Diagnostic and Prognostic Significance of MiR-150 in Colorectal Cancer: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources and Search Strategy
2.2. Study Selection
2.3. Inclusion and Exclusion Criteria
2.4. Data Extraction
2.5. Quality Assessment
2.6. Meta-Analysis Assessment
3. Results
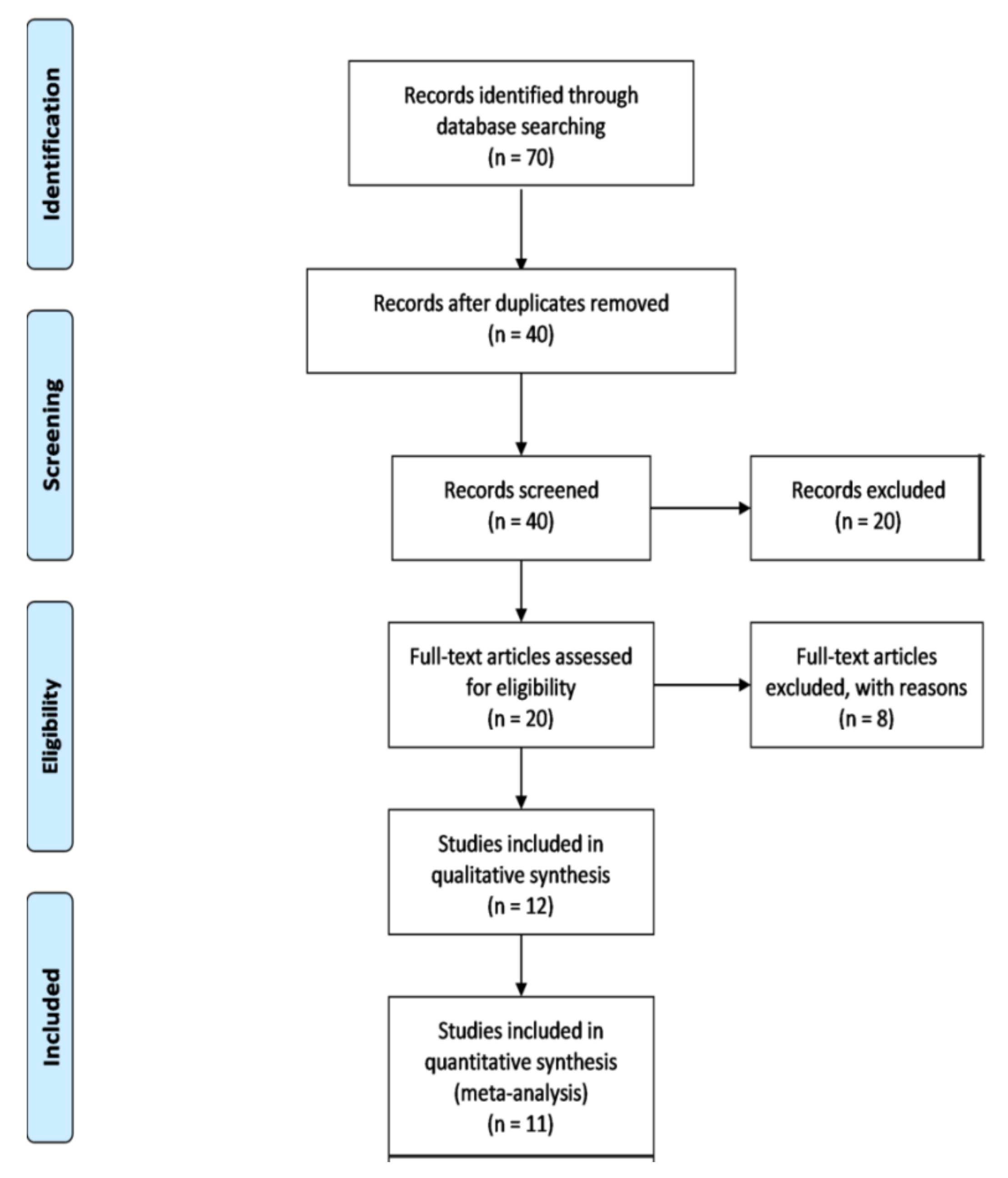
3.1. Search Results
3.2. Characteristics of the Selected Studies
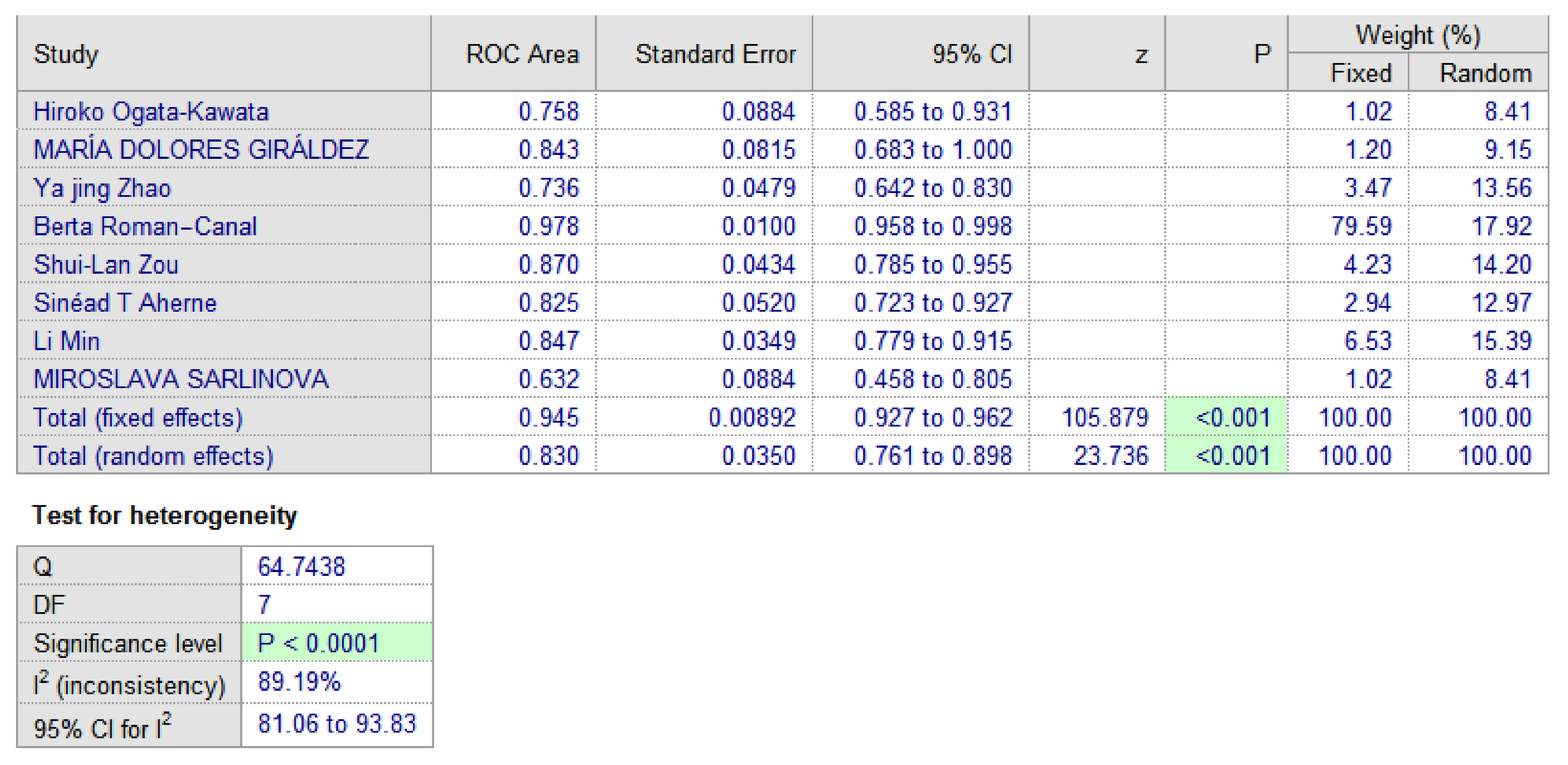
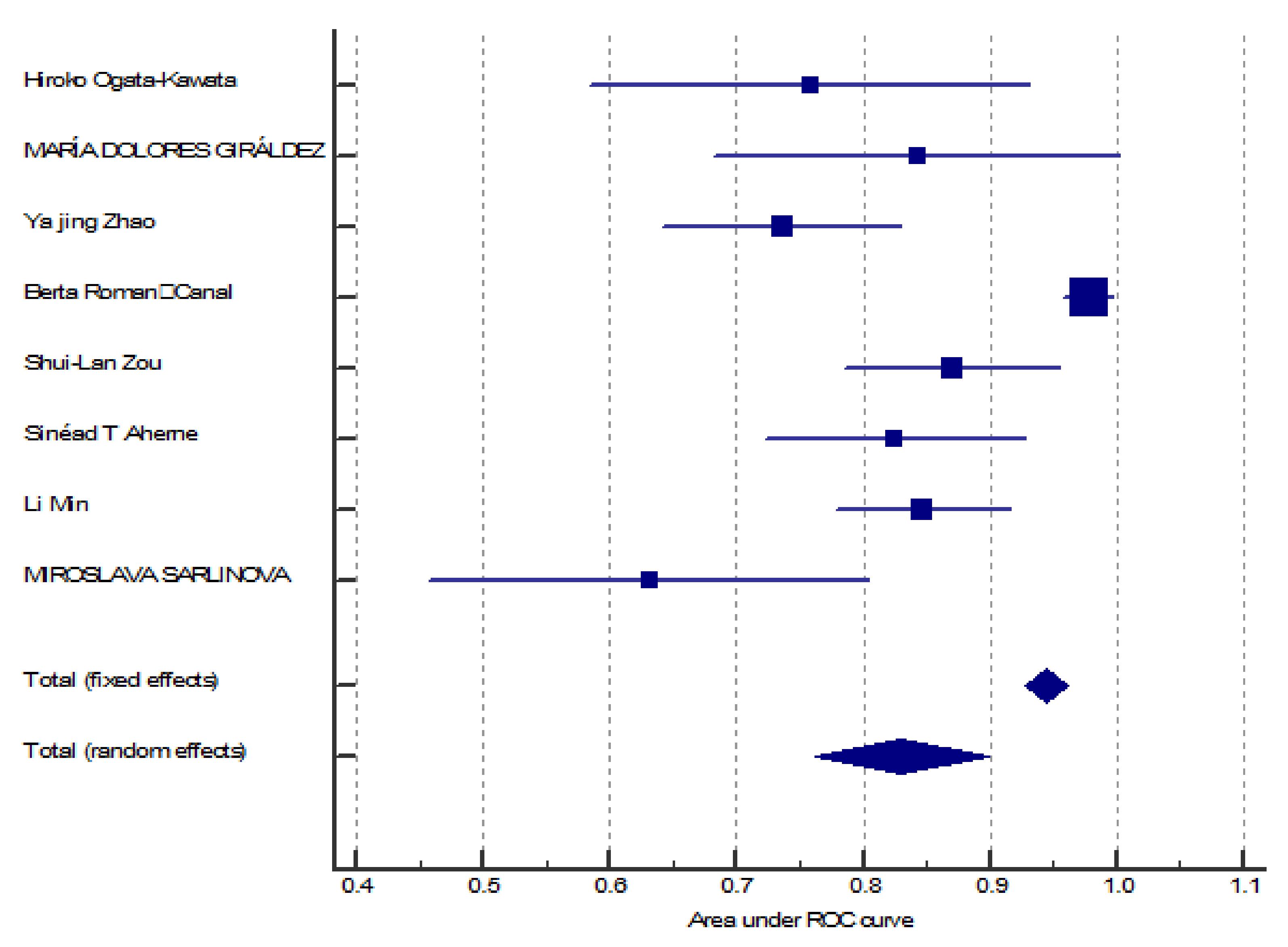
3.3. MiR-150 as A Diagnostic Biomarker
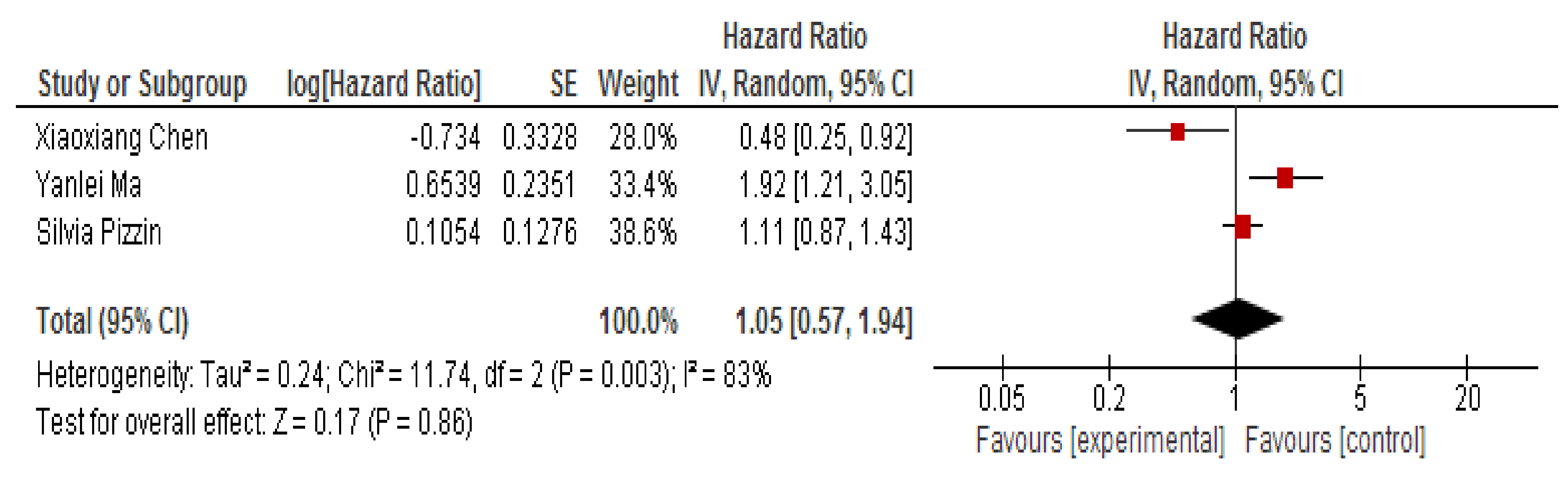
3.4. MiR-150 as A Prognostic Biomarker
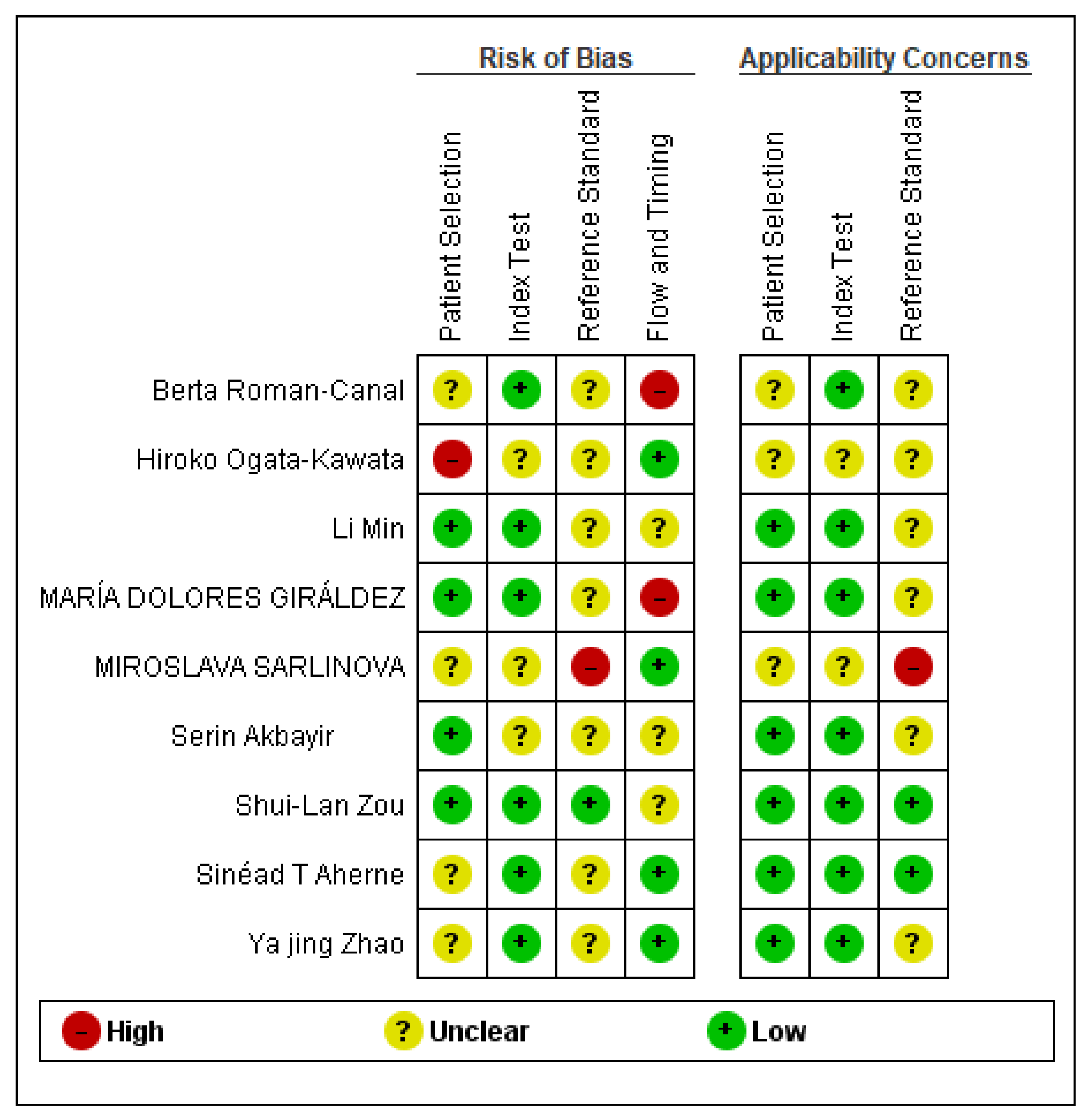
3.5. Bias Assessment and Applicability Judgments
4. Discussion
Study Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F.; Thomas, A. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Argiles, G.; Arnold, D.; Prager, G.; Sobrero, A.F.; Van Cutsem, E. Maximising clinical benefit with adequate patient management beyond the second line in mCRC. ESMO Open 2019, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bekaii-Saab, T.S.; Ou, F.S.; Ahn, D.H.; Boland, P.M.; Ciombor, K.K.; Heying, E.N.; Dockter, T.J.; Jacobs, N.L.; Pasche, B.C.; Cleary, J.M.; et al. Regorafenib dose-optimisation in patients with refractory metastatic colorectal cancer (ReDOS): A randomised, multicentre, open-label, phase 2 study. Lancet Oncol. 2019, 20, 1070–1082. [Google Scholar] [CrossRef]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Cederquist, L.; Chen, Y.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Engstrom, P.F.; et al. NCCN Guidelines® Insights Colon Cancer, Version 2.2018 Featured Updates to the NCCN Guidelines. J. Natl. Compr. Cancer Netw. 2018, 16, 359–369. [Google Scholar] [CrossRef]
