This section describes the dataset used in the study and provides information about the methodology applied. To ensure the reliability and generalizability of the results of the study, a strategic approach was followed.
2.1. Data and Data Preprocessing
The CirrMRI600+ dataset was used in the study, the first dataset specifically designed for liver cirrhosis research [
24]. This dataset contains both T1w and T2w MR images. The dataset contains 310 T1 and 318 T2 abdominal MR images from different stages of cirrhosis. These images were manually segmented. This dataset includes scans exhibiting a variety of morphological changes, reflecting the various complexities associated with real-world complications of cirrhosis, such as contour nodularity, hepatic segment atrophy or hypertrophy, ascites, varices, and splenomegaly. This complexity is crucial for training robust and generalizable DL models that perform effectively on unseen data. The dataset also includes a wide range of disease presentations. Thus, it is also compatible with real clinical scenarios. When creating the dataset, the researchers adopted the following strategy [
24]:
Three different scanners (Achieva, Philips (1.5T and 3T) (Amsterdam, Netherlands), and Symphony, Siemens 1.5 T (Erlangen, Germany) scanners with full anonymization protocols) were obtained to maintain heterogeneity in the MRI scans. To address the variability stemming from the use of different MRI scanners, all images were preprocessed using a standard pipeline: the intensity values were min–max normalized to a [0–1] range, image dimensions were resized to 224 × 224 pixels, and formats were unified using Nibabel. While scanner harmonization techniques were not explicitly applied, the robust cross-validation performance suggests that the ensemble architecture and transfer learning strategies provided sufficient generalization. Future studies could explore domain adaptation methods to further mitigate device-dependent biases;
To ensure image quality, participating radiologists selected and annotated “good enough” images. The rest were excluded.
This study included patients with liver cirrhosis and a non-cirrhotic control group (the healthy folder). It aimed to capture comprehensive samples of liver cirrhosis with different etiologies and stages, emphasizing variability and various complications. The CirrMRI600+ dataset includes metadata on patient age, gender, and scanner type. In terms of sex distribution, the healthy group consisted of 34 females and 21 males, while the cirrhosis group included 128 females and 190 males. The mean age of healthy individuals was 62.78 years (±14.93), and for cirrhotic patients, it was 60.10 years (±13.76). This demographic spread contributes to the diversity of the training data. In addition, MRI images were acquired from three different scanner types (Philips 1.5T, Philips 3T, and Siemens 1.5T), enhancing heterogeneity. The details are provided in
Table 1. This demographic spread contributes to the generalizability of the proposed model.
In this research, T2-weighted 2D images in the dataset were used. To use these images, a series of data preprocessing steps was performed. First, the following information can be given about the data downloaded from the database. T2-weighted images were analyzed in the study. The T2 folders in the healthy subjects folder in the database were considered. There are 55 images with the .nii extension in this folder. Files with .nii extension are files belonging to the Neuroimaging Informatics Technology Initiative (NIfTI) format used to store medical imaging data, and these files usually contain imaging sequences. These sequences are volumetric data obtained by medical imaging techniques such as MRI, collected with specific protocols. All 2D slices from each 3D volume were included in the training process to preserve spatial variability and reduce overfitting. No slice selection filtering (e.g., central slice prioritization) was applied. Spatial resampling or alignment was not required, as the dataset had undergone prior quality control by radiologists. The sequences of the images obtained from these healthy subjects were first extracted using the Nibabel library in the Python programming language. Since the study aims to perform a fully automatic detection and leveling process, all sequences are included in the training and testing processes. In this way, it was also aimed to prevent the overfitting problem. As a result of extracting the sequences of the .nii file, a total of 2143 images of healthy subjects were obtained.
Within the Cirrhosis T2-2D folder in the database, the images are divided into three different folders: train, test, and valid. Since the 10-fold cross-validation (CV) technique was applied in this study to ensure the reliability and generalizability of the results, these folders were brought together, the subjects were gathered, and a total of 318 patient folders were obtained. The MetaData file of the data in the database contains tags such as Patient ID, Age, Gender, and Radiological Evaluation. Using the Patient ID and Radiological Evaluation tags, the data were grouped into “1 = mild”, “2 = moderate”, and “3 = severe” folders. As a result of this process, subfolders were created for 131 patients in the mild folder, 114 patients in the moderate folder, and 73 patients in the severe folder. These subfolders contain different numbers of images for all patients. Therefore, the images were grouped in patientID_images_imageNo format. This resulted in 2838 images in the mild folder, 2391 images in the moderate folder, and 1473 images in the severe folder.
Table 2 presents the information on the data status as a result of these data preprocessing stages.
