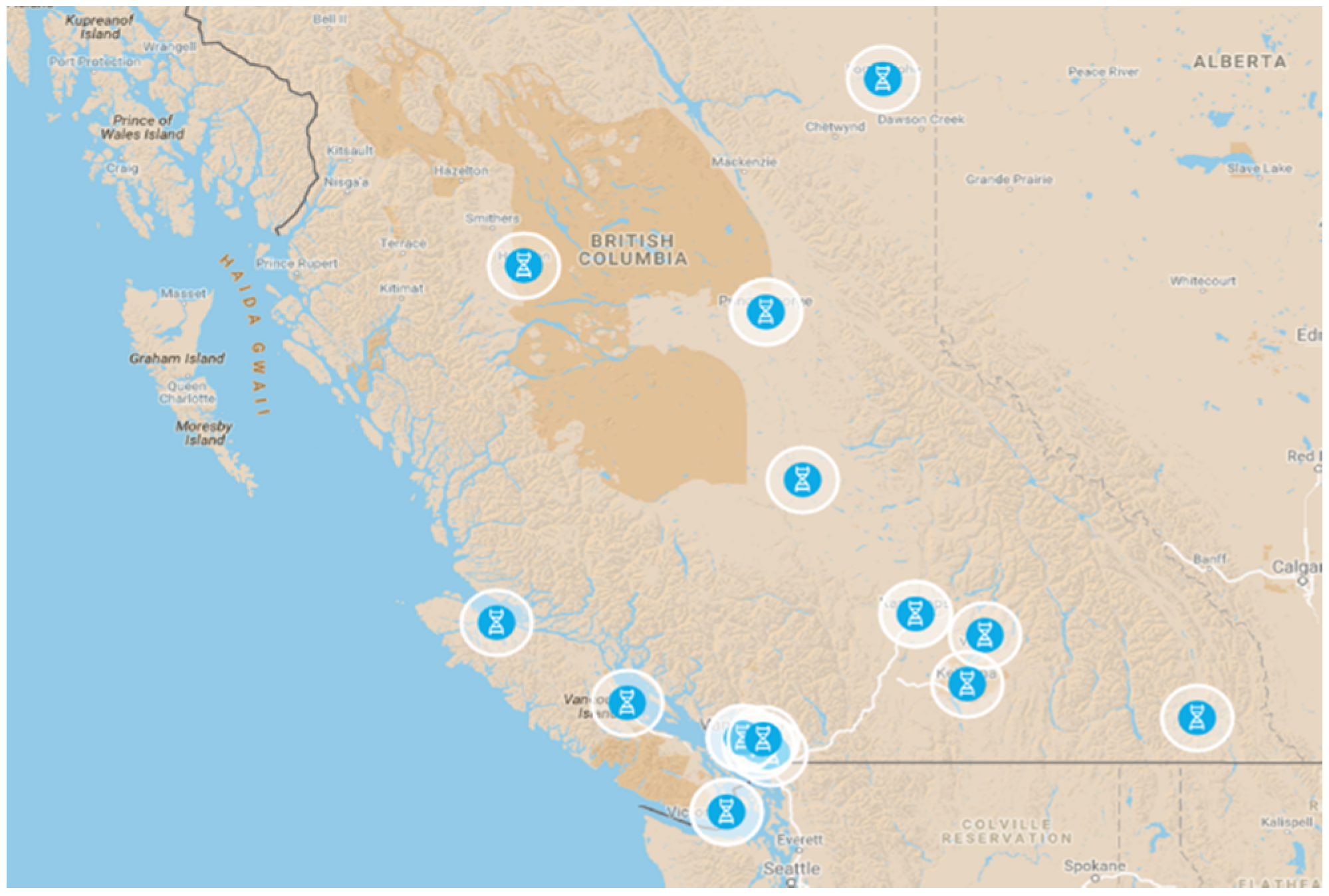
2.1. Pharmacy Selection
2.2. Pharmacist Training
2.3. Operations Logistics & Report Interpretation
2.4. Quality Control (QC)
2.5. Participant Selection and Consent
2.6. Data & Sample Collection
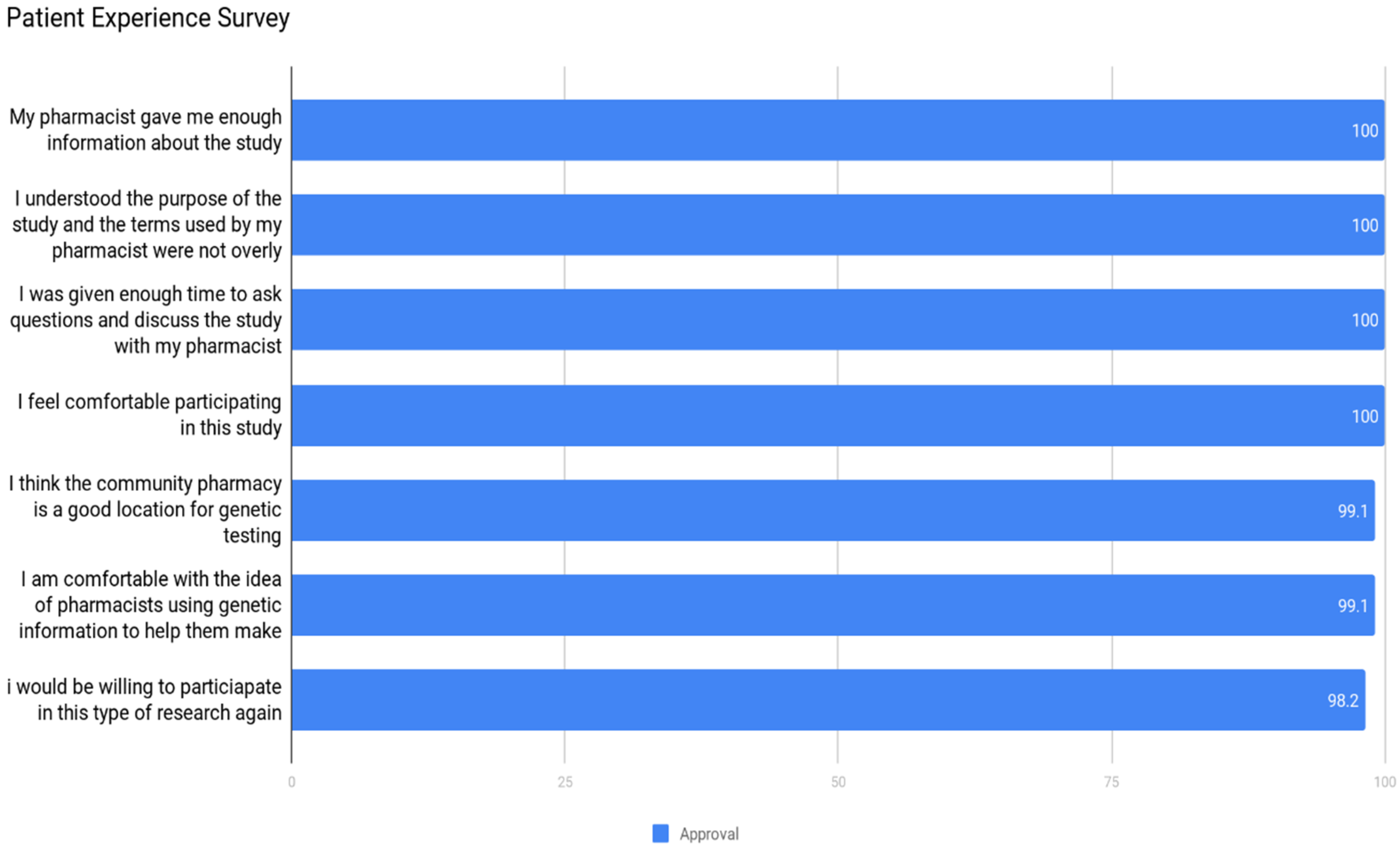
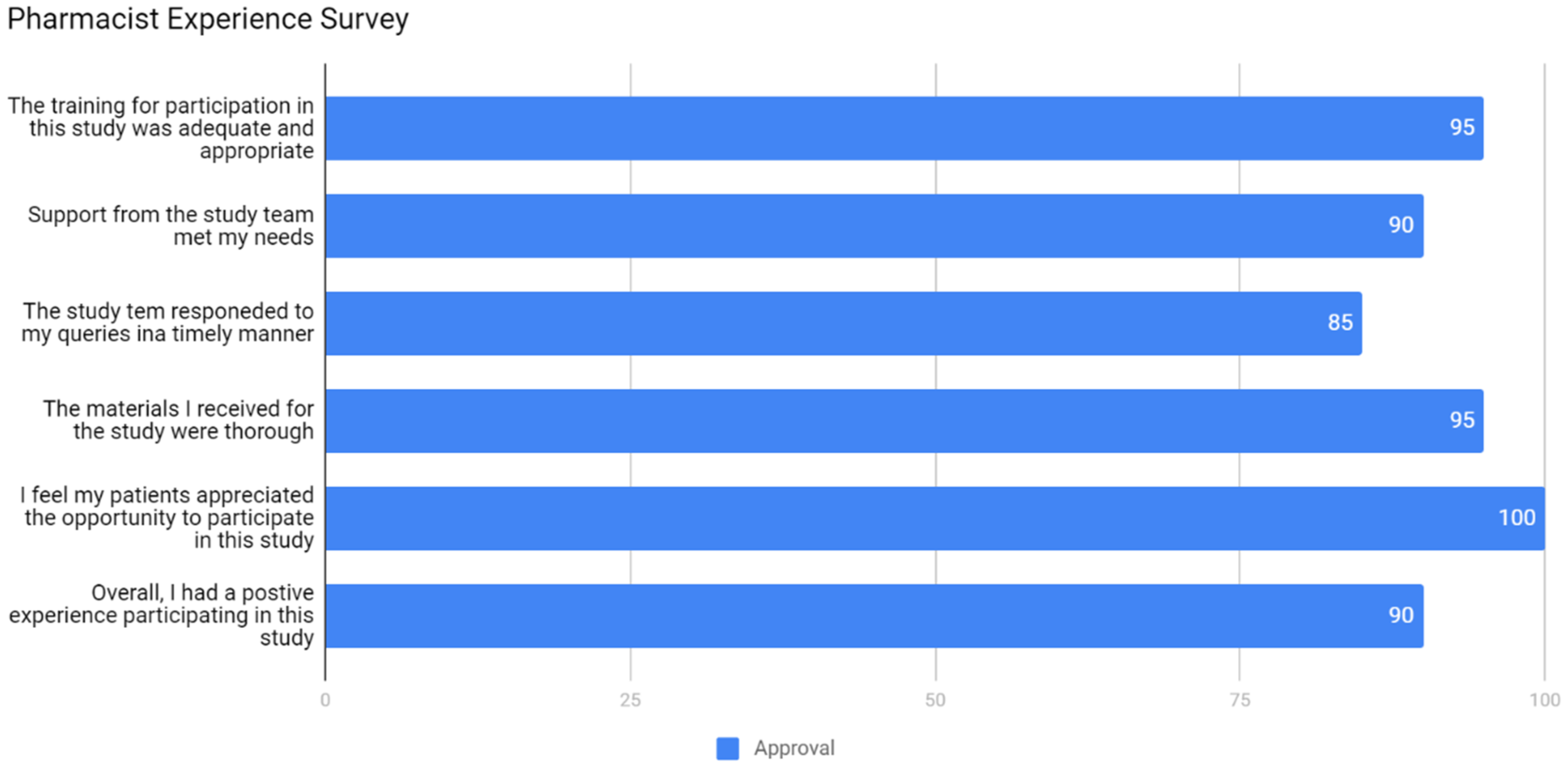
2.7. Experience Survey
2.8. Transport of Samples & Participant Information
2.9. Sample Processing
2.10. TargetRich Sequencing (TRS)
2.11. Genotyping
2.12. Data Reporting
2.13. Patient Consults at the Pharmacy
2.14. Data Collection & Analysis
2.15. Research Ethics Board Approval & Legal Compliance
- The Personal Information Protection Act, The Freedom of Information and Protection of Privacy Act, The Health Professions Act and its Bylaws, The Health Care (Consent) and Care Facility (Admission) Act, and The Pharmacy Operations and Drug Scheduling Act. These laws lay out the obligations of the pharmacist, the pharmacy and the University of British Columbia with respect to personal and health information.