- Mizukami, T.; Izawa, N.; Nakajima, T.E.; Sunakawa, Y. Targeting EGFR and RAS/RAF Signaling in the Treatment of Metastatic Colorectal Cancer: From Current Treatment Strategies to Future Perspectives. Drugs 2019, 79, 633–645. [Google Scholar] [CrossRef]
- Sveen, A.; Kopetz, S.; Lothe, R.A. Biomarker-guided therapy for colorectal cancer: Strength in complexity. Nat. Rev. Clin. Oncol. 2020, 17, 11–32. [Google Scholar] [CrossRef]
- Esmaeili, M.; Keshani, M.; Vakilian, M.; Esmaeili, M.; Peymani, M.; Seyed Forootan, F.; Chau, T.L.; Göktuna, S.I.; Zaker, S.R.; Nasr Esfahani, M.H.; et al. Role of non-coding RNAs as novel biomarkers for detection of colorectal cancer progression through interaction with the cell signaling pathways. Gene 2020, 753, 144796. [Google Scholar] [CrossRef]
- Herrera, M.; Galindo-Pumariño, C.; García-Barberán, V.; Peña, C. A snapshot of the tumor microenvironment in colorectal cancer: The liquid biopsy. Int. J. Mol. Sci. 2019, 20, 6016. [Google Scholar] [CrossRef] [PubMed]
- Advani, S.; Kopetz, S. Ongoing and future directions in the management of metastatic colorectal cancer: Update on clinical trials. J. Surg. Oncol. 2019, 119, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Linnekamp, J.F.; Wang, X.; Medema, J.P.; Vermeulen, L. Colorectal cancer heterogeneity and targeted therapy: A case for molecular disease subtypes. Cancer Res. 2015, 75, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Berindan-neagoe, I.; Monroig, P.; Pasculli, B.; Calin, G.A. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014, 64, 311–336. [Google Scholar] [CrossRef] [PubMed]