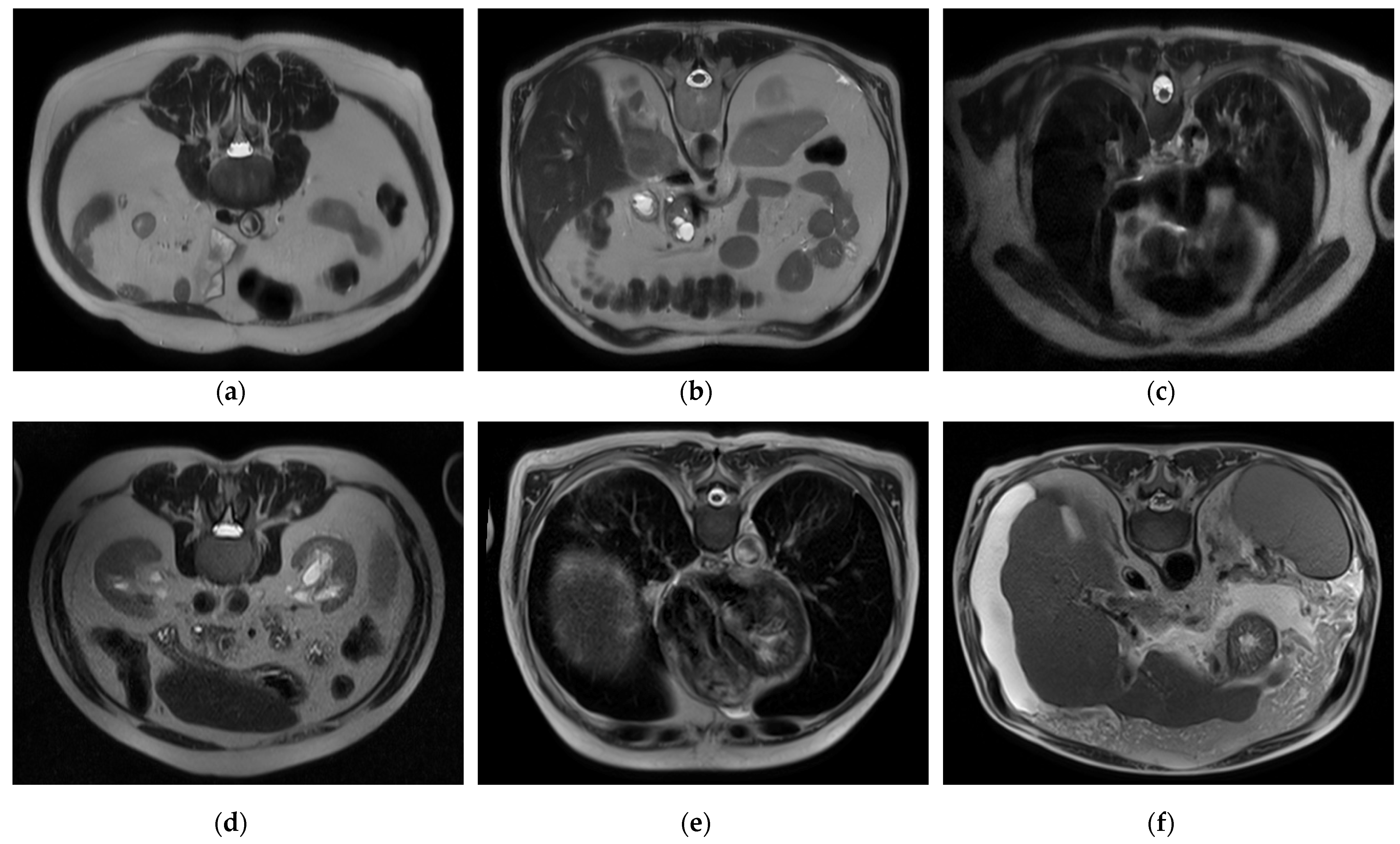
To handle a class imbalance in the binary classification task, a balanced subset was created by randomly selecting 2143 images from the cirrhosis group to match the healthy class. This undersampling strategy ensured balanced class distribution without introducing synthetic data. For the staging task involving mild, moderate, and severe categories, no artificial oversampling or data augmentation was applied. This was a deliberate methodological choice to restrict the model’s learning exclusively to real, clinically validated MRI slices. While this may affect minority class performance, it also avoids introducing synthetic variability that may compromise clinical interpretability. Future work may investigate augmentation techniques as a means of improving severe-class representation. Sample images for each class and level are presented in
Figure 1, respectively.
2.2. Hybrid CNN, Stacked Ensemble, and XAI-Based Multilayer AI Approach for Automatic Cirrhosis Diagnosis and Stage Classification
In the study, four different tasks were performed, and a fully automated, comprehensive diagnosis and diagnostic process for cirrhosis was created. These tasks are as follows:
Classification of healthy individuals and individuals with cirrhosis based on image data;
Automatic detection of the stage (e.g., mild, moderate, or severe) of individuals with cirrhosis;
