Skin Lesion Segmentation in Dermoscopic Images with Combination of YOLO and GrabCut Algorithm
Abstract
1. Introduction
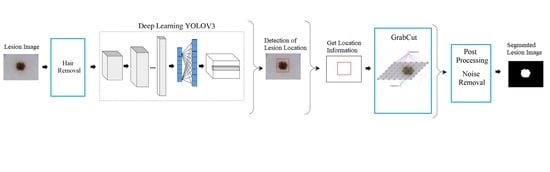
- Yolov3 [13], which is a deep convolutional neural network that has been trained for the detection of lesion location in the image and it has been used to automate segmentation algorithm GrabCut, which is also known as a semi-automatic algorithm, for segmenting skin lesions for the first time in literature.
- An alternative real-time skin lesion segmentation method has been proposed with the incorporation of different techniques into a pipeline.
- Deep learning-based segmentation methods process fixed-size images and produce low-resolution segmentation images. The present method processes dimension-independent images and produces high-resolutions segmentation results.
2. Related Works
3. Materials and Methods
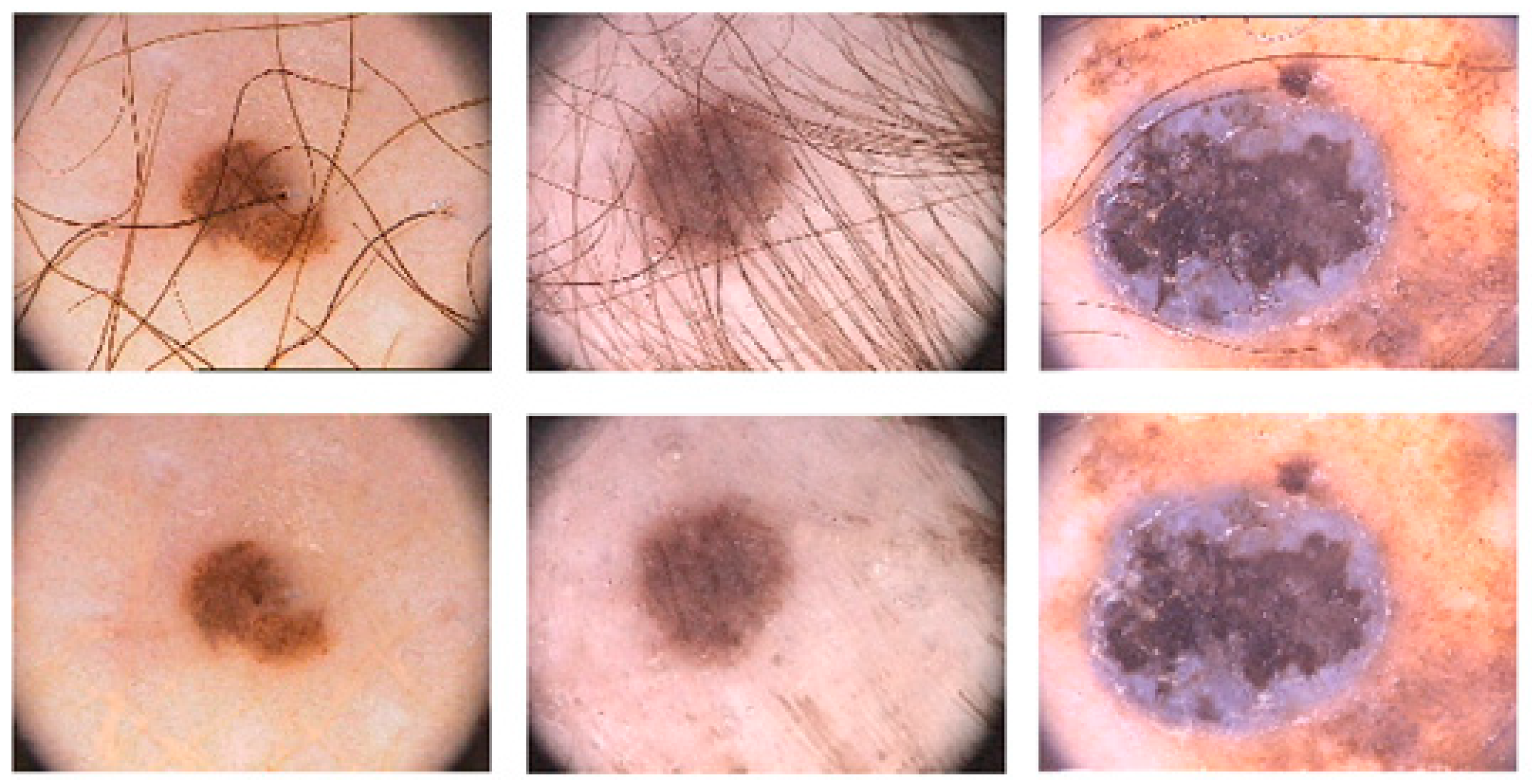
3.1. Datasets
3.2. Prepossessing and Labelling
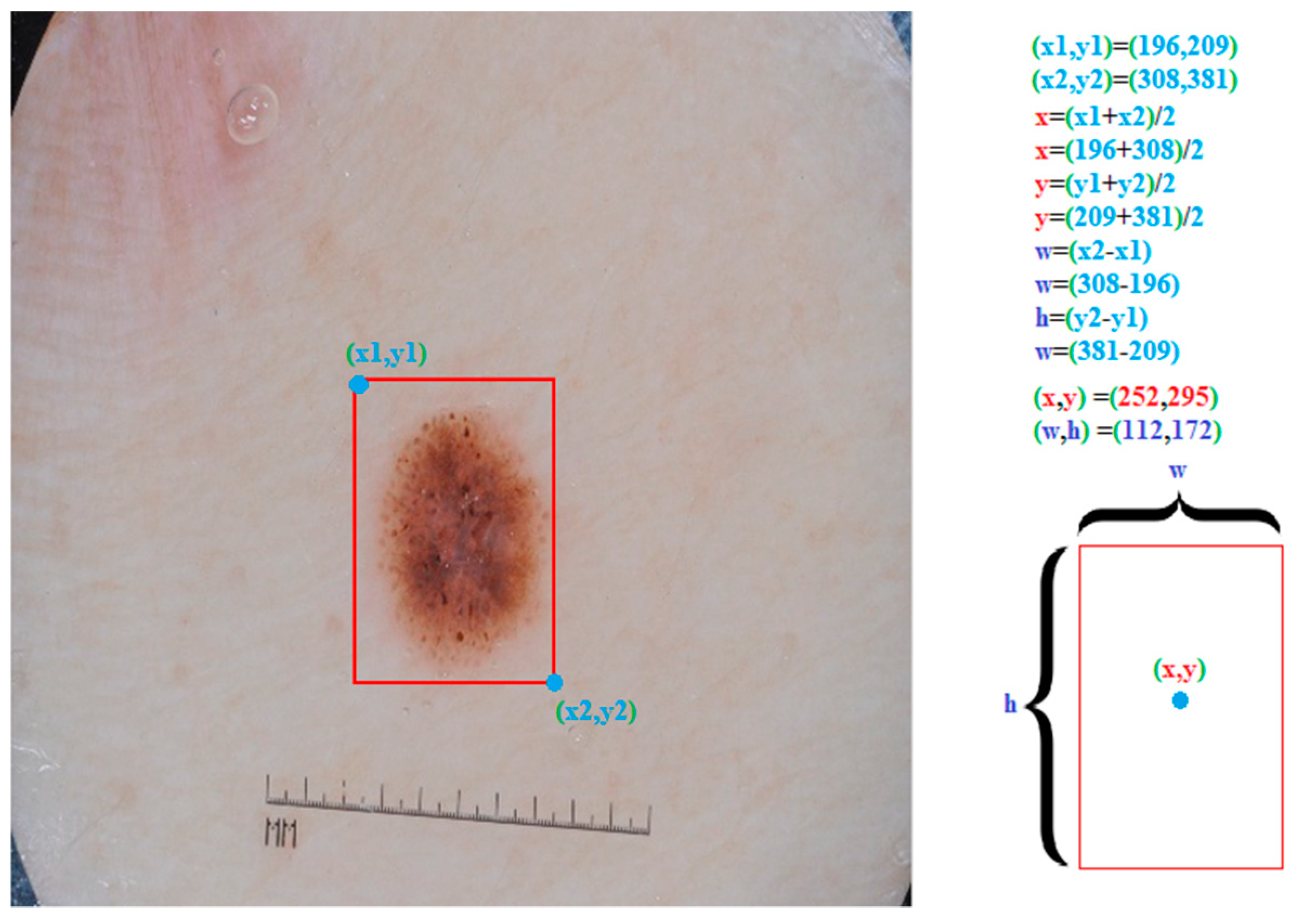
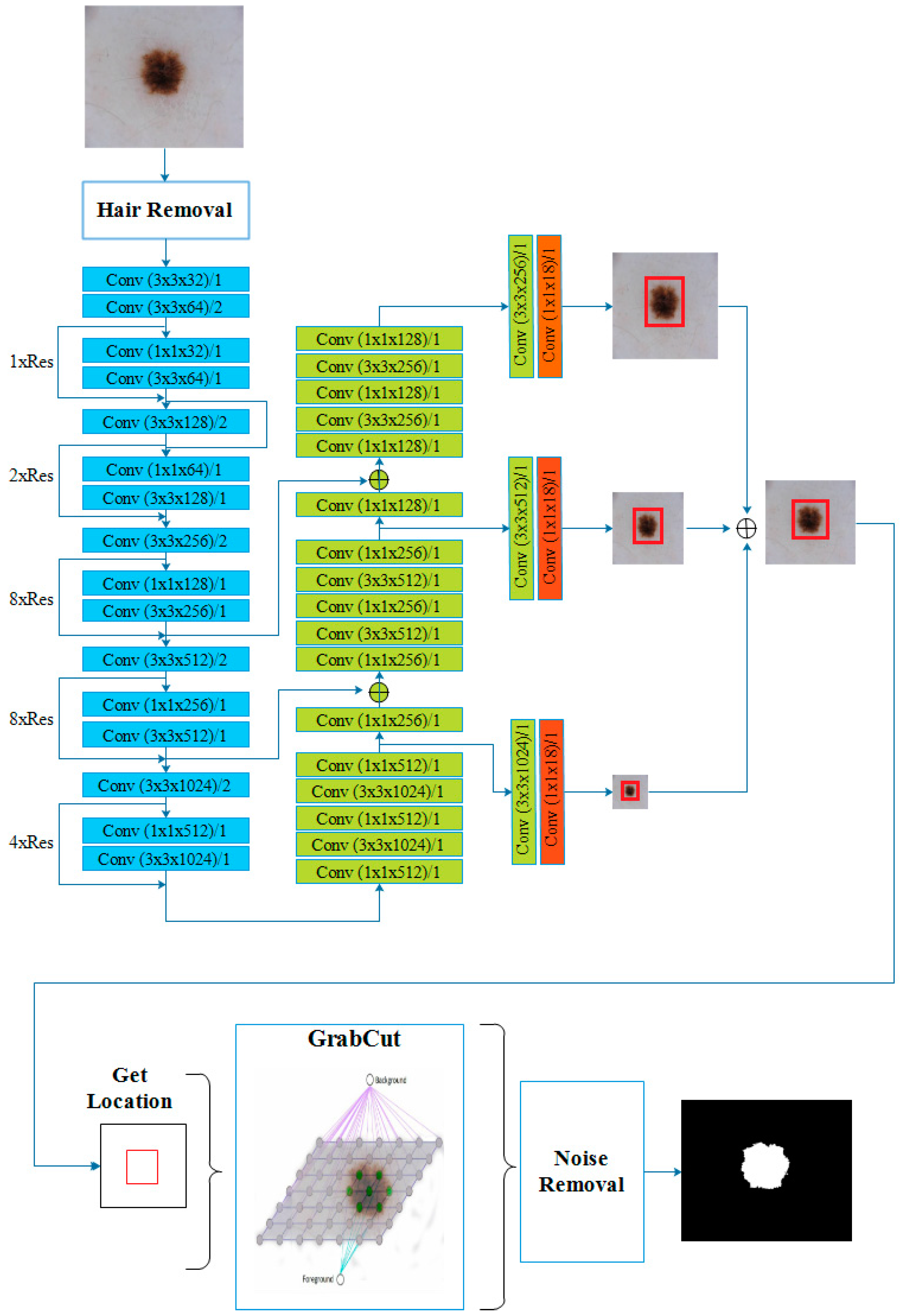
3.3. Yolo Architecture
- Yolov3 uses logistic regression as loss function while predicting the confidence scores of bounding boxes.