- Redis, R.S.; Berindan-Neagoe, I.; Pop, V.I.; Calin, G.A. Non-coding RNAs as Theranostics in Human Cancers Roxana. J. Cell. Biochem. 2012, 113, 1451–1459. [Google Scholar] [PubMed]
- Xu, Y.; Luo, X.; He, W.; Chen, G.; Li, Y.; Li, W.; Wang, X.; Lai, Y.; Ye, Y. Long Non-Coding RNA PVT1/miR-150/HIG2 Axis Regulates the Proliferation, Invasion and the Balance of Iron Metabolism of Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2018, 49, 1403–1419. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Li, J.; Wang, X.; Song, W. MicroRNA-150 promotes cell proliferation, migration, and invasion of cervical cancer through targeting PDCD4. Biomed. Pharmacother. 2018, 97, 511–517. [Google Scholar] [CrossRef]
- Li, H.; Ouyang, R.; Wang, Z.; Zhou, W.; Chen, H.; Jiang, Y.; Zhang, Y.; Liao, M.; Wang, W.; Ye, M.; et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci. Rep. 2016, 6, 39001. [Google Scholar] [CrossRef]
- He, Z.; Dang, J.; Song, A.; Cui, X.; Ma, Z.; Zhang, Y. The involvement of miR-150/β-catenin axis in colorectal cancer progression. Biomed. Pharmacother. 2020, 121, 109495. [Google Scholar] [CrossRef]
- Fan, H.; Liu, X.; Zheng, W.W.; Zhuang, Z.H.; Wang, C.D. MiR-150 alleviates EMT and cell invasion of colorectal cancer through targeting Gli1. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4853–4859. [Google Scholar]
- Zhao, Y.J.; Song, X.; Niu, L.; Tang, Y.; Song, X.; Xie, L. Circulating exosomal mir-150-5p and mir-99b-5p as diagnostic biomarkers for colorectal cancer. Front. Oncol. 2019, 9, 1129. [Google Scholar] [CrossRef] [PubMed]
