Association of an APBA3 Missense Variant with Risk of Premature Ovarian Failure in the Korean Female Population
Abstract
1. Introduction
2. Results
2.1. Patients and Kinship Analysis
2.2. GWAS
2.3. Predicted Influence of rs55941146 on Protein Structure
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment
4.2. Whole Genome Genotyping Using the PMRA Chip
4.3. GWAS Process
4.4. Statistical Analysis
4.5. Protein Structure Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Coulam, C.B.; Adamson, S.C.; Annegers, J.F. Incidence of premature ovarian failure. Obstet. Gynecol. 1986, 67, 604. [Google Scholar] [CrossRef] [PubMed]
- Fortuño, C.; Labarta, E. Genetics of primary ovarian insufficiency: A review. J. Assist. Reprod. Genet. 2014, 31, 1573–1585. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Jiao, X.; Simpson, J.L.; Chen, Z.-J. Genetics of primary ovarian insufficiency: New developments and opportunities. Hum. Reprod. Updat. 2015, 21, 787–808. [Google Scholar] [CrossRef]
- Jiao, X.; Zhang, H.; Ke, H.; Zhang, J.; Cheng, L.; Liu, Y.; Qin, Y.; Chen, Z.-J. Premature Ovarian Insufficiency: Phenotypic Characterization Within Different Etiologies. J. Clin. Endocrinol. Metab. 2017, 102, 2281–2290. [Google Scholar] [CrossRef] [PubMed]
- Goswami, D.; Conway, G.S. Premature Ovarian Failure. Horm. Res. Paediatr. 2007, 68, 196–202. [Google Scholar] [CrossRef]
- Kalantaridou, S.N.; Naka, K.K.; Papanikolaou, E.; Kazakos, N.; Kravariti, M.; Calis, K.A.; Paraskevaidis, E.A.; Sideris, D.A.; Tsatsoulis, A.; Chrousos, G.P.; et al. Impaired Endothelial Function in Young Women with Premature Ovarian Failure: Normalization with Hormone Therapy. J. Clin. Endocrinol. Metab. 2004, 89, 3907–3913. [Google Scholar] [CrossRef]
- Eshre Guideline Group on POI; Webber, L.; Davies, M.; Anderson, R.; Bartlett, J.; Braat, D.; Cartwright, B.; Cifkova, R.; Keizer-Schrama, S.D.M.; Hogervorst, E.; et al. ESHRE Guideline: Management of women with premature ovarian insufficiency. Hum. Reprod. 2016, 31, 926–937. [Google Scholar]
- Smitz, J.; Cortvrindt, R. The earliest stages of folliculogenesis in vitro. Reproduction 2002, 123, 185–202. [Google Scholar] [CrossRef]
- Rimon-Dahari, N.; Yerushalmi-Heinemann, L.; Alyagor, L.; Dekel, N. Ovarian Folliculogenesis. Results Probl. Cell Differ. 2016, 58, 167–190. [Google Scholar]