- The Health Professions Act and its Bylaws and The Personal Information Protection Act. These laws governed the pharmacist with respect to the collection, use, disclosure and security of personal and health information.
- The Freedom of Information and Protection of Privacy Act and the policies of UBC and its Research Ethics Board.
2.16. Timeline
3. Results
3.1. Comparison of Genotypes
3.2. Community Acceptance
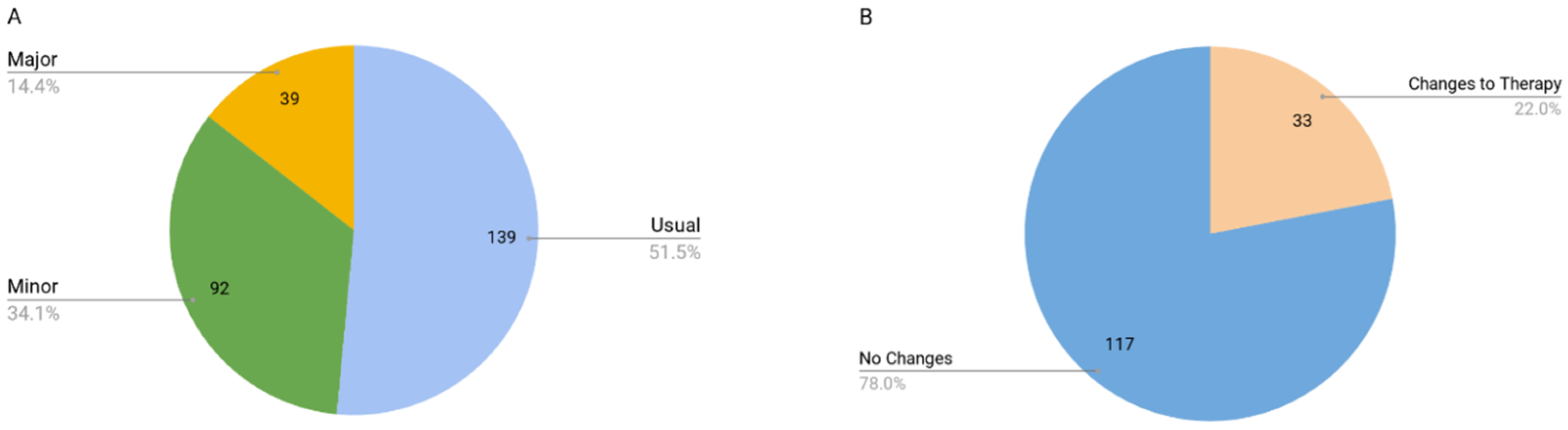
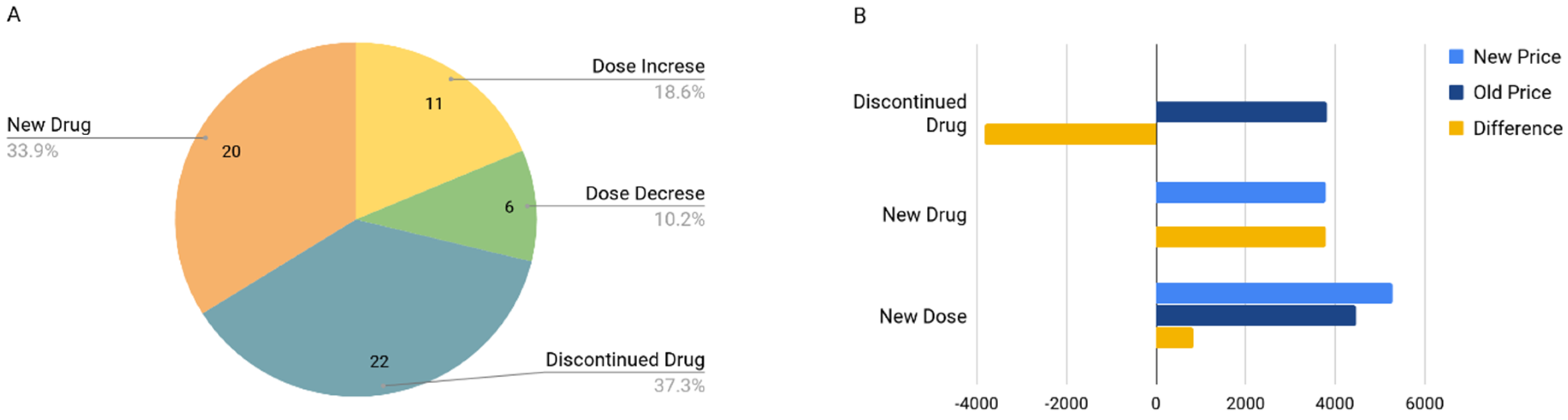
3.3. Drug Cost Analysis
4. Discussion
- The public perceives pharmacists/pharmacies as a very appropriate healthcare professional/venue to deliver pharmacogenomic services.