- Saberinia, A.; Alinezhad, A.; Jafari, F.; Soltany, S.; Akhavan Sigari, R. Oncogenic miRNAs and target therapies in colorectal cancer. Clin. Chim. Acta 2020, 508, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; Altman, D.G.; Booth, A.; et al. Preferred reporting items for systematic review and meta-analysis protocols (prisma-p) 2015: Elaboration and explanation. BMJ 2015, 349, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, V.A. (Eds.) Cochrane Handbook for Systematic Reviews of Interventions Version 6.0 (Updated July 2019); Cochrane: London, UK, 2019. [Google Scholar]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M.; The QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Giráldez, M.D.; Lozano, J.J.; Ramírez, G.; Hijona, E.; Bujanda, L.; Castells, A.; Gironella, M. Circulating MicroRNAs as biomarkers of colorectal cancer: Results from a genome-wide profiling and validation study. Clin. Gastroenterol. Hepatol. 2013, 11, 681–688. [Google Scholar] [CrossRef]
- Roman-Canal, B.; Tarragona, J.; Moiola, C.P.; Gatius, S.; Bonnin, S.; Ruiz-Miró, M.; Sierra, J.E.; Rufas, M.; González, E.; Porcel, J.M.; et al. EV-associated miRNAs from peritoneal lavage as potential diagnostic biomarkers in colorectal cancer. J. Transl. Med. 2019, 17, 1–14. [Google Scholar] [CrossRef]
- Zou, S.L.; Chen, Y.L.; Ge, Z.Z.; Qu, Y.Y.; Cao, Y.; Kang, Z.X. Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomark. 2019, 26, 69–77. [Google Scholar] [CrossRef]
- Aherne, S.T.; Madden, S.F.; Hughes, D.J.; Pardini, B.; Naccarati, A.; Levy, M.; Vodicka, P.; Neary, P.; Dowling, P.; Clynes, M. Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression. BMC Cancer 2015, 15, 329. [Google Scholar] [CrossRef]