Learning the distinctive features of cirrhosis stages with MTL and deep transfer learning (DTL) approaches and improving the classification performance specific to these stages;
Using XAI techniques to justify the diagnostic decisions made by the model and increase the reliability of the model.
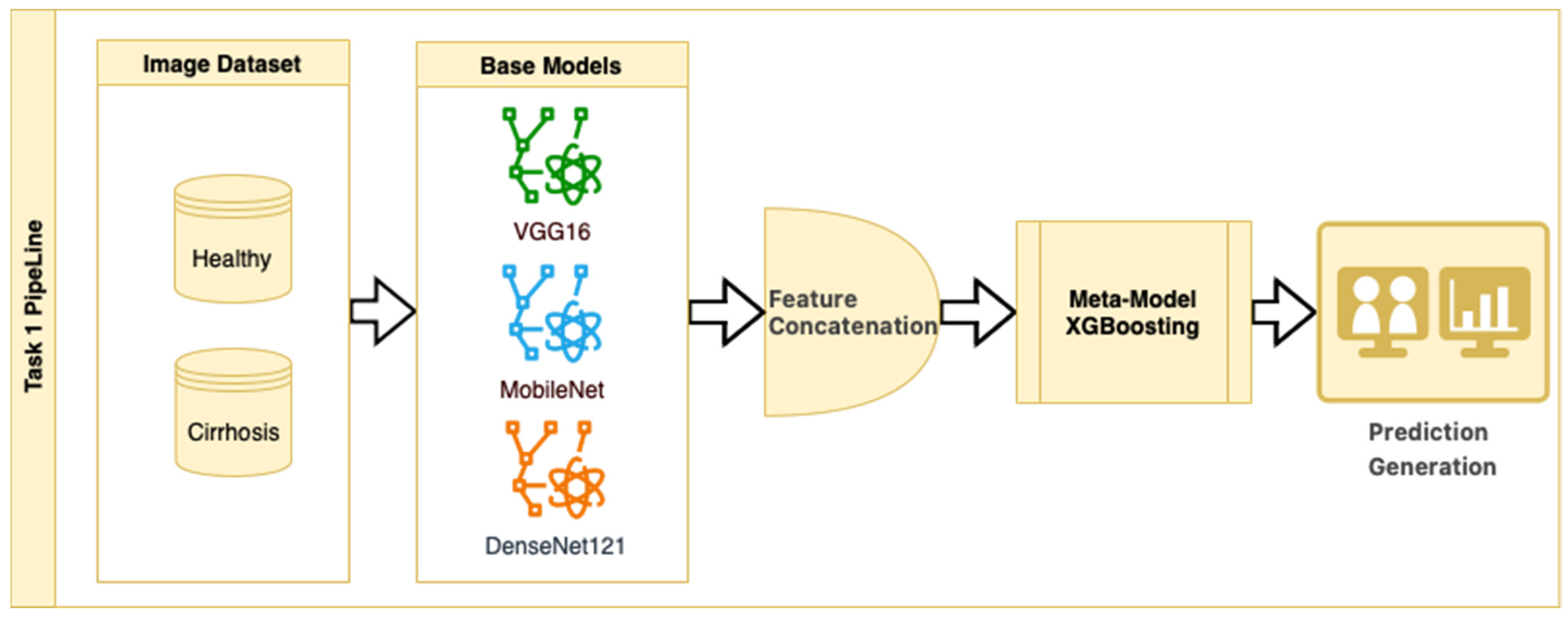
In Task 1, a hybrid approach for classifying the stages of liver cirrhosis in medical images is presented. The method consists of a multilayered pipeline that includes feature extraction with DTL (VGG16, MobileNet, and DenseNet121), combining these features to form a stacked ensemble structure and using XGBoost as a meta-level classifier. Stacked ensemble learning is an ML approach that aims to create a more powerful learning system by combining the insights of multiple models [
25]. In this structure, there are usually two layers (a base layer and a meta-layer). The base layer consists of models that operate on different features or representations. In this study, we use DTL models to obtain pre-trained robust representations. The meta-layer is an algorithm that combines the predictions of the base layer models to produce the final predictions. Here, the XGBoost ensemble learning algorithm was chosen as the meta-layer. XGBoost is characterized by its capacity to control overlearning while improving prediction accuracy [