- Frequencies of alleles, interactions, and clinically actionable results are consistent with other studies published in the scientific literature.
- Changes in drug therapy based on PGx test results represent an inconsequential change in annual drug therapy cost. While drug therapy changes may result in a small cost increase, it is just as likely that costs may decrease.
- Any cost increase due to drug therapy changes is likely to be small and is justified on the basis that the patient will be taking the most appropriate drug and dose for them as an individual based on their phenotype.
- Pharmacogenomic testing is appropriate and affordable for certain patient populations.
- Pharmacogenomic services offered by pharmacists are ready for primetime wide commercial implementation.
4.1. Selection of Antidepressants/Antipsychotics as Inclusion Criteria
4.2. Pharmacist- & Pharmacy-Specific Considerations
4.3. Potential Impact of S-201 and Other Pending Legislation
4.4. Drug Cost Consequences
5. Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| GENE | myDNA Genotype | TRS Genotype | Phenotype | myDNA Genotype Frequency | TRS Genotype Frequency | myDNA Genotype Frequency % n = 150 | TRS Genotype Frequency %, n = 37 | Population Level |
|---|---|---|---|---|---|---|---|---|
| CYP2C19 | *1/*1 | CYP2C19 *1/*1 | Normal metaboliser | 53 | 10 | 35.33 | 27 | 39.7 |
| CYP2C19 | *1/*17 | CYP2C19 *1/*17 | Rapid metaboliser | 50 | 14 | 33.33 | 37.8 | 25.80% |
| CYP2C19 | *1/*2 | CYP2C19 *1/*2 | Intermediate metaboliser | 21 | 7 | 14 | 18.9 | 20.70% |
| CYP2C19 | *17/*17 | CYP2C19 *17/*17 | Ultrarapid metaboliser | 4 | 1 | 2.67 | 2.7 | 0 |
| CYP2C19 | *2/*17 | CYP2C19 *2/*17 | High intermediate metaboliser | 12 | 3 | 8 | 8.1 | 6.20% |
| CYP2C19 | *2/*2 | CYP2C19 *2/*2 | Poor metaboliser | 10 | 1 | 6.67 | 2.7 | 2.90% |
| CYP2C19 | NA | CYP2C19 *XX/*XX | NA | NA | 1 | NA | 2.7 | NA |
| CYP2C9 | *1/*1 | CYP2C9 *1/*1 | Normal metaboliser | 104 | 23 | 69.33 | 62.2 | 64.84% |
| CYP2C9 | *1/*2 | CYP2C9 *1/*2 | High intermediate metaboliser | 23 | 8 | 15.33 | 21.6 | 20.38% |
| CYP2C9 | *1/*3 | CYP2C9 *1/*3 | Intermediate metaboliser | 15 | 4 | 10 | 10.8 | 10.60% |
| CYP2C9 | *2/*2 | CYP2C9 *2/*2 | Poor metaboliser | 4 | 1 | 2.67 | 2.7 | 1.65% |
| CYP2C9 | *2/*3 | CYP2C9 *2/*3 | Poor metaboliser | 3 | 1 | 2 | 2.7 | 1.87% |
| CYP2C9 | *3/*3 | CYP2C9 *3/*3 | Poor metaboliser | 1 | NA | 0.67 | NA | 0.67% |
| CYP2D6 | *1/*1 | CYP2D6 *1/*1 | Normal metaboliser | 23 | 4 | 15.33 | 10.8 | 14.37% |
| CYP2D6 | *1/*10 | CYP2D6 *1/*10 | Normal metaboliser | 3 | NA | 2 | NA | 2.02% |
| CYP2D6 | *1/*1 × 2 | CYP2D6 *1/*1 × 2 | Ultrarapid metaboliser | 1 | NA | 0.67 | NA | 0.54% |
| CYP2D6 | *1/*2 | CYP2D6 *1/*2 | Normal metaboliser | 32 | 9 | 21.33 | 24.3 | 14.76% |
| CYP2D6 | *1/*2 × 3 | CYP2D6 *1/*2 × 3 | Ultrarapid metaboliser | 1 | NA | 0.67 | NA | 0.89% |
| CYP2D6 | *1/*3 | CYP2D6 *1/*3 | Low normal metaboliser | 2 | 1 | 1.33 | 2.7 | 1.24% |
| CYP2D6 | *1/*36 | CYP2D6 *1/*36 | Low normal metaboliser | 1 | NA | 0.67 | NA | 0.04% |
| CYP2D6 | *1/*4 | CYP2D6 *1/*4 | Low normal metaboliser | 17 | 3 | 11.33 | 8.1 | 13.79% |
| CYP2D6 | *1/*41 | CYP2D6 *1/*41 | Normal metaboliser | 12 | 2 | 8 | 5.4 | 7.93% |
| CYP2D6 | *1/*5 | CYP2D6 *1/*5 | Low normal metaboliser | 3 | 1 | 2 | 2.7 | 2.22% |
| CYP2D6 | *1/*6 | CYP2D6 *1/*6 | Low normal metaboliser | 1 | NA | 0.67 | NA | 0.46% |
| CYP2D6 | *1/*9 | CYP2D6 *1/*9 | Normal metaboliser | 1 | 1 | 0.67 | 2.7 | 1.70% |
| CYP2D6 | *10/*10 | CYP2D6 *10/*10 | Intermediate metaboliser | 1 | NA | 0.67 | NA | 0.92% |
| CYP2D6 | *2/*10 | CYP2D6 *2/*10 | Normal metaboliser | 2 | 1 | 1.33 | 2.7 | 0.91% |
| CYP2D6 | *2/*2 | CYP2D6 *2/*2 | Normal metaboliser | 9 | 3 | 6 | 8.1 | 5.09% |
| CYP2D6 | *2/*2 × 2 | CYP2D6 *2/*2 × 2 | Ultrarapid metaboliser | 1 | NA | 0.67 | NA | 0.02% |