- Min, L.; Zhu, S.; Chen, L.; Liu, X.; Wei, R.; Zhao, L.; Yang, Y.; Zhang, Z.; Kong, G.; Li, P.; et al. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: A comparison with plasma total miRNAs. J. Extracell. Vesicles 2019, 8, 1643670. [Google Scholar] [CrossRef] [PubMed]
- Sarlinova, M.; Halasa, M.; Mistuna, D.; Musak, L.; Iliev, R.; Slaby, O.; Mazuchova, J.; Valentova, V.; Plank, L.; Halasova, E. Mir-21, mir-221 and mir-150 are deregulated in peripheral blood of patients with colorectal cancer. Anticancer Res. 2016, 36, 5449–5454. [Google Scholar] [CrossRef] [PubMed]
- Pizzini, S.; Bisognin, A.; Mandruzzato, S.; Biasiolo, M.; Facciolli, A.; Perilli, L.; Rossi, E.; Esposito, G.; Rugge, M.; Pilati, P.; et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genom. 2013, 14, 589. [Google Scholar] [CrossRef] [PubMed]
- Yanlei, M.; Zhang, P.; Wang, F.; Zhang, H.; Yang, J.; Peng, J.; Liu, W.; Qin, H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut 2012, 61, 1447–1453. [Google Scholar]
- Chen, X.; Xu, X.; Pan, B.; Zeng, K.; Xu, M.; Liu, X.; He, B.; Pan, Y.; Sun, H.; Wang, S. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging 2018, 10, 3421–3437. [Google Scholar] [CrossRef]
- Akbayir, S.; Unal, N.D.; Balcı, S.; Gorur, A.; Yaroglu, H.Y.; Ayaz, L.; Gundogdu, R.; Dırlık, M.; Serın, M.S.; Tamer, L. Evaluation of Plasma microRNA Expression Levels in Early Diagnosis of Colorectal Cancer. J. Clin. Exp. Oncol. 2014, 3, 1–6. [Google Scholar]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Wang, W.; Wang, X.; Zhang, Y.; Wang, D.; Gao, H.; Wang, L.; Gao, S. Prognostic role of microRNA-150 in various carcinomas: A meta-analysis. Onco Targets Ther. 2016, 9, 1371–1379. [Google Scholar] [CrossRef]