26]. This structure combines the powerful representation learning capabilities of DTL and the efficient generalization power of XGBoost to provide high performance on both complex datasets and limited data environments. The stacked ensemble procedure is as follows: “Base models (CNNs) extract high-level features from images, the extracted features are combined to create a meta-feature vector, and the meta-model (XGBoost) performs the final classification on this vector”.
In the stacked ensemble structure, three pre-trained CNN architectures (VGG16, MobileNet, and DenseNet121) were used as feature extractors, each initialized with ImageNet weights (include_top = False). Feature maps were obtained from the last convolutional layers of each model and then flattened into 1D vectors. These flattened feature vectors were concatenated along the feature dimension to form a single composite feature vector for each image. No additional feature normalization or decorrelation techniques (e.g., z-score scaling or PCA) were applied, as the pre-trained models include internal batch normalization, and the chosen XGBoost meta-classifier is robust to scale differences and performs its own feature selection and regularization. The meta-model was trained on these concatenated vectors to perform the final classification. While the relative contribution of each CNN model to the ensemble output was not explicitly analyzed in this study, this is a promising area for future ablation and explainability work.
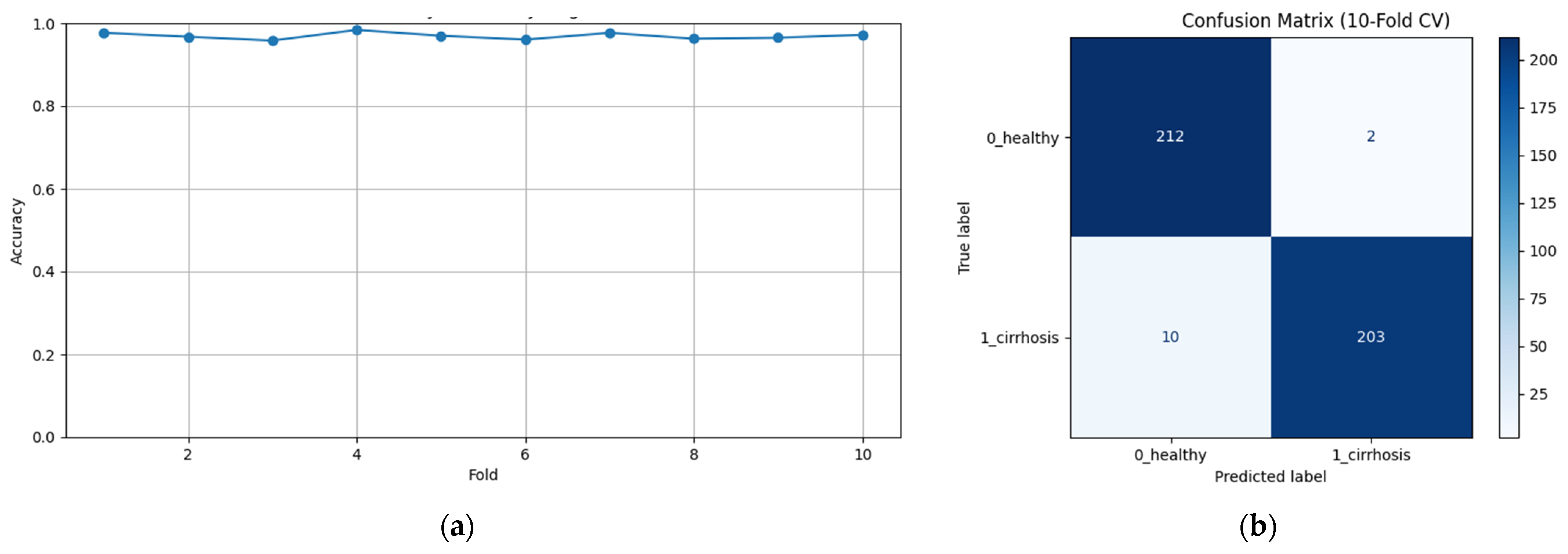
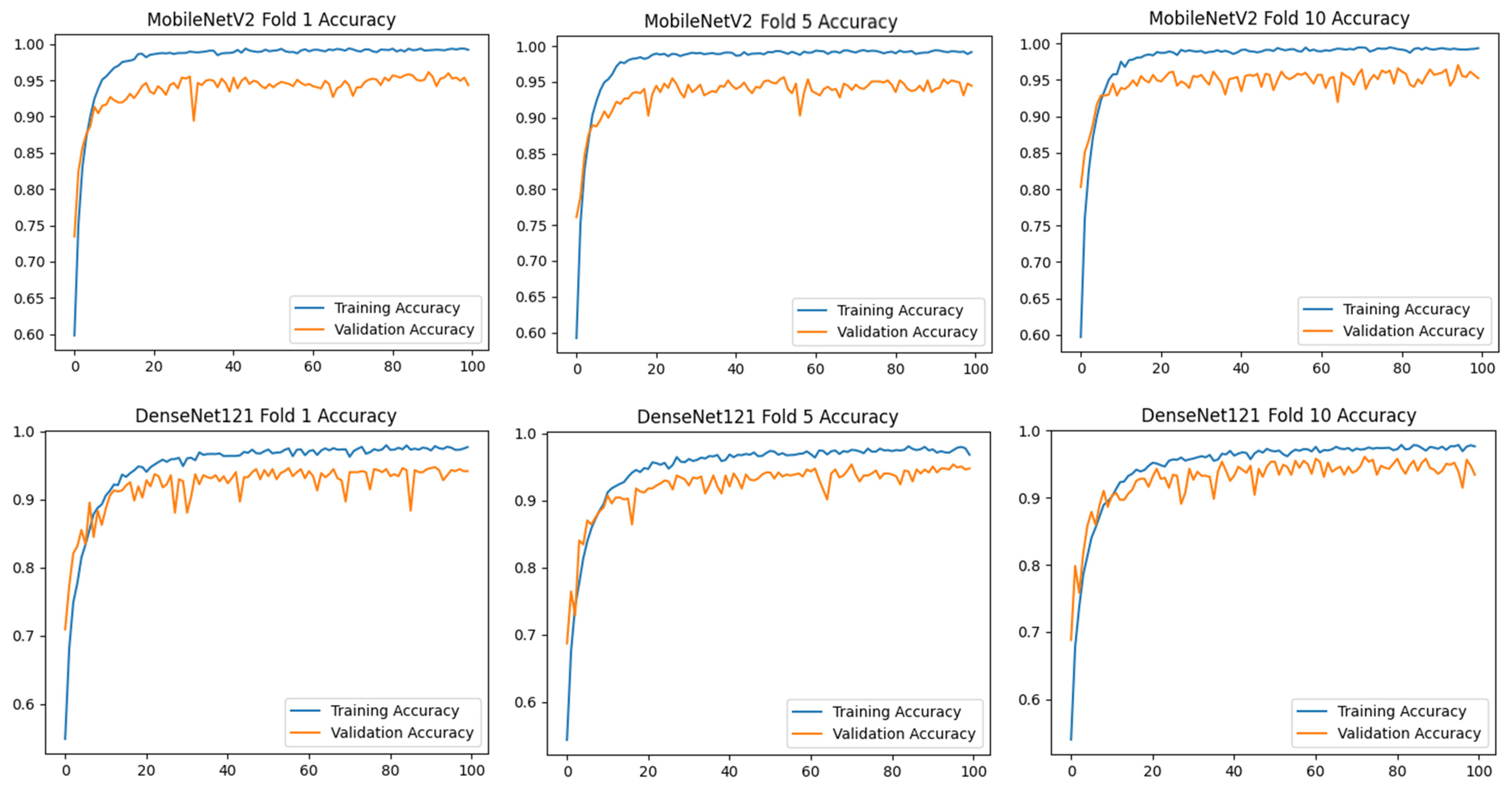
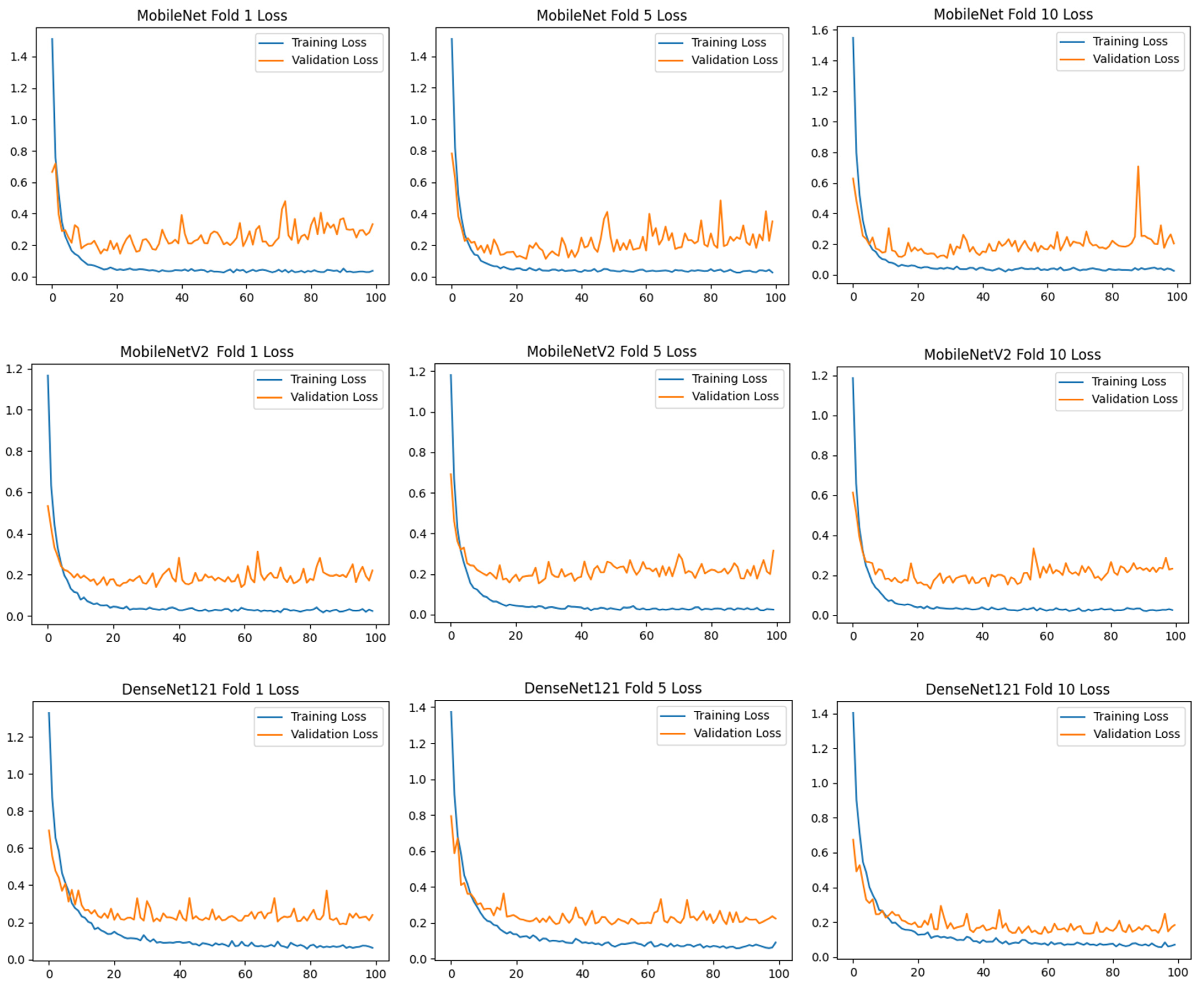
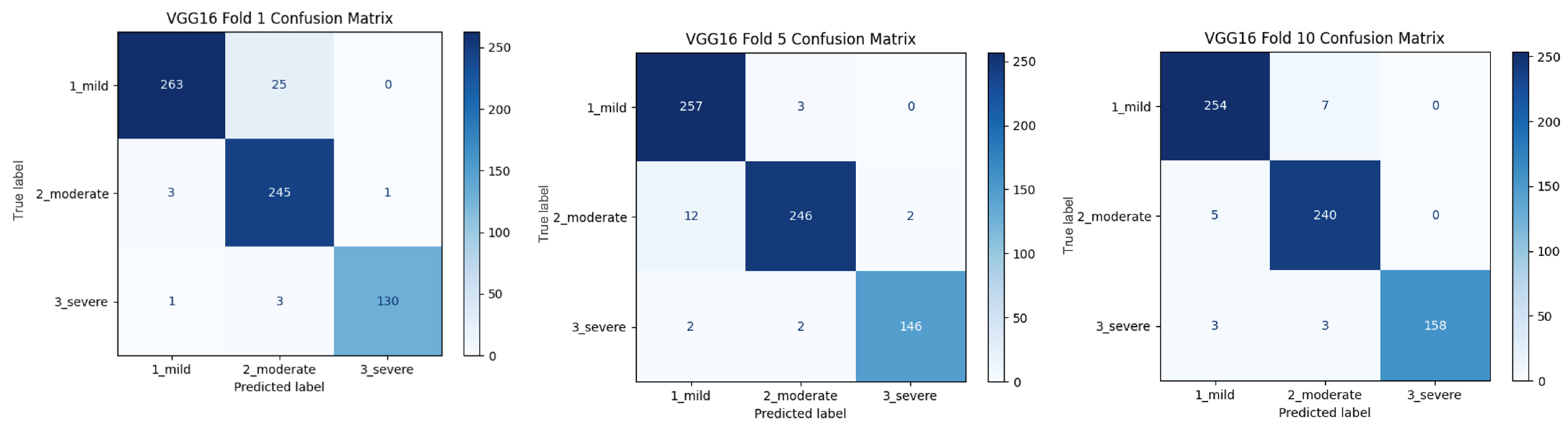
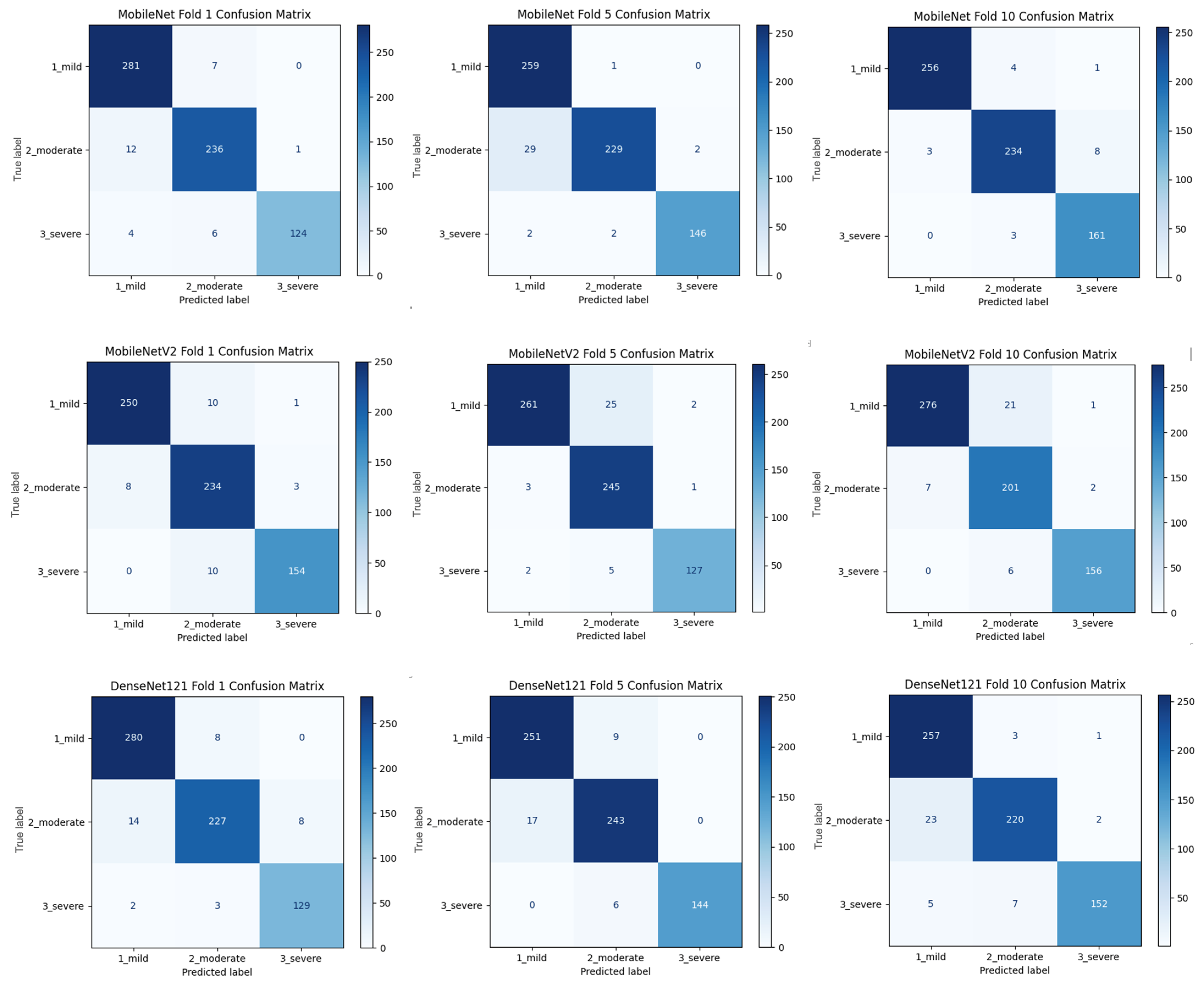
To assess the generalizability of the model, a 10-fold CV was applied, and the statistical reliability of the results (mean ± standard deviation) was reported. To prevent overtraining and enhance model generalization, several strategies were implemented during the training process. First, an early stopping mechanism was employed based on validation loss monitoring, with a patience parameter of 10 epochs. This ensured that training was halted if the model ceased to improve on the validation set, thereby avoiding unnecessary weight updates and overfitting. Second, a 10-fold CV approach was utilized, whereby the model was trained on 90% of the data and evaluated on the remaining 10% across folds. Although no external held-out test set was used, the validation subset in each fold functioned as an unseen test partition for performance estimation. This methodology enabled robust evaluation of model generalization across diverse data splits and minimized the risk of performance inflation due to overfitting.