| CYP2D6 | *2/*3 | CYP2D6 *2/*3 | Low normal metaboliser | 1 | NA | 0.67 | NA | 0.67% |
| CYP2D6 | *2/*4 | CYP2D6 *2/*4 | Low normal metaboliser | 11 | 1 | 7.33 | 2.7 | 7.23% |
| CYP2D6 | *2/*41 | CYP2D6 *2/*41 | Normal metaboliser | 5 | 1 | 3.33 | 2.7 | 4.46% |
| CYP2D6 | *2/*5 | CYP2D6 *2/*5 | Low normal metaboliser | 4 | 1 | 2.67 | 2.7 | 1.24% |
| CYP2D6 | *2/*6 | CYP2D6 *2/*6 | Low normal metaboliser | 1 | 1 | 0.67 | 2.7 | 0.41% |
| CYP2D6 | *3/*3 | CYP2D6 *3/*3 | Poor metaboliser | 1 | NA | 0.67 | NA | 0.02% |
| CYP2D6 | *3/*41 | CYP2D6 *3/*41 | Intermediate metaboliser | 1 | NA | 0.67 | NA | 0.33% |
| CYP2D6 | NA | CYP2D6 *35/*5 | NA | NA | 1 | NA | 2.7 | NA |
| CYP2D6 | *4/*10 | CYP2D6 *4/*10 | Intermediate metaboliser | 2 | NA | 1.33 | NA | 0.70% |
| CYP2D6 | NA | CYP2D6 *4/*35A | NA | NA | 1 | NA | 2.7 | NA |
| CYP2D6 | *4/*4 | CYP2D6 *4/*4 | Poor metaboliser | 4 | 3 | 2.67 | 8.1 | 3.42% |
| CYP2D6 | *4/*41 | CYP2D6 *4/*41 | Intermediate metaboliser | 5 | 2 | 3.33 | 5.4 | 3.59% |
| CYP2D6 | *4/*6 | CYP2D6 *4/*6 | Poor metaboliser | 1 | NA | 0.67 | NA | 0.28% |
| CYP2D6 | *4/*9 | CYP2D6 *4/*9 | Intermediate metaboliser | 1 | NA | 0.67 | NA | 0.76% |
| CYP2D6 | *5/*41 | CYP2D6 *5/*41 | Intermediate metaboliser | 1 | NA | 0.67 | NA | 0.48% |
| CYP2D6 | *9/*41 | CYP2D6 *9/*41 | Intermediate metaboliser | 2 | 1 | 1.33 | 2.7 | 0.50% |
| CYP3A4 | *1/*1 | CYP3A4 *1/*1 | Normal metaboliser | 141 | 33 | 94 | 89.2 | 93.70% |
| CYP3A4 | *1/*22 | CYP3A4 *1/*22 | Normal metaboliser | 8 | 3 | 5.33 | 8.1 | 6.20% |
| CYP3A4 | NA | CYP3A4 *1/*8 | NA | NA | 1 | NA | 2.7 | 0 |
| CYP3A5 | *1/*3 | CYP3A5 *1/*3 | Low normal metaboliser | 14 | 1 | 9.33 | 2.7 | 38.80% |
| CYP3A5 | *3/*3 | CYP3A5 *3/*3 | Poor metaboliser | 136 | 36 | 90.67 | 97.3 | 54.30% |
| OPRM1 | AA | NA | Normal metaboliser | 107 | NA | 71.33 | NA | 77.10% |
| OPRM1 | AG | NA | Low normal metaboliser | 35 | NA | 23.33 | NA | 21.40% |
| OPRM1 | GG | NA | Reduced metaboliser | 8 | NA | 5.33 | NA | 1.50% |
| SLCO1B1 | CC | rs4149056:C/C Hom | Reduced metaboliser | 3 | NA | 2 | NA | 1.80% |
| SLCO1B1 | TC | rs4149056:T/C Het | Low normal metaboliser | 37 | 10 | 24.67 | 27 | 23.10% |
| SLCO1B1 | TT | rs4149056:T/T Wild | Normal metaboliser | 110 | 27 | 73.33 | 73 | 75.20% |
| CYP1A2 | *1F/*1F | rs762551:A/A Hom | Rapid metaboliser | 69 | 12 | 46 | 32.4 | 45% |
| CYP1A2 | *1A/*1F | rs762551:C/A Het | Normal metaboliser | 69 | 23 | 46 | 62.2 | 44.20% |
| CYP1A2 | *1A/*1A | rs762551:C/C Wild | Normal metaboliser | 12 | 2 | 8 | 5.4 | 10.80% |
| VKORC1 | GG | rs9923231:C/C Wild | Normal warfarin sensitivity | 57 | 15 | 38 | 40.5 | 35.80% |
| VKORC1 | AG | rs9923231:C/T Het | Increased warfarin sensitivity | 59 | 13 | 39.33 | 35.1 | 47.90% |
| VKORC1 | AA | rs9923231:T/T Hom | High warfarin sensitivity | 34 | 9 | 22.67 | 24.3 | 16.30% |
References
- Tolstoi, L.G.; Smith, C.L. Human genome project and cystic fibrosis-a symbiotic relationship. J. Am. Diet. Assoc. 1999, 99, 1421–1427. [Google Scholar] [CrossRef]
- Van Ommen, G.; Bakker, E.; den Dunnen, J. The human genome project and the future of diagnostics, treatment, and prevention. Lancet 1999, 354, S5–S10. [Google Scholar] [CrossRef]
- Dobson, R.T.; Taylor, J.G.; Henry, C.J.; Lachaine, J.; Zello, G.A.; Keegan, D.L.; Forbes, D.A. Taking the lead: Community pharmacists’ perception of their role potential within the primary care team. Res. Soc. Adm. Pharm. 2009, 5, 327–336. [Google Scholar] [CrossRef]
- Smith, S.R.; Clancy, C.M. Medication therapy management programs: Forming a new cornerstone for quality and safety in medicare. Am. J. Med. Qual. 2006, 21, 276–279. [Google Scholar] [CrossRef] [PubMed]
- University of British Columbia. Adverse Drug Events Costly to Health Care System, Canadian Study Shows. 2011. Available online: https://www.sciencedaily.com/releases/2011/02/110225082932.htm (accessed on 4 June 2020).