- Yolov3 utilizes multiple independent logistic classifiers instead of softmax function for the prediction of the class confidence probabilities. This improvement is very important when there are multiple objects in the image.
- Yolov3 benefits from a powerful feature extractor network named DarkNet-53 with 53 convolutional layers and residual blocks. In addition to the DarkNet-53, 53 more layers have been added for the detection task, thus making a 106 layered fully convolutional architecture. Figure 9 illustrates DarkNet-53 architecture.
- Yolov3 employs strided convolution for down sampling instead of max-pooling.
- Yolov3 is made up several convolutional layers in addition to the base feature extractor layers, enhancing its capability of multiscale predictions at three different sizes. This allows the system to make more accurate detections on small objects in the image.
- The last improvement in Yolov3 is cross-layer connections between the prediction layers. The feature maps obtained from the up-sampling operation have been combined with the feature maps of with the previous layers by using concatenation operation. This combination provided more accurate detection performance on small objects.
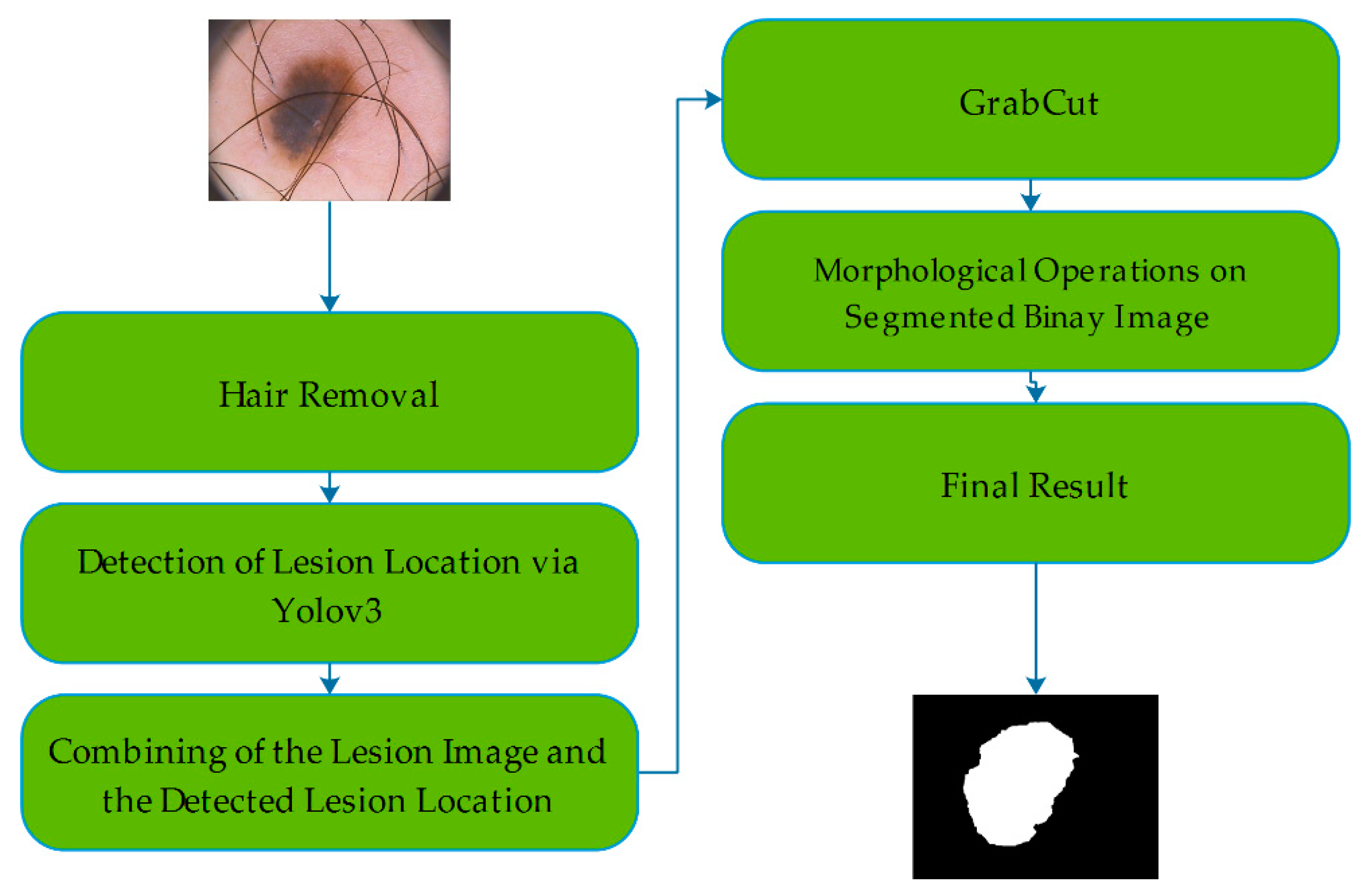
3.4. GrabCut Algorithm
- The initial step starts with a rectangle surrounding the object of interest that is drawn by the user manually. This step gives information about background and foreground of the interested area. The pixels inside of the rectangle considered as unknown while the pixels outside of the rectangle are considered as background. Based on this information, the algorithm creates a model to determine whether the unknown pixels belong to the foreground or background.