- Jin, M.; Yang, Z.; Ye, W.; Xu, H.; Hua, X. MicroRNA-150 predicts a favorable prognosis in patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS ONE 2014, 9, e103965. [Google Scholar] [CrossRef]
- Amirkhah, R.; Schmitz, U.; Linnebacher, M.; Wolkenhauer, O.; Farazmand, A. MicroRNA–mRNA Interactions in Colorectal Cancer and Their Role in Tumor Progression. Genes Chromosom. Cancer 2015, 54, 129–141. [Google Scholar] [CrossRef]
- Li, C.; Du, X.; Xia, S.; Chen, L. MicroRNA-150 inhibits the proliferation and metastasis potential of colorectal cancer cells by targeting iASPP. Oncol. Rep. 2018, 40, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, M.; Wang, M.; Yan, D.; Feng, G.; An, G. The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer. BMC Cancer 2014, 14, 714. [Google Scholar] [CrossRef] [PubMed]
- Moody, L.; Dvoretskiy, S.; An, R.; Mantha, S.; Pan, Y.-X. The Efficacy of miR-20a as a Diagnostic and Prognostic Biomarker for Colorectal Cancer: A Systematic Review and Meta-Analysis. Cancers 2019, 11, 1111. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Shen, Y.; Zhao, P.; Cheng, M.; Zhu, Y.; Xu, B. Biomarker roles identification of miR-106 family for predicting the risk and poor survival of colorectal cancer. BMC Cancer 2020, 20, 506. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yan, G.; Yin, L.; Liu, T.; Li, C.; Wang, L. Prognostic roles of microRNA 143 and microRNA 145 in colorectal cancer: A meta-analysis. Int. J. Biol. Markers 2019, 34, 6–14. [Google Scholar] [CrossRef]