- Patiño, R.; Sullivan, C.V. Ovarian follicle growth, maturation, and ovulation in teleost fish. Fish Physiol. Biochem. 2002, 26, 57–70. [Google Scholar] [CrossRef]
- Baerwald, A.R.; Adams, G.P.; Pierson, R.A. Ovarian antral folliculogenesis during the human menstrual cycle: A review. Hum. Reprod. Updat. 2011, 18, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015, 94, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Kaipia, A.; Hsueh, A.J.W. Regulation of ovarian follicle atresia. Annu. Rev. Physiol. 1997, 59, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Eppig, J.J. Intercommunication between mammalian oocytes and companion somatic cells. BioEssays 1991. [Google Scholar] [CrossRef] [PubMed]
- Alam, H.; Miyano, T. Interaction between growing oocytes and granulosa cells in vitro. Reprod. Med. Biol. 2019, 19, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, A.J.; Billig, H.; Tsafriri, A. Ovarian follicle atresia: A hormonally controlled apoptotic process. Endocr. Rev. 1994, 15, 707–724. [Google Scholar]
- McGee, E.; Spears, N.; Minami, S.; Hsu, S.-Y.; Chun, S.-Y.; Billig, H.; Hsueh, A.J. Preantral ovarian follicles in serum-free culture: Suppression of apoptosis after activation of the cyclic guanosine 3′, 5′-monophosphate pathway and stimulation of growth and differentiation by follicle-stimulating hormone. Endocrinology 1997, 138, 2417–2424. [Google Scholar] [CrossRef]
- Richards, J.S. Hormonal control of gene expression in the ovary. Endocr. Rev. 1994, 15, 725–751. [Google Scholar] [CrossRef]
- Ma, L.; Chen, Y.; Mei, S.; Liu, C.; Ma, X.; Li, Y.; Jiang, Y.; Ha, L.; Xu, X. Single nucleotide polymorphisms in premature ovarian failure-associated genes in a Chinese Hui population. Mol. Med. Rep. 2015, 12, 2529–2538. [Google Scholar] [CrossRef][Green Version]
- Qin, Y.; Sun, M.; You, L.; Wei, D.; Sun, J.; Liang, X.; Zhang, B.; Jiang, H.; Xu, J.; Chen, Z.-J. ESR1, HK3 and BRSK1 gene variants are associated with both age at natural menopause and premature ovarian failure. Orphanet J. Rare Dis. 2012, 7, 5. [Google Scholar] [CrossRef]