- An initial segmentation model is created in which the unknown pixels are regarded as the foreground class and all the other except are seen as the background.
- The initial background and foreground classes are created by using Gaussian Mixture Models, by creating piece GMM components for two regions.
- Each pixel in the background class is designated to the most probable Gaussian component in the background GMM. The same process is performed for the foreground pixels designated to the most probable foreground Gaussian component.
- New GMM are attained by using the sets of pixels created in the previous step.
- A graph with n nodes is built and the weight values between the connections are determined. After that, a minimum cut algorithm is used to determine foreground and background pixels.
- Steps of 4–6 are repeated until attaining the final segmentation result.
3.5. Proposed System by Using Yolov3 and GrabCut
3.6. Training Yolov3
3.7. Performance Evaluation Metrics
4. Results
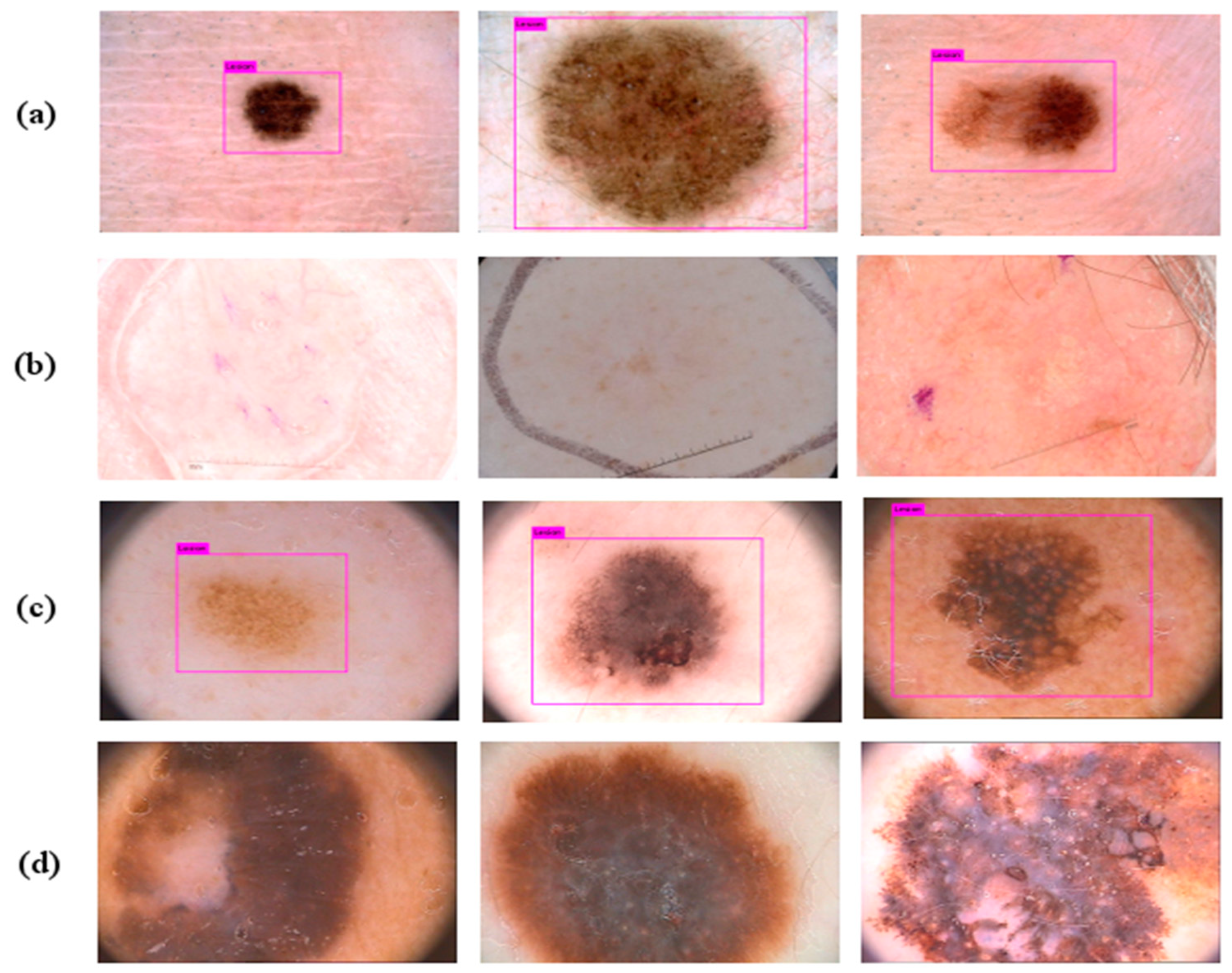
4.1. Detection Results on the PH2 and the ISBI 2017 Datasets
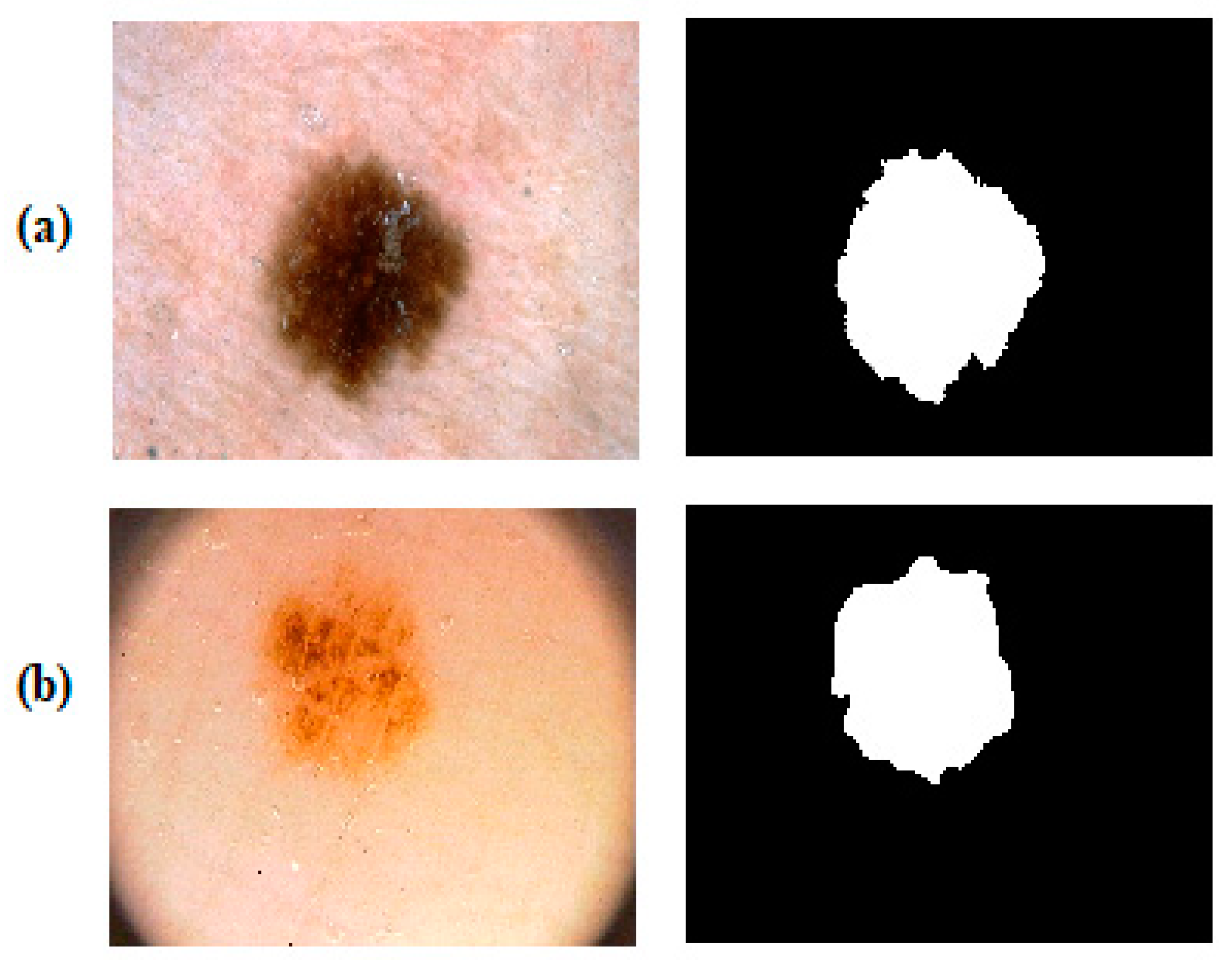
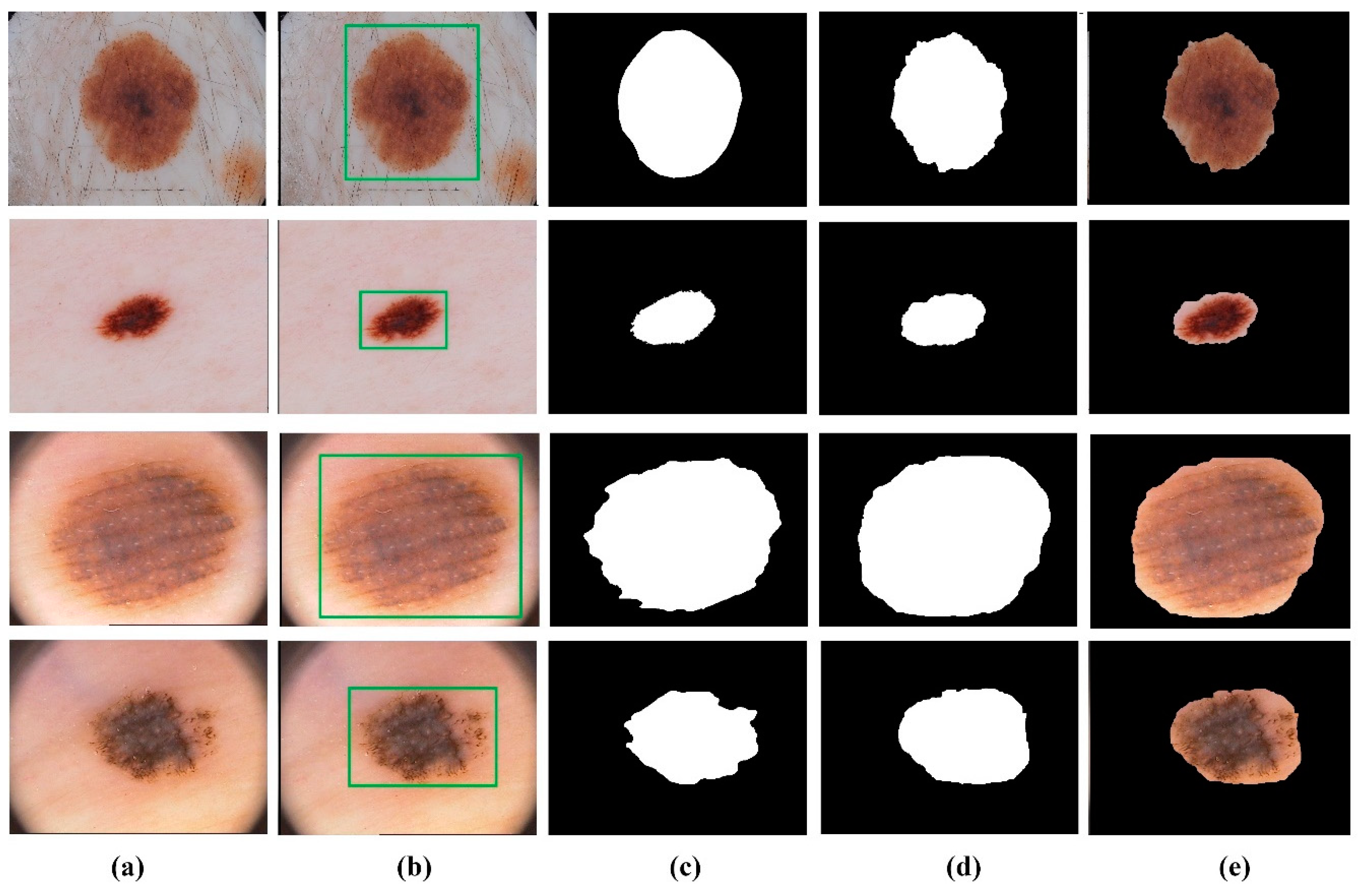
4.2. Segmentation Results on the PH2 and the ISBI 2017 Datasets
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Karimkhani, C.; Green, A.; Nijsten, T.; Weinstock, M.; Dellavalle, R.; Naghavi, M.; Fitzmaurice, C. The global burden of melanoma: results from the Global Burden of Disease Study 2015. Br. J. Dermatol. 2017, 177, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.A.; Kampp, J. Skin Cancer Epidemiology, Detection, and Management. Med Clin. N. Am. 2015, 99, 1323–1335. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Isern, N.G.; Burton, S.D.; Hu, J.Z. Studies of secondary melanoma on C57BL/6J mouse liver using 1H NMR metabolomics. Metabolites 2013, 3, 1011–1035. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar]
- Tarver, T.; American Cancer Society. Cancer facts and figures 2014. J. Consum. Health Internet 2012, 16, 366–367. [Google Scholar] [CrossRef]
- Siegel, R.; Miller, K.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2017, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Pellacani, G.; Seidenari, S. Comparison between morphological parameters in pigmented skin lesion images acquired by means of epiluminescence surface microscopy and polarized-light videomicroscopy. Clin. Dermatol. 2002, 20, 222–227. [Google Scholar] [CrossRef]
- Ali, A.-R.A.; Deserno, T.M. A systematic review of automated melanoma detection in dermatoscopic images and its ground truth data. In Medical Imaging 2012: Image Perception, Observer Performance, and Technology Assessment; International Society for Optics and Photonics: Bellingham, WA, USA, 2012; p. 8318. [Google Scholar]
- Sinz, C.; Tschandl, P.; Rosendahl, C.; Akay, B.N.; Argenziano, G.; Blum, A.; Braun, R.P.; Cabo, H.; Gourhant, J.-Y.; Kreusch, J.; et al. Accuracy of dermatoscopy for the diagnosis of nonpigmented cancers of the skin. J. Am. Acad. Dermatol. 2017, 77, 1100–1109. [Google Scholar] [CrossRef]