- Guraya, S. Prognostic significance of circulating microRNA-21 expression in esophageal, pancreatic and colorectal cancers; a systematic review and meta-analysis. Int. J. Surg. 2018, 60, 41–47. [Google Scholar] [CrossRef]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Das, V.; Kalita, J.; Pal, M. Predictive and prognostic biomarkers in colorectal cancer: A systematic review of recent advances and challenges. Biomed. Pharmacother. 2017, 87, 8–19. [Google Scholar] [CrossRef]
- Masuda, T.; Hayashi, N.; Kuroda, Y.; Ito, S.; Eguchi, H.; Mimori, K. MicroRNAs as biomarkers in colorectal cancer. Cancers 2017, 9, 124. [Google Scholar] [CrossRef]
- Sabarimurugan, S.; Kumarasamy, C.; Royam, M.M.; Lakhotiya, K.; Muthukaliannan, G.K.; Ramalingam, S.; Jayaraj, R. Validation of miRNA prognostic significance in stage II colorectal cancer: A protocol for systematic review and meta-analysis of observational clinical studies. Medicine 2019, 98, e14570. [Google Scholar] [CrossRef]
| ID | Year | Country | No. of Patients | No. of Controls | MiRNA | Sample | Method for Detection | Reported Value | Expression Status |
|---|---|---|---|---|---|---|---|---|---|
| Hiroko Ogata-Kawata [27] | 2014 | Japan | 88 | 11 | 150 | Serum | RT-PCR | AUC, 95% CI | Up-regulated |
| MARÍA DOLORES GIRÁLDEZ [28] | 2013 | Spain | 63 | 73 | 150 | Plasma | RT-PCR | AUC, 95% CI | Up-regulated |
| Ya Jing Zhao [21] | 2019 | China | 169 | 155 | 150-5p | Serum | RT-PCR | AUC, 95% CI | Down-regulated |
| Berta Roman-Canal [29] | 2019 | Spain | 25 | 25 | 150-5p | Peritoneal lavages | RT-PCR | AUC, 95% CI | Up-regulated |
| Shui-Lan Zou [30] | 2019 | China | 133 | 60 | 150-5p | Serum | QRT-PCR | AUC, 95% CI | Down-regulated |
| Sinéad T Aherne [31] | 2015 | Czech Republic | 52 | 82 | 150 | Serum | QRT-PCR | AUC, 95% CI | Down-regulated |