- Knauff, E.A.H.; Franke, L.; Van Es, M.A.; Berg, L.H.V.D.; Van Der Schouw, Y.T.; Laven, J.S.; Lambalk, C.B.; Hoek, A.; Goverde, A.J.; Christin-Maitre, S.; et al. Genome-wide association study in premature ovarian failure patients suggests ADAMTS19 as a possible candidate gene. Hum. Reprod. 2009, 24, 2372–2378. [Google Scholar] [CrossRef]
- Sakamoto, T.; Seiki, M. Mint3 Enhances the Activity of Hypoxia-inducible Factor-1 (HIF-1) in Macrophages by Suppressing the Activity of Factor Inhibiting HIF-1. J. Biol. Chem. 2009, 284, 30350–30359. [Google Scholar] [CrossRef]
- Tanahashi, H.; Tabira, T. Genomic organization of the human X11L2 gene (APBA3), a third member of the X11 protein family interacting with Alzheimerʼs β-amyloid precursor protein. NeuroReport 1999, 10, 2575–2578. [Google Scholar] [CrossRef]
- Denko, N.C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 2008, 8, 705–713. [Google Scholar] [CrossRef]
- Lando, D.; Peet, D.J.; Gorman, J.J.; Whelan, D.A.; Whitelaw, M.L.; Bruick, R.K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002, 16, 1466–1471. [Google Scholar] [CrossRef]
- Lando, D.; Peet, D.J.; Whelan, D.A.; Gorman, J.J.; Whitelaw, M.L. Asparagine Hydroxylation of the HIF Transactivation Domain: A Hypoxic Switch. Science 2002, 295, 858–861. [Google Scholar] [CrossRef]
- Mahon, P.C. FIH-1: A novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001, 15, 2675–2686. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.-W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nat. Cell Biol. 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Zhou, J.; Yao, W.; Li, C.; Wu, W.; Li, Q.; Liu, H. Administration of follicle-stimulating hormone induces autophagy via upregulation of HIF-1α in mouse granulosa cells. Cell Death Dis. 2017, 8, e3001. [Google Scholar] [CrossRef] [PubMed]
- Peluso, J.J.; Steger, R.W. Role of FSH in regulating granulosa cell division and follicular atresia in rats. Reproduction 1978, 54, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.Y.; Eisenhauer, K.M.; Minami, S.; Billig, H.; Perlas, E.; Hsueh, A.J. Hormonal regulation of apoptosis in early antral follicles: Follicle-stimulating hormone as a major survival factor. Endocrinology 1996, 137, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. HIF-1 and tumor progression: Pathophysiology and therapeutics. Trends Mol. Med. 2002, 8, S62–S67. [Google Scholar] [CrossRef]