- Bi, L.; Kim, J.; Ahn, E.; Kumar, A.; Fulham, M.; Feng, D. Dermoscopic image segmentation via multi-stage fully convolutional networks. IEEE Trans. Biomed. Eng. 2017, 64, 2065–2074. [Google Scholar] [CrossRef]
- Okur, E.; Turkan, M. A survey on automated melanoma detection. Eng. Appl. Artif. Intell. 2018, 73, 50–67. [Google Scholar] [CrossRef]
- Rother, C.; Kolmogorov, V.; Blake, A. Grabcut: Interactive foreground extraction using iterated graph cuts. In ACM Transactions on Graphics (TOG); ACM: New York, NY, USA, 2004. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Ganster, H.; Pinz, P.; Rohrer, R.; Wildling, E.; Binder, M.; Kittler, H. Automated melanoma recognition. IEEE Trans. Med Imaging 2001, 20, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, G.; Krawczyk, B.; Celebi, M.E.; Iyatomi, H. An ensemble classification approach for melanoma diagnosis. Memetic Comput. 2014, 6, 233–240. [Google Scholar] [CrossRef]
- Celebi, M.E.; Iyatomi, H.; Schaefer, G.; Stoecker, W.V. Lesion border detection in dermoscopy images. Comput. Med. Imaging Graph. 2009, 33, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Korotkov, K.; Garcia, R. Computerized analysis of pigmented skin lesions: A review. Artif. Intell. Med. 2012, 56, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Filho, M.; Ma, Z.; Tavares, J.M.R.S. A Review of the Quantification and Classification of Pigmented Skin Lesions: From Dedicated to Hand-Held Devices. J. Med. Syst. 2015, 39, 177. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.B.; Filho, M.E.; Ma, Z.; Papa, J.P.; Pereira, A.S.; Tavares, J.M.R. Withdrawn: Computational methods for the image segmentation of pigmented skin lesions: A Review. Comput. Methods Programs Biomed. 2016, 131, 127–141. [Google Scholar] [CrossRef]
- Emre Celebi, M.; Wen, Q.; Hwang, S.; Iyatomi, H.; Schaefer, G. Lesion border detection in dermoscopy images using ensembles of thresholding methods. Skin Res. Technol. 2013, 19, e252–e258. [Google Scholar] [CrossRef]
- Yuksel, M.; Borlu, M. Accurate Segmentation of Dermoscopic Images by Image Thresholding Based on Type-2 Fuzzy Logic. IEEE Trans. Fuzzy Syst. 2009, 17, 976–982. [Google Scholar] [CrossRef]
- Peruch, F.; Bogo, F.; Bonazza, M.; Cappelleri, V.-M.; Peserico, E. Simpler, Faster, More Accurate Melanocytic Lesion Segmentation Through MEDS. IEEE Trans. Biomed. Eng. 2014, 61, 557–565. [Google Scholar] [CrossRef]
- Møllersen, K.; Kirchesch, H.M.; Schopf, T.G.; Godtliebsen, F. Unsupervised segmentation for digital dermoscopic images. Ski. Res. Technol. 2010, 16, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Bovik, A.C. Automatic segmentation of dermoscopy images using self-generating neural networks seeded by genetic algorithm. Pattern Recognit. 2013, 46, 1012–1019. [Google Scholar] [CrossRef]
- Zhou, H.; Schaefer, G.; Sadka, A.H.; Celebi, M.E. Anisotropic mean shift based fuzzy c-means segmentation of deroscopy images. IEEE J. Sel. Top. Signal Process. 2009, 3, 26–34. [Google Scholar] [CrossRef]
- Kockara, S.; Mete, M.; Yip, V.; Lee, B.; Aydin, K. A soft kinetic data structure for lesion border detection. Bioinformatics 2010, 26, i21–i28. [Google Scholar] [CrossRef] [PubMed]