This strategy is an example of a heterogeneous stacked ensemble that aims to optimize performance by exploiting the synergy of transfer learning and stacking, especially when working with limited medical data. The pipeline for Task 1 is presented in
Figure 2.
Figure 2 shows that pre-trained models in ImageNet, such as VGG16, MobileNet, and DenseNet121, are used only as feature extractors by removing the last layers. These models were used to extract high-level features (edges, textures, shapes, etc.) from images. Within the scope of multi-model integration (ensemble learning), features extracted from three different CNN models are combined to form a single feature vector. Since each model captures different features in this method, the combination provides a stronger representation. A gradient boosting algorithm was trained on the extracted features using the XGBoost Classifier as the meta-model. The XGBoost algorithm is effective in learning non-linear relationships on the features extracted by CNNs and is preferred because it is resistant to overfitting. In addition, the class distribution was preserved while dividing the dataset into 10 parts with a 10-fold CV. Statistical reliability was ensured by calculating the Mean and Standard Deviation over the 10-fold results. Accuracy, precision, recall, and F1-score were calculated for each class, and a confusion matrix and accuracy graph were obtained. To maintain the balance between classes while performing these operations, since the number of images in the healthy class is 2143, 2143 images from the cirrhosis class were also used by the random selection method. It should be noted that random sampling was only applied in the binary classification task to ensure a class balance between healthy and cirrhosis images. For the cirrhosis staging task, no data were discarded: all available images from the mild, moderate, and severe stages were included. Although class imbalance remains a challenge, especially for the severe class, this strategy ensured maximum use of the dataset’s diagnostic variety. Future studies may explore complementary balancing techniques such as class weighting or augmentation.
Within the scope of Task 2, a comprehensive stage detection was performed using the TensorFlow Keras Applications module with a transfer learning technique. In this module, various pre-trained model architectures are available, including ConvNeXt, DenseNet, EfficientNet, EfficientNet, EfficientNet_V2, Inception_ResNet_V2, Inception_V3, MobileNet, MobileNet_V2, MobileNet_V3, NASNet, ResNet, ResNet_V2, VGG16, and Xception. For each module, there are different pre-trained models with different numbers of layers (functions/models). In this study, the base model of each module was transferred and used. Thus, successful models provide a basis for future analysis where more complex models can be explored with additional layers from the same successful module. The models used in the study are listed in
Table 3, which provides an overview of the different pre-trained architectures used in the transfer learning process.
As shown in
Table 3, 14 CNN architectures were compared using the 10-fold CV technique. The models were initialized with ImageNet weights, and transfer learning was applied by freezing the convolution layers. All models had the same classifier scheme (
). A fixed input resolution (
) and batch size (
) were used as standard evaluation protocols. Adam optimization (LR =
) and categorical cross-entropy loss were applied. Accuracy and loss values were calculated, and confusion matrices of the models were created. Training curves and metrics were recorded for 100 epochs for each fold.
An MTL process was implemented for Task 3. MTL is an ML approach that allows multiple related tasks to be learned simultaneously. This method aims to increase the generalization capability of the model by sharing common features between different tasks [
27]. In contrast to traditional single-task learning approaches, MTL increases generalization ability by encouraging knowledge sharing across related tasks. This approach is particularly advantageous for tasks that require learning common representations. Especially in DL models, MTL produces multiple outputs using the same underlying network structure, thus eliminating the need to train separate models for each task. This approach can improve performance even when source data are limited [
28]. One of the most important advantages of MTL is the increase in learning efficiency through knowledge transfer between tasks. For example, by performing object recognition and segmentation tasks simultaneously, an image classification model can achieve better results in both tasks [
29]. Another advantage of MTL is that it improves data efficiency. Sharing knowledge from related tasks can improve the performance of the model, especially when labeled data are limited. It helps discover relationships between tasks by learning a common representation and ensures that the transferred knowledge contributes to the learning process of each task. Furthermore, MTL reduces the risk of overfitting, making the model more robust, more balanced, and generalizable. This offers an important advantage, especially when working on small datasets [
30]. Due to these properties, MTL is widely used in various fields, such as image processing and biomedical data analysis.
When combined with DTL, MTL becomes even more powerful. While transfer learning transfers the knowledge of a pre-trained model to a new task, MTL provides a more efficient learning process by sharing this knowledge across multiple tasks [
31]. In particular, models trained on large-scale datasets (e.g., VGG16, MobileNet, etc.), when combined with MTL, make it possible to achieve high performance with fewer data [
32]. This combination saves time and allows the model to produce more consistent results across multiple tasks.