- Krishnamachary, B.; Berg-Dixon, S.; Kelly, B.; Agani, F.; Feldser, D.; Ferreira, G.; Iyer, N.; LaRusch, J.; Pak, B.; Taghavi, P.; et al. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 2003, 63, 1138–1143. [Google Scholar] [PubMed]
- Seagroves, T.N.; Ryan, H.E.; Lu, H.; Wouters, B.G.; Knapp, M.; Thibault, P.; Laderoute, K.; Johnson, R.S. Transcription Factor HIF-1 Is a Necessary Mediator of the Pasteur Effect in Mammalian Cells. Mol. Cell. Biol. 2001, 21, 3436–3444. [Google Scholar] [CrossRef] [PubMed]
- Tacchini, L.; Bianchi, L.; Bernelli-Zazzera, A.; Cairo, G. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific post-transcriptional regulation. J. Biol. Chem. 1999, 274, 24142–24146. [Google Scholar] [CrossRef] [PubMed]
- Heiden, M.G.V.; Plas, D.R.; Rathmell, J.C.; Fox, C.J.; Harris, M.H.; Thompson, C.B. Growth Factors Can Influence Cell Growth and Survival through Effects on Glucose Metabolism. Mol. Cell. Biol. 2001, 21, 5899–5912. [Google Scholar] [CrossRef]
- Alam, H.; Weck, J.; Maizels, E.; Park, Y.; Lee, E.J.; Ashcroft, M.; Hunzicker-Dunn, M. Role of the Phosphatidylinositol-3-Kinase and Extracellular Regulated Kinase Pathways in the Induction of Hypoxia-Inducible Factor (HIF)-1 Activity and the HIF-1 Target Vascular Endothelial Growth Factor in Ovarian Granulosa Cells in Response to Follicle-Stimulating Hormone. Endocrinology 2009, 150, 915–928. [Google Scholar]
- Alam, H.; Maizels, E.T.; Park, Y.; Ghaey, S.; Feiger, Z.J.; Chandel, N.S.; Hunzicker-Dunn, M. Follicle-stimulating hormone activation of hypoxia-inducible factor-1 by the phosphatidylinositol 3-kinase/AKT/Ras homolog enriched in brain (Rheb)/mammalian target of rapamycin (mTOR) pathway is necessary for induction of select protein markers of follicular differentiation. J. Biol. Chem. 2004, 279, 19431–19440. [Google Scholar]
- Huang, Y.; Hua, K.; Zhou, X.; Jin, H.; Chen, X.; Lü, X.; Yu, Y.; Zha, X.; Feng, Y. Activation of the PI3K/AKT pathway mediates FSH-stimulated VEGF expression in ovarian serous cystadenocarcinoma. Cell Res. 2008, 18, 780–791. [Google Scholar] [CrossRef]
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular Growth and Atresia in Mammalian Ovaries: Regulation by Survival and Death of Granulosa Cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef]