| Li Min [32] | 2019 | China | 15 | 10 | 150-3p | Serum | QRT-PCR | AUC, 95% CI | Down- and up-regulated |
| MIROSLAVA SARLINOVA [33] | 2016 | Slovakia | 71 | 80 | 150 | Plasma | QRT-PCR | AUC, 95% CI | Down-regulated |
| Serin Akbayir [37] | 2013 | Turkey | 37 | 238 | 150-5p | Plasma | QRT-PCR | - | Down-regulated |
| Silvia Pizzin [34] | 2013 | Italy | 46 | - | 150 | Tissue | QRT-PCR | HR, 95% CI | Down-regulated |
| Yanlei Ma [35] | 2012 | China | 239 | - | 150 | Tissue | QRT-PCR | HR, 95% CI | Down-regulated |
| Xiaoxiang Chen [36] | 2018 | China | 112 | - | 150-5p | Tissue | QRT-PCR | HR, 95% CI | Down-regulated |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sur, D.; Burz, C.; Sabarimurugan, S.; Irimie, A. Diagnostic and Prognostic Significance of MiR-150 in Colorectal Cancer: A Systematic Review and Meta-Analysis. J. Pers. Med. 2020, 10, 99. https://doi.org/10.3390/jpm10030099
Sur D, Burz C, Sabarimurugan S, Irimie A. Diagnostic and Prognostic Significance of MiR-150 in Colorectal Cancer: A Systematic Review and Meta-Analysis. Journal of Personalized Medicine. 2020; 10(3):99. https://doi.org/10.3390/jpm10030099
Chicago/Turabian StyleSur, Daniel, Claudia Burz, Shanthi Sabarimurugan, and Alexandru Irimie. 2020. "Diagnostic and Prognostic Significance of MiR-150 in Colorectal Cancer: A Systematic Review and Meta-Analysis" Journal of Personalized Medicine 10, no. 3: 99. https://doi.org/10.3390/jpm10030099
APA StyleSur, D., Burz, C., Sabarimurugan, S., & Irimie, A. (2020). Diagnostic and Prognostic Significance of MiR-150 in Colorectal Cancer: A Systematic Review and Meta-Analysis. Journal of Personalized Medicine, 10(3), 99. https://doi.org/10.3390/jpm10030099