Within the scope of MTL, this study first performed triple classification on the same data and then applied binary classifications for each cirrhosis stage. The VGG16 model was preferred for this task, and the rationale is presented in the Results section. The implementation procedure was as follows:
Multi-classification:
Binary classification:
Binary classification:
Binary classification:
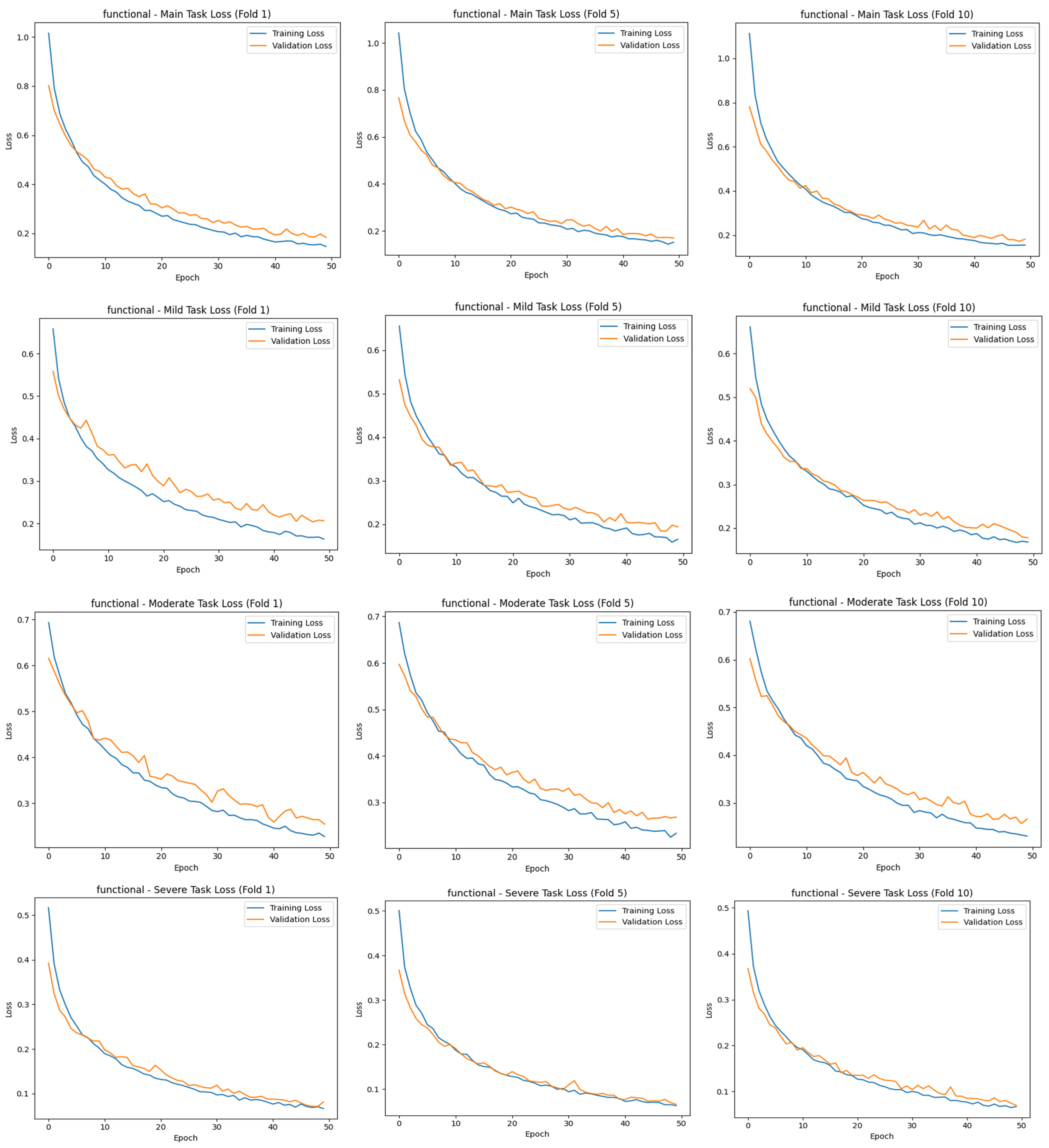
The “main output” here is the multiclass classification task, where the model mainly tries to distinguish between three stages (mild, moderate, and severe) simultaneously. This contributes to the identification of the stages of cirrhosis, referred to as “task 2”. The total loss value of this study was calculated using Equation (1):
The main task, the classification of cirrhosis stages, is a multiclass classification problem with mutually exclusive classes, where each image belongs to only one class (e.g., mild, moderate, or severe). In such tasks, the model is expected to probabilistically predict the correct class for each instance. In this context, categorical cross-entropy is used as the