- Yu, Y.S.; Sui, H.S.; Bin Han, Z.; Li, W.; Luo, M.J.; Tan, J.-H. Apoptosis in Granulosa cells during follicular atresia: Relationship with steroids and insulin-like growth factors. Cell Res. 2004, 14, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jo, M.; Lee, E.; Choi, D. Induction of apoptotic cell death via accumulation of autophagosomes in rat granulosa cells. Fertil. Steril. 2011, 95, 1482–1486. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.-H.; Yu, H.-Y.; Wang, P.; Mao, G.-K.; Liu, W.-X.; Li, M.-N.; Wang, H.-N.; Shang, Y.-L.; Liu, C.; Xu, Z.-L.; et al. Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice. Cell Death Dis. 2015, 6, e1589. [Google Scholar] [CrossRef]
- Wu, J.; Carlock, C.; Zhou, C.; Nakae, S.; Hicks, J.; Adams, H.P.; Lou, Y. IL-33 is required for disposal of unnecessary cells during ovarian atresia through regulation of autophagy and macrophage migration. J. Immunol. 2015, 194, 2140–2147. [Google Scholar] [CrossRef]
- Gawriluk, T.R.; Hale, A.N.; Flaws, J.A.; Dillon, C.P.; Green, D.R.; Rucker, I.E.B. Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction 2011, 141, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Baddela, V.S.; Sharma, A.; Michaelis, M.; Vanselow, J. HIF1 driven transcriptional activity regulates steroidogenesis and proliferation of bovine granulosa cells. Sci. Rep. 2020, 10, 3906. [Google Scholar] [CrossRef]
- Xu, B.; Hua, J.; Zhang, Y.; Jiang, X.; Zhang, H.; Ma, T.; Zheng, W.; Sun, R.; Shen, W.; Sha, J.; et al. Proliferating Cell Nuclear Antigen (PCNA) Regulates Primordial Follicle Assembly by Promoting Apoptosis of Oocytes in Fetal and Neonatal Mouse Ovaries. PLoS ONE 2011, 6, e16046. [Google Scholar] [CrossRef]
- Hernández-Ochoa, I.; Barnett-Ringgold, K.R.; Dehlinger, S.L.; Gupta, R.K.; Leslie, T.C.; Roby, K.F.; Flaws, J.A. The Ability of the Aryl Hydrocarbon Receptor to Regulate Ovarian Follicle Growth and Estradiol Biosynthesis in Mice Depends on Stage of Sexual Maturity1. Biol. Reprod. 2010, 83, 698–706. [Google Scholar] [CrossRef]
- Christenson, L.K.; Gunewardena, S.; Hong, X.; Spitschak, M.; Baufeld, A.; Vanselow, J. Research Resource: Preovulatory LH Surge Effects on Follicular Theca and Granulosa Transcriptomes. Mol. Endocrinol. 2013, 27, 1153–1171. [Google Scholar] [CrossRef]