- Hazell, L.; Shakir, S.A.W. Under-reporting of adverse drug reactions: A systematic review. Drug Saf. 2006, 29, 385–396. [Google Scholar] [CrossRef]
- Owen, J.A. Integrating pharmacogenomics into pharmacy practice via medication therapy management. J. Am. Pharm. Assoc. 2011, 51, e64–e74. [Google Scholar] [CrossRef]
- Goldberger, J.J.; Buxton, A.E. Personalized medicine vs. guideline-based medicine. JAMA 2013, 309, 2559. [Google Scholar] [CrossRef]
- Blalock, S.J.; Roberts, A.W.; Lauffenburger, J.C.; Thompson, T.; O’Connor, S.K. The effect of community pharmacy-based interventions on patient health outcomes: A systematic review. Med. Care Res. Rev. 2013, 70, 235–266. [Google Scholar] [CrossRef] [Green Version]
- Korf, B.R.; Berry, A.B.; Limson, M.; Marian, A.J.; Murray, M.F.; O’Rourke, P.P.; Passamani, E.R.; Relling, M.V.; Tooker, J.; Tsongalis, G.J.; et al. Framework for development of physician competencies in genomic medicine: Report of the Competencies Working Group of the Inter-Society Coordinating Committee for Physician Education in Genomics. Genet. Med. 2014, 16, 804–809. [Google Scholar] [CrossRef]
- Shin, J.; Kayser, S.R.; Langaee, T.Y. Pharmacogenetics: From discovery to patient care. Am. J. Health Syst. Pharm. 2009, 66, 625–637. [Google Scholar] [CrossRef]
- Klein, M.E.; Parvez, M.M.; Shin, J.-G. Clinical implementation of pharmacogenomics for personalized precision medicine: Barriers and solutions. J. Pharm. Sci. 2017, 106, 2368–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor, S.K.; Ferreri, S.P.; Michaels, N.M.; Chater, R.W.; Viera, A.J.; Faruki, H.; McLeod, H.L.; Roederer, M. Making pharmacogenetic testing a reality in a community pharmacy. J. Am. Pharm. Assoc. 2012, 52, e259–e265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padgett, L.; O’Connor, S.; Roederer, M.; McLeod, H.; Ferreri, S. Pharmacogenomics in a community pharmacy: ACT now. J. Am. Pharm. Assoc. 2011, 51, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Valgus, J.; Weitzel, K.W.; Peterson, J.F.; Crona, D.J.; Formea, C.M. Current practices in the delivery of pharmacogenomics: Impact of the recommendations of the Pharmacy Practice Model Summit. Am. J. Health. Syst. Pharm. 2019, 76, 521–529. [Google Scholar] [CrossRef]
- Nislow, C.; Kunzli, M.; Spinelli, J.; Neira, M.; Sinha, S. Desrosiers D. Pharmacogenomics at the point of care: Phase 1. Unpublished.
- Hippman, C.; Nislow, C. Pharmacogenomic Testing: Clinical Evidence and Implementation Challenges. J. Pers. Med. 2019, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Agena Bioscience. Clinical Approval. Agenabio. Agena Bioscience. 2014. Available online: https://agenabio.com/company/clinical-approval/ (accessed on 4 June 2020).
- Johansen, P.; Andersen, J.D.; Børsting, C.; Morling, N. Evaluation of the iPLEX® Sample ID Plus Panel designed for the Sequenom MassARRAY® system. A SNP typing assay developed for human identification and sample tracking based on the SNPforID panel. Forensic Sci. Int. Genet. 2013, 7, 482–487. [Google Scholar] [CrossRef] [Green Version]
- Syrmis, M.W.; Moser, R.J.; Whiley, D.M.; Vaska, V.; Coombs, G.W.; Nissen, M.D.; Sloots, T.P.; Nimmo, G.R. Comparison of a multiplexed MassARRAY system with real-time allele-specific PCR technology for genotyping of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2011, 17, 1804–1810. [Google Scholar] [CrossRef] [Green Version]
- Bray, M.S.; Boerwinkle, E.; Doris, P.A. High-throughput multiplex SNP genotyping with MALDI-TOF mass spectrometry: Practice, problems and promise. Hum. Mutat. 2001, 17, 296–304. [Google Scholar] [CrossRef]
- Nunes, A.P.; Oliveira, I.O.; Santos, B.R.; Millech, C.; Silva, L.P.; González, D.A.; Hallal, P.C.; Menezes, A.M.B.; Araújo, C.L.; Barros, F.C. Quality of DNA extracted from saliva samples collected with the Oragene™ DNA self-collection kit. BMC Med. Res. Methodol. 2012, 12, 65. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, G.M.; Artino, A.R. Analyzing and interpreting data from likert-type scales. J. Grad. Med. Educ. 2013, 5, 541–542. [Google Scholar] [CrossRef] [Green Version]
- Kailos Genetics. TargetRichTM UMI/IndexAdapters & Sequencing User Manual for PGX. 2017. Available online: https://d10u8wcbc2zotv.cloudfront.net/media/Kailos_TargetRich_UMI-Sample_Adapter_PGX_Seq_Protocol_August2017.pdf (accessed on 4 June 2020).