appropriate cost (loss) function for the main task. This function measures the deviation between the predicted class distribution and the true distribution (usually one-hot coded) and evaluates the distance between probabilities with a logarithmic measure [
33]. The categorical cross entropy formula is defined, as in Equation (2):
Here,
is the number of instances,
is the number of classes,
is the one-hot representation of the true labels, and
is the probability predicted by the model. The Softmax activation function used in the output layer of the model guarantees that the sum of these probabilities is
and reflects the confidence level of each class. This method allows the model not only to predict the correct class but also to optimize the confidence in its prediction. Thus, the model can perform more stable and meaningful classifications in multiclass tasks [
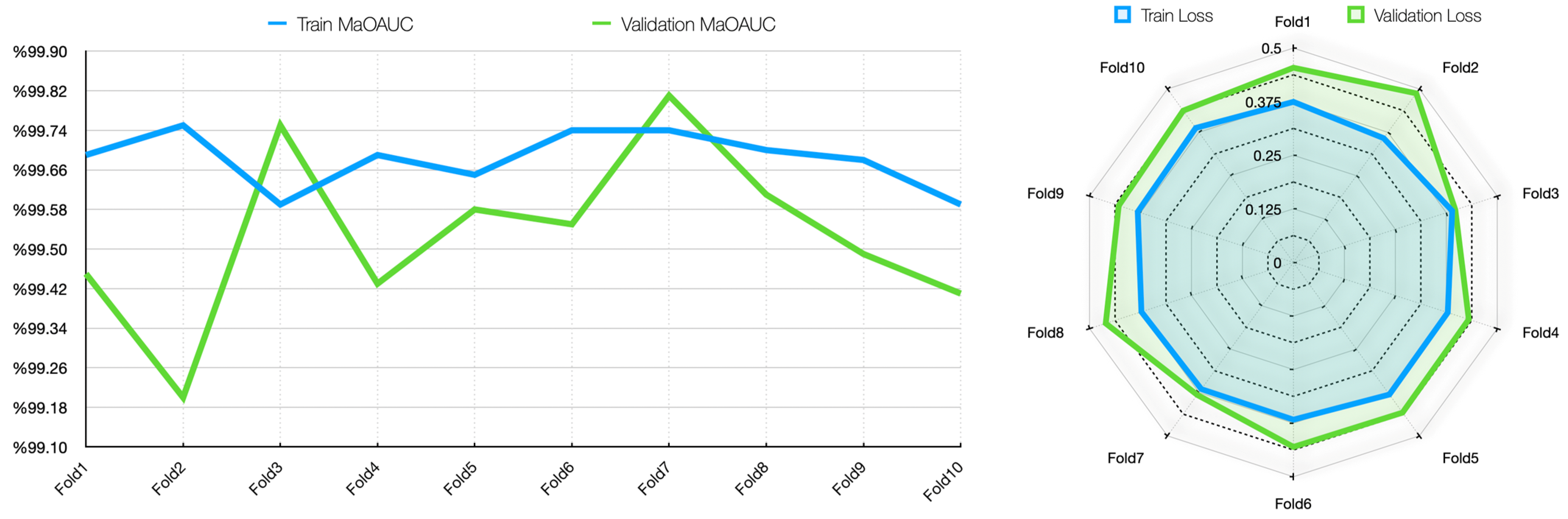
34]. The results of the study were trained for 50 epochs using the 10-fold CV method, evaluated with different metrics, and contributed to the correct detection of each stage of cirrhosis.
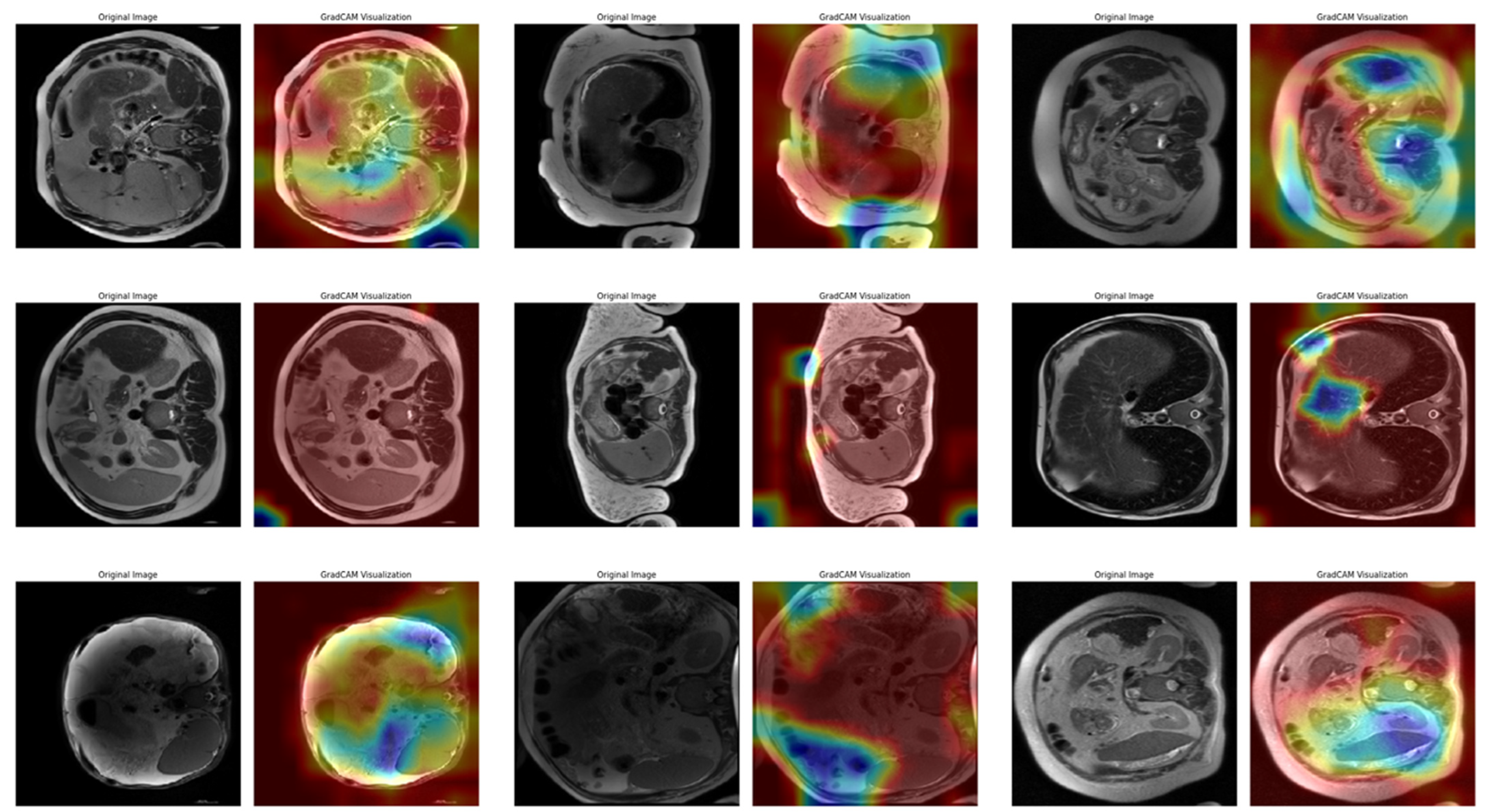