- Suer, S.; Kockara, S.; Mete, M. An improved border detection in dermoscopy images for density-based clustering. In BMC Bioinformatics; BioMed Central: London, UK, 2011. [Google Scholar]
- Abbas, Q.; Celebi, M.; García, I.F. Skin tumor area extraction using an improved dynamic programming approach. Skin Res. Technol. 2012, 18, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Ashour, A.S.; Hawas, A.R.; Guo, Y.; Wahba, M.A. A novel optimized neutrosophic k-means using genetic algorithm for skin lesion detection in dermoscopy images. Signal Image Video Process. 2018, 12, 1311–1318. [Google Scholar] [CrossRef]
- Abbas, Q.; Celebi, M.E.; García, I.F.; Rashid, M. Lesion border detection in dermoscopy images using dynamic programming. Ski. Res. Technol. 2011, 17, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Celebi, M.E.; Kingravi, H.A.; Iyatomi, H.; Aslandogan, Y.A.; Stoecker, W.V.; Moss, R.H.; Malters, J.M.; Grichnik, J.M.; Marghoob, A.A.; Rabinovitz, H.S.; et al. Border detection in dermoscopy images using statistical region merging. Ski. Res. Technol. 2008, 14, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Silveira, M.; Nascimento, J.C.; Marques, J.S.; Marcal, A.R.S.; Mendonca, T.; Yamauchi, S.; Maeda, J.; Rozeira, J. Comparison of Segmentation Methods for Melanoma Diagnosis in Dermoscopy Images. IEEE J. Sel. Top. Signal Process. 2009, 3, 35–45. [Google Scholar] [CrossRef]
- Erkol, B.; Moss, R.H.; Stanley, R.J.; Stoecker, W.V.; Hvatum, E. Automatic lesion boundary detection in dermoscopy images using gradient vector flow snakes. Ski. Res. Technol. 2005, 11, 17–26. [Google Scholar] [CrossRef]
- Mete, M.; Sirakov, N.M. Lesion detection in demoscopy images with novel density-based and active contour approaches. In BMC Bioinformatics; BioMed Central: London, UK, 2010. [Google Scholar]
- Wang, H.; Moss, R.H.; Chen, X.; Stanley, R.J.; Stoecker, W.V.; Celebi, M.E.; Malters, J.M.; Grichnik, J.M.; Marghoob, A.A.; Rabinovitz, H.S.; et al. Modified watershed technique and post-processing for segmentation of skin lesions in dermoscopy images. Comput. Med. Imaging Graph. 2011, 35, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Çelebi, M.; Wen, Q.; Iyatomi, H.; Shimizu, K.; Zhou, H.; Schaefer, G. A State-of-the-Art Survey on Lesion Border Detection in Dermoscopy Images. Dermoscopy Image Anal. 2015, 10, 97–129. [Google Scholar]
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P. Techniques and algorithms for computer aided diagnosis of pigmented skin lesions—A review. Biomed. Signal Process. Control. 2018, 39, 237–262. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Cambridge, MA, USA, 2012; pp. 1097–1105. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin, Germany, 2015. [Google Scholar]