- Yenuganti, V.R.; Viergutz, T.; Vanselow, J. Oleic acid induces specific alterations in the morphology, gene expression and steroid hormone production of cultured bovine granulosa cells. Gen. Comp. Endocrinol. 2016, 232, 134–144. [Google Scholar] [CrossRef]
- Hubbi, M.E.; Semenza, G.L. Regulation of cell proliferation by hypoxia-inducible factors. Am. J. Physiol. Physiol. 2015, 309, C775–C782. [Google Scholar] [CrossRef] [PubMed]
- Hodge, S.E.; Greenberg, D.A. How Can We Explain Very Low Odds Ratios in GWAS? I. Polygenic Models. Hum. Hered. 2016, 81, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. Biorxiv 2018, 3, 731. [Google Scholar] [CrossRef]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef]
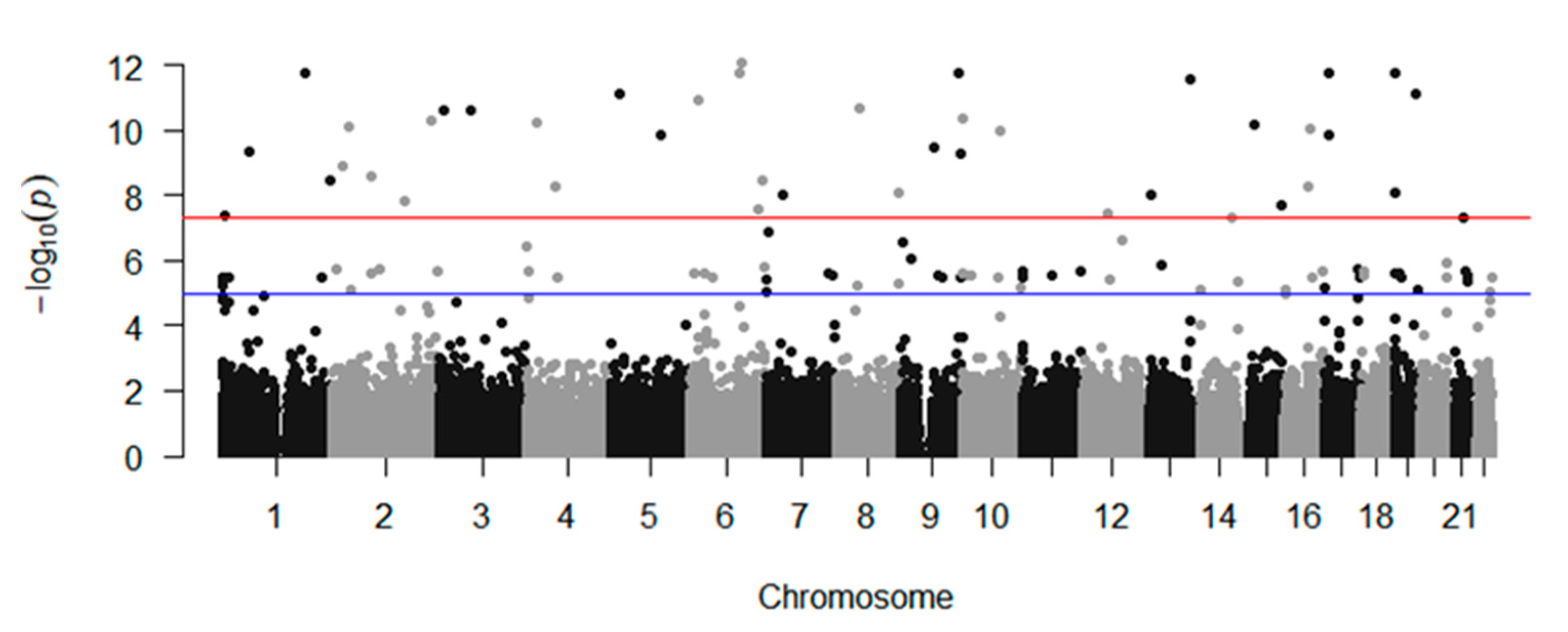
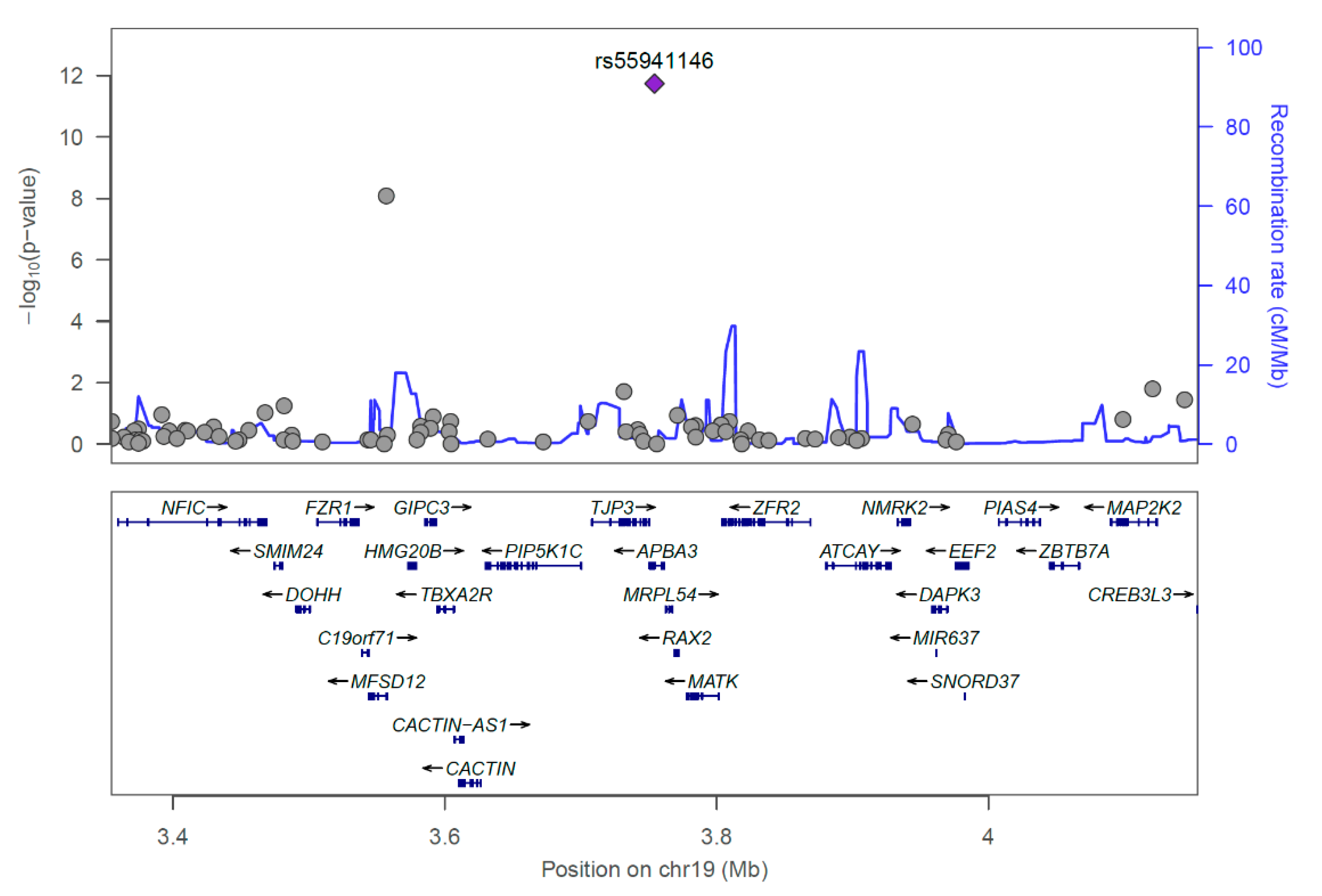
| Information | Discovery | Replication | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CHR | SNP | BP | A1 | TEST | OR | STAT | p | OR | STAT | p |
| 1 | rs3007782 | 4,472,736 | T | ADD | 4.287 | 5.488 | 4.06 × 10−8 | 1.061 | 0.1557 | 0.8763 |
| 1 | rs74918369 | 61,457,396 | C | ADD | 8.071 | 6.242 | 4.31 × 10−10 | 2.133 | 1.283 | 0.1994 |
| 1 | rs115143460 | 1.9 × 108 | A | ADD | 13.33 | 7.047 | 1.83 × 10−12 | 3.37 × 10−9 | 0.0009 | 0.9993 |
| 1 | rs12026894 | 2.45 × 108 | A | ADD | 6.483 | 5.903 | 3.57 × 10−9 | 1.33 | 1.01 | 0.3125 |
| 2 | rs12615054 | 22,223,766 | G | ADD | 7.526 | 6.069 | 1.29 × 10−9 | 0.9204 | −0.2382 | 0.8117 |
| 2 | rs141292341 | 37,310,903 | T | ADD | 10.14 | 6.5 | 8.05 × 10−11 | 1.38 | 0.4903 | 0.6239 |
| 2 | rs145263938 | 87,842,313 | T | ADD | 5.618 | 5.956 | 2.58 × 10−9 | 0.5837 | −1.115 | 0.2648 |
| 2 | rs12692712 | 1.65 × 108 | C | ADD | 4.92 | 5.649 | 1.61 × 10−8 | 1.668 | 1.819 | 0.06891 |
| 2 | rs79371157 | 2.28 × 108 | T | ADD | 9.601 | 6.56 | 5.38 × 10−11 | 0.812 | −0.3447 | 0.7303 |
| 3 | rs28630998 | 13,245,725 | G | ADD | 8.74 | 6.678 | 2.42 × 10−11 | 4.312 | 3.007 | 0.00264 |
| 3 | rs2323277 | 74,110,248 | A | ADD | 11.5 | 6.684 | 2.33 × 10−11 | 1.209 | 0.4123 | 0.6801 |
| 4 | rs141002338 | 24,750,774 | C | ADD | 10.45 | 6.536 | 6.32 × 10−11 | 0.5075 | −1.032 | 0.3019 |
| 4 | rs1510746 | 67,469,201 | C | ADD | 6.439 | 5.839 | 5.25 × 10−9 | 0.6251 | −0.9697 | 0.3322 |