- Kailos Genetics. Long-Range PCR for CYP2D6 CNV Analysis. Supplementary procedure for TargetRichTM PGx Assay. 2017. Available online: https://d10u8wcbc2zotv.cloudfront.net/media/Kailos_2D6CNV_LR-PCR_Protocol2017-RevB.pdf (accessed on 4 June 2020).
- Ellis, J.A.; Ong, B. The Massarray® System for Targeted Snp Genotyping. 2017. Available online: http://link.springer.com/10.1007/978-1-4939-6442-0_5 (accessed on 4 June 2020).
- Mostafa, S.; Kirkpatrick, C.M.J.; Byron, K.; Sheffield, L. An analysis of allele, genotype and phenotype frequencies, actionable pharmacogenomic (Pgx) variants and phenoconversion in 5408 Australian patients genotyped for CYP2D6, CYP2C19, CYP2C9 and VKORC1 genes. J. Neural Transm. 2019, 126, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Aristarán, M.; Tigas, M. Introducing Tabula. Available online: https://source.opennews.org/articles/introducing-tabula/ (accessed on 4 June 2020).
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 4 June 2020).
- Del Tredici, A.L.; Malhotra, A.; Dedek, M.; Espin, F.; Roach, D.; Zhu, G.U.; Voland, J.; Moreno, T.A. Frequency of cyp2d6 alleles including structural variants in the united states. Front. Pharmacol. 2018, 9, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. 2019. Available online: http://biorxiv.org/lookup/doi/10.1101/531210 (accessed on 4 June 2020).
- Hardy, G.H. Mendelian proportions in a mixed population. Science 1908, 28, 49–50. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. JOSS 2019, 4, 1686. [Google Scholar] [CrossRef]
- Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Stetsenko, P.; Short, T.; Lianoglou, S.; Antonyan, E.; Bonsch, M.; Parsonage, H. Data. Table: Extension of “Data. Frame”. 2019. Available online: https://CRAN.R-project.org/package=data.table (accessed on 4 June 2020).
- Wickham, H. Reshaping Data with the reshape Package. J. Stat. Softw. 2007, 21, 1–20. [Google Scholar] [CrossRef]
- Murrell, P. Compare: Comparing Objects for Differences. 2015. Available online: https://CRAN.R-project.org/package=compare (accessed on 4 June 2020).
- Wickham, H. The split-apply-combine strategy for data analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Varrichio, C. Rowr: Row-Based Functions for R Objects. 2016. Available online: https://cran.r-project.org/web/packages/rowr/index.html (accessed on 4 June 2020).
- Van der Wouden, C.H.; Bank, P.C.D.; Özokcu, K.; Swen, J.J.; Guchelaar, H.J. Pharmacist-Initiated Pre-Emptive Pharmacogenetic Panel Testing with Clinical Decision Support in Primary Care: Record of PGx Results and Real-World Impact. Genes 2019, 10, 416. [Google Scholar] [CrossRef] [Green Version]
- Hiemke, C.; Bergemann, N.; Clement, H.W.; Conca, A.; Deckert, J.; Domschke, K.; Eckermann, G.; Egberts, K.; Gerlach, M.; Greiner, C.; et al. Consensus Guidelines for Therapeutic Drug Monitoring in Neuropsychopharmacology: Update 2017. Pharmacopsychiatry 2018, 51, 9–62. [Google Scholar]
- Van Westrhenen, R.; Aitchison, K.J.; Ingelman-Sundberg, M.; Jukić, M.M. Pharmacogenomics of Antidepressant and Antipsychotic Treatment: How Far Have We Got and Where Are We Going? Front. Psychiatry 2020, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Corponi, F.; Fabbri, C.; Serretti, A. Pharmacogenetics in Psychiatry. Adv. Pharmacol. 2018, 83, 297–331. [Google Scholar]
- Rush, A.J.; Trivedi, M.H.; Wisniewski, S.R.; Nierenberg, A.A.; Stewart, J.W.; Warden, D.; Niederehe, G.; Thase, M.E.; Lavori, P.W.; Lebowitz, B.D.; et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: A STAR*D report. Am. J. Psychiatry 2006, 163, 1901–1917. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.M.; Dunnenberger, H.M.; Hicks, J.K.; Caudle, K.E.; Carrillo, M.W.; Freimuth, R.R.; Williams, M.S.; Klein, T.E.; Peterson, J.F. Developing knowledge resources to support precision medicine: Principles from the Clinical Pharmacogenetics Implementation Consortium (Cpic). J. Am. Med. Inform. Assoc. 2016, 23, 796–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buermans, H.P.; Vossen, R.H.; Anvar, S.Y.; Allard, W.G.; Guchelaar, H.J.; White, S.J.; van der Straaten, T. Flexible and Scalable Full-Length CYP2D6 Long Amplicon PacBio Sequencing. Hum. Mutat. 2017, 38, 310–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical Annotation for CYP3A4*1, CYP3A4*20, CYP3A4*8; Paclitaxel; Breast Neoplasms and Ovarian Neoplasms (Level 3 Toxicity/ADR). 2015. Available online: https://www.pharmgkb.org/variant/PA166157507/clinicalAnnotation/1444666533 (accessed on 4 June 2020).