- Cireşan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Mitosis detection in breast cancer histology images with deep neural networks. In Proceedings of the International Conference on Medical Image Computing and Computer-assisted Intervention; Springer: Berlin, Germany, 2013. [Google Scholar]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C. Brain Tumor Segmentation using Convolutional Neural Networks in MRI Images. IEEE Trans. Med Imaging 2016, 35, 1. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Ruiz, A.; Mordang, J.J.; Karssemeijer, N.; Sechopoulos, I.; Mann, R.M. Can radiologists improve their breast cancer detection in mammography when using a deep learning-based computer system as decision support? In Society of Photo-Optical Instrumentation Engineers (SPIE) Conference Series; International Society for Optics and Photonics: Bellingham, WA, USA, 2018. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. arXiv 2015, arXiv:1511.00561. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- García-García, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martínez, V.; García-Rodríguez, J. A review on deep learning techniques applied to semantic segmentation. arXiv 2017, arXiv:1704.06857. [Google Scholar]
- Yu, Z.; Jiang, X.; Zhou, F.; Qin, J.; Ni, D.; Chen, S.; Lei, B.; Wang, T. Melanoma Recognition in Dermoscopy Images via Aggregated Deep Convolutional Features. IEEE Trans. Biomed. Eng. 2018, 66, 1006–1016. [Google Scholar] [CrossRef]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018. [Google Scholar]
- Yuan, Y.; Chao, M.; Lo, Y.-C. Automatic Skin Lesion Segmentation Using Deep Fully Convolutional Networks with Jaccard Distance. IEEE Trans. Med. Imaging 2017, 36, 1876–1886. [Google Scholar] [CrossRef]
- Mendonça, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.; Rozeira, J. PH 2-A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013. [Google Scholar]
- Goyal, M.; Yap, M.H. Multi-class semantic segmentation of skin lesions via fully convolutional networks. arXiv 2017, arXiv:1711.10449. [Google Scholar]
- Lin, B.S.; Michael, K.; Kalra, S.; Tizhoosh, H.R. Skin lesion segmentation: U-nets versus clustering. In Proceedings of the 2017 IEEE Symposium Series on Computational Intelligence (SSCI), Honolulu, HI, USA, 27 November–1 December 2017. [Google Scholar]
- Yuan, Y.; Chao, M.; Lo, Y.-C. Automatic skin lesion segmentation with fully convolutional-deconvolutional networks. arXiv 2017, arXiv:1703.05165. [Google Scholar]
- Al-Masni, M.A.; Al-Antari, M.A.; Choi, M.-T.; Han, S.-M.; Kim, T.-S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; He, X.; Zhou, F.; Yu, Z.; Ni, D.; Chen, S.; Wang, T.; Lei, B. Dense Deconvolutional Network for Skin Lesion Segmentation. IEEE J. Biomed. Health Inform. 2018, 23, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, N.; Wang, Y.; Wang, M. Segmentation of dermoscopy image using adversarial networks. Multimed. Tools Appl. 2018, 78, 10965–10981. [Google Scholar] [CrossRef]
- Yuan, Y.; Lo, Y.C. Improving dermoscopic image segmentation with enhanced convolutional-deconvolutional networks. IEEE J. Biomed. Health Inform. 2019, 23, 519–526. [Google Scholar] [CrossRef] [PubMed]
- ISIC. Skin Lesion Analysis Towards Melanoma Detection 2017. Available online: https://challenge.kitware.com/#challenge/n/ISIC_2017%3A_Skin_Lesion_Analysis_Towards_Melanoma_Detection (accessed on 29 May 2019).