| 5 | rs144522971 | 22,316,095 | T | ADD | 11.99 | 6.853 | 7.22 × 10−12 | 2.813 | 1.338 | 0.1811 |
| 5 | rs296486 | 1.13 × 108 | A | ADD | 8.526 | 6.41 | 1.45 × 10−10 | 2.628 | 2.506 | 0.01222 |
| 6 | rs117716146 | 16,032,432 | T | ADD | 11.65 | 6.789 | 1.13 × 10−11 | 0.8826 | −0.1782 | 0.8585 |
| 6 | rs111721931 | 1.1 × 108 | G | ADD | 13.21 | 7.047 | 1.83 × 10−12 | 1.26 × 10−9 | −0.0019 | 0.9985 |
| 6 | rs137945470 | 1.18 × 108 | A | ADD | 14.85 | 7.153 | 8.51 × 10−13 | 2.078 | 0.5152 | 0.6064 |
| 6 | rs11759078 | 1.53 × 108 | G | ADD | 4.917 | 5.565 | 2.62 × 10−8 | 0.9774 | −0.0669 | 0.9467 |
| 6 | rs113416075 | 1.66 × 108 | A | ADD | 7.197 | 5.911 | 3.39 × 10−9 | 0.577 | −1.466 | 0.1428 |
| 7 | rs11761631 | 6,932,435 | A | ADD | 3.595 | 5.276 | 1.32 × 10−7 | 0.9426 | −0.2598 | 0.795 |
| 7 | rs3807170 | 37,898,032 | T | ADD | 6.065 | 5.735 | 9.76 × 10−9 | 0.721 | −0.7404 | 0.4591 |
| 8 | rs140245000 | 55,592,196 | A | ADD | 10.09 | 6.695 | 2.16 × 10−11 | 2.11 | 1.041 | 0.298 |
| 8 | rs137854443 | 1.45 × 108 | C | ADD | 0.208 | −5.769 | 7.99 × 10−9 | NA | NA | NA |
| 9 | rs77096227 | 8,010,136 | C | ADD | 3.712 | 5.139 | 2.76 × 10−7 | 0.9232 | −0.3035 | 0.7615 |
| 9 | rs149748677 | 78,256,026 | A | ADD | 8.312 | 6.272 | 3.57 × 10−10 | 0.5173 | −0.5861 | 0.5578 |
| 9 | rs146186513 | 1.31 × 108 | G | ADD | 13.33 | 7.047 | 1.83 × 10−12 | NA | NA | NA |
| 9 | rs77609276 | 1.38 × 108 | T | ADD | 8.534 | 6.201 | 5.60 × 10−10 | 0.7848 | −0.4485 | 0.6538 |
| 10 | rs78014704 | 1,842,302 | T | ADD | 9.568 | 6.59 | 4.39 × 10−11 | 0.5369 | −1.34 | 0.1803 |
| 10 | rs34498403 | 86,799,865 | T | ADD | 8.2 | 6.455 | 1.09 × 10−10 | 1.373 | 0.3439 | 0.7309 |
| 12 | rs10877123 | 58,881,273 | A | ADD | 3.971 | 5.498 | 3.84 × 10−8 | 1.111 | 0.4515 | 0.6516 |
| 12 | rs74650809 | 92,513,269 | A | ADD | 3.578 | 5.176 | 2.27 × 10−7 | 1.05 | 0.2121 | 0.8321 |
| 13 | rs56238336 | 23,029,868 | C | ADD | 4.942 | 5.737 | 9.65 × 10−9 | 1.546 | 1.448 | 0.1476 |
| 13 | rs146581261 | 1.11 × 108 | A | ADD | 14.3 | 6.992 | 2.71 × 10−12 | 1.682 | 0.9786 | 0.3278 |
| 14 | rs1957293 | 92,708,417 | A | ADD | 4.209 | 5.448 | 5.10 × 10−8 | 1.068 | 0.2128 | 0.8315 |
| 15 | rs78879506 | 37,133,840 | A | ADD | 10.6 | 6.534 | 6.39 × 10−11 | 1.125 | 0.3918 | 0.6952 |