- Cousin, M.A.; Matey, E.T.; Blackburn, P.R.; Boczek, N.J.; McAllister, T.M.; Kruisselbrink, T.M.; Babovic-Vuksanovic, D.; Lazaridis, K.N.; Klee, E.W. Pharmacogenomic findings from clinical whole exome sequencing of diagnostic odyssey patients. Mol. Genet. Genom. Med. 2017, 5, 269–279. [Google Scholar] [CrossRef] [Green Version]
- Krebs, K.; Milani, L. Translating pharmacogenomics into clinical decisions: Do not let the perfect be the enemy of the good. Hum. Genom. 2019, 13, 39. [Google Scholar] [CrossRef] [Green Version]
- Rahman, T.; Ash, D.M.; Lauriello, J.; Rawlani, R. Misleading Guidance from Pharmacogenomic Testing. Am. J. Psychiatry 2017, 174, 922–924. [Google Scholar] [CrossRef]
- Keks, N.; Hope, J.; Keogh, S. Switching and stopping antidepressants. Aust. Prescr. 2016, 39, 76–83. [Google Scholar]
- Pratt, L.A.; Brody, D.J.; Gu, Q. Antidepressant use among persons aged 12 and over: United States, 2011–2014. NCHS Data Brief 2017, 283, 1–8. [Google Scholar]
- Proulx, J. Drug use among seniors in Canada, 2016. Value Health 2018, 21, S146. [Google Scholar] [CrossRef]
- Walker, J. Legislative Summary of Bill S-201: An Act to Prohibit and Prevent Genetic Discrimination. 2016. Available online: https://lop.parl.ca/sites/PublicWebsite/default/en_CA/ResearchPublications/LegislativeSummaries/421S201E (accessed on 4 June 2020).
- Winner, J.G.; Carhart, J.M.; Altar, C.A.; Goldfarb, S.; Allen, J.D.; Lavezzari, G.; Parsons, K.K.; Marshak, A.G.; Garavaglia, S.; Dechairo, B.M. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr. Med. Res. Opin. 2015, 31, 1633–1643. [Google Scholar] [CrossRef]
- Tanner, J.-A.; Brown, L.C.; Yu, K.; Li, J.; Dechairo, B.M. Canadian medication cost savings associated with combinatorial pharmacogenomic guidance for psychiatric medications. Clin. Outcomes Res. 2019, 11, 779–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Antidepressants | Usage | Antidepressants | Usage | Antipsychotics | Usage |
|---|---|---|---|---|---|
| Agomelatine | 0 | Mianserin | 0 | Aripiprazole | 9 |
| Amitriptyline | 12 | Mirtazapine | 12 | Clozapine | 0 |
| Citalopram | 26 | Moclobemide | 2 | Haloperidol | 0 |
| Clomipramine | 0 | Nortriptyline | 6 | Olanzapine | 5 |
| Dothiepin | 0 | Paroxetine | 2 | Quetiapine | 24 |
| Duloxetine | 10 | Sertraline | 17 | Risperidone | 4 |
| Escitalopram | 27 | Trimipramine | 0 | Zuclopenthixol | 0 |
| Fluoxetine | 12 | Vanlafaxine | 23 | ||
| Fluvoxamine | 1 | Vortioxetine | 2 | ||
| Imipramine | 1 | Total: 195 |
| GENE | TRS Genotype | myDNA Genotype | Comparison |
|---|---|---|---|
| CYP2C19 | *XX/*XX | *1/*17 | Kailos no call |
| CYP2C19 | CYP2C19 *1/*2 | *2/*2 | different |
| CYP2C9 | *1/*3 | *3/*3 | different |
| CYP2D6 | *2/*2 | *2/*5 | different |
| CYP2D6 | *35A/*5 | *2/*5 | Kailos only allele *35A |
| CYP2D6 | *35A/*4 | *2/*4 | Kailos only allele *35A |
| CYP3A4 | *1/*8 | *1/*1 | Kailos only allele *8 |
| SLCO1B1 | T/C Het | T/T Wild | different |
| SLCO1B1 | T/C Het | T/T Wild | different |
| GENE | myDNA Genotype | TRS Genotype | Phenotype | myDNA Genotype Frequency % n = 150 | TRS Genotype Frequency % n = 37 | Population Level Frequency % |
|---|---|---|---|---|---|---|
| CYP2C19 | *1/*1 | *1/*1 | Normal metabolizer | 35.33 | 27 | 39.7 |
| CYP2C19 | *1/*17 | *1/*17 | Rapid metabolizer | 33.33 | 37.8 | 25.80% |
| CYP2C19 | *1/*2 | *1/*2 | Intermediate metabolizer | 14 | 18.9 | 20.70% |
| CYP2C19 | *17/*17 | *17/*17 | Ultrarapid metabolizer | 2.67 | 2.7 | 0 |
| CYP2C19 | *2/*17 | *2/*17 | High intermediate metabolizer | 8 | 8.1 | 6.20% |
| CYP2C19 | *2/*2 | *2/*2 | Poor metabolizer | 6.67 | 2.7 | 2.90% |
| CYP2C19 | NA | *XX/*XX | NA | NA | 2.7 | NA |
| CYP2C9 | *1/*1 | *1/*1 | Normal metabolizer | 69.33 | 62.2 | 64.84% |
| CYP2C9 | *1/*2 | *1/*2 | High intermediate metabolizer | 15.33 | 21.6 | 20.38% |
| CYP2C9 | *1/*3 | *1/*3 | Intermediate metabolizer | 10 | 10.8 | 10.60% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Breaux, S.; Desrosiers, F.A.D.; Neira, M.; Sinha, S.; Nislow, C. Pharmacogenomics at the Point of Care: A Community Pharmacy Project in British Columbia. J. Pers. Med. 2021, 11, 11. https://doi.org/10.3390/jpm11010011
Breaux S, Desrosiers FAD, Neira M, Sinha S, Nislow C. Pharmacogenomics at the Point of Care: A Community Pharmacy Project in British Columbia. Journal of Personalized Medicine. 2021; 11(1):11. https://doi.org/10.3390/jpm11010011
Chicago/Turabian StyleBreaux, Samantha, Francis Arthur Derek Desrosiers, Mauricio Neira, Sunita Sinha, and Corey Nislow. 2021. "Pharmacogenomics at the Point of Care: A Community Pharmacy Project in British Columbia" Journal of Personalized Medicine 11, no. 1: 11. https://doi.org/10.3390/jpm11010011