- Lee, T.; Ng, V.; Gallagher, R.; Coldman, A.; McLean, D. Dullrazor®: A software approach to hair removal from images. Comput. Boil. Med. 1997, 27, 533–543. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 24–27 June 2014; pp. 580–587. [Google Scholar]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. arXiv 2017, arXiv:1612.08242. [Google Scholar]
- Zivkovic, Z. Improved adaptive Gaussian mixture model for background subtraction. In Proceedings of the 17th International Conference on ICPR Pattern Recognition, Cambridge, UK, 26–26 August 2004. [Google Scholar]
- Wang, D. The Experimental Implementation of GrabCut for Hardcode Subtitle Extraction. In Proceedings of the 2018 IEEE/ACIS 17th International Conference on Computer and Information Science (ICIS), Singapore, 6–8 June 2018. [Google Scholar]
- Redmon, J. Darknet: Open Source Neural Networks in C. 2013–2019. Available online: http://pjreddie.com/darknet/ (accessed on 29 May 2019).
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M.; Hoo-Chang, S. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Powers, D.M. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Li, Y.; Shen, L. Skin Lesion Analysis towards Melanoma Detection Using Deep Learning Network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Kim, J.; Ahn, E.; Feng, D. Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. arXiv 2017, arXiv:1703.04197. [Google Scholar]
- Burdick, J.; Marques, O.; Weinthal, J.; Furht, B. Rethinking skin lesion segmentation in a convolutional classifier. J. Digit. Imaging 2018, 31, 435–440. [Google Scholar] [CrossRef]






| Dataset. | Training Data | Validation Data | Test Data | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Label | B | M | SK | Total | B | M | SK | Total | B | M | SK | AT | Total | |
| ISBI 2017 | 1375 | 374 | 254 | 2000 | 78 | 30 | 42 | 150 | 393 | 197 | 90 | * | 600 | 2750 |
| PH2 | * | * | * | * | * | * | * | * | 80 | 40 | * | 80 | 200 | 200 |
| Total | 2000 | 150 | 800 | 2950 | ||||||||||
| Datasets | Detection Accuracy | IOU | Total Undetectable |
|---|---|---|---|
| PH2 | 94.40 | 90 | 6 images in 200 |
| ISBI 2017 | 96 | 86 | 22 images in 600 |
| Datasets | Acc | Sen | Spe | Jac | Dic |
|---|---|---|---|---|---|
| PH2 | 92.99 | 83.63 | 94.02 | 79.54 | 88.13 |
| ISBI 2017 | 93.39 | 90.82 | 92.68 | 74.81 | 84.26 |
| References | Acc | Sen | Spe | Jac | Dic |
|---|---|---|---|---|---|
| Yuan et al. (CDNN) [55] | 93.40 | 82.50 | 97.50 | 76.50 | 84.90 |
| Li et al. [76] | 93.20 | 82.00 | 97.80 | 76.20 | 84.70 |
| Bi et al. (ResNets) [77] | 93.40 | 80.20 | 98.50 | 76.00 | 84.40 |
| Lin et al. (U-Net) [54] | - | - | - | 62.00 | 77.00 |
| Al-Masni et. al. [56] | 94.03 | 85.40 | 96.69 | 77.11 | 87.08 |
| Proposed Method | 93.39 | 90.82 | 92.68 | 74.81 | 84.26 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ünver, H.M.; Ayan, E. Skin Lesion Segmentation in Dermoscopic Images with Combination of YOLO and GrabCut Algorithm. Diagnostics 2019, 9, 72. https://doi.org/10.3390/diagnostics9030072
Ünver HM, Ayan E. Skin Lesion Segmentation in Dermoscopic Images with Combination of YOLO and GrabCut Algorithm. Diagnostics. 2019; 9(3):72. https://doi.org/10.3390/diagnostics9030072
Chicago/Turabian StyleÜnver, Halil Murat, and Enes Ayan. 2019. "Skin Lesion Segmentation in Dermoscopic Images with Combination of YOLO and GrabCut Algorithm" Diagnostics 9, no. 3: 72. https://doi.org/10.3390/diagnostics9030072
APA StyleÜnver, H. M., & Ayan, E. (2019). Skin Lesion Segmentation in Dermoscopic Images with Combination of YOLO and GrabCut Algorithm. Diagnostics, 9(3), 72. https://doi.org/10.3390/diagnostics9030072