| 15 | rs57049930 | 93,855,283 | T | ADD | 5.072 | 5.615 | 1.97 × 10−8 | 0.9163 | −0.2447 | 0.8067 |
| 16 | rs77141302 | 54,570,512 | C | ADD | 4.301 | 5.826 | 5.68 × 10−9 | 1.138 | 0.4584 | 0.6466 |
| 16 | rs149428 | 58,668,566 | A | ADD | 8.74 | 6.479 | 9.21 × 10−11 | 0.2768 | −1.683 | 0.09247 |
| 17 | rs146234219 | 9,038,490 | T | ADD | 13.33 | 7.047 | 1.83 × 10−12 | 3.36 × 10−9 | 0.0009 | 0.9993 |
| 17 | rs17773918 | 11,399,739 | G | ADD | 8.535 | 6.425 | 1.32 × 10−10 | 3.569 | 1.718 | 0.08571 |
| 19 | rs1715092 | 3,557,005 | A | ADD | 4.858 | 5.762 | 8.31 × 10−9 | 0.9295 | −0.2799 | 0.7796 |
| 19 | rs55941146 | 3,754,338 | C | ADD | 13.33 | 7.047 | 1.83 × 10−12 | 4.75 | 3.885 | 0.000102 |
| 19 | rs918371 | 48,808,545 | T | ADD | 11.69 | 6.854 | 7.17 × 10−12 | 1.29 × 10−9 | −0.0008 | 0.9993 |
| 21 | rs16991683 | 35,800,134 | A | ADD | 4.26 | 5.47 | 4.49 × 10−8 | 0.7194 | −1.104 | 0.2696 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Park, Y.; Koh, I.; Kim, N.K.; Baek, K.-H.; Yun, B.-S.; Lee, K.J.; Song, J.Y.; Lee, E.; Kwack, K. Association of an APBA3 Missense Variant with Risk of Premature Ovarian Failure in the Korean Female Population. J. Pers. Med. 2020, 10, 193. https://doi.org/10.3390/jpm10040193
Park J, Park Y, Koh I, Kim NK, Baek K-H, Yun B-S, Lee KJ, Song JY, Lee E, Kwack K. Association of an APBA3 Missense Variant with Risk of Premature Ovarian Failure in the Korean Female Population. Journal of Personalized Medicine. 2020; 10(4):193. https://doi.org/10.3390/jpm10040193
Chicago/Turabian StylePark, JeongMan, YoungJoon Park, Insong Koh, Nam Keun Kim, Kwang-Hyun Baek, Bo-Seong Yun, Kyung Ju Lee, Jae Yun Song, Eunil Lee, and KyuBum Kwack. 2020. "Association of an APBA3 Missense Variant with Risk of Premature Ovarian Failure in the Korean Female Population" Journal of Personalized Medicine 10, no. 4: 193. https://doi.org/10.3390/jpm10040193
APA StylePark, J., Park, Y., Koh, I., Kim, N. K., Baek, K.-H., Yun, B.-S., Lee, K. J., Song, J. Y., Lee, E., & Kwack, K. (2020). Association of an APBA3 Missense Variant with Risk of Premature Ovarian Failure in the Korean Female Population. Journal of Personalized Medicine, 10(4), 193. https://doi.org/10.3390/jpm10040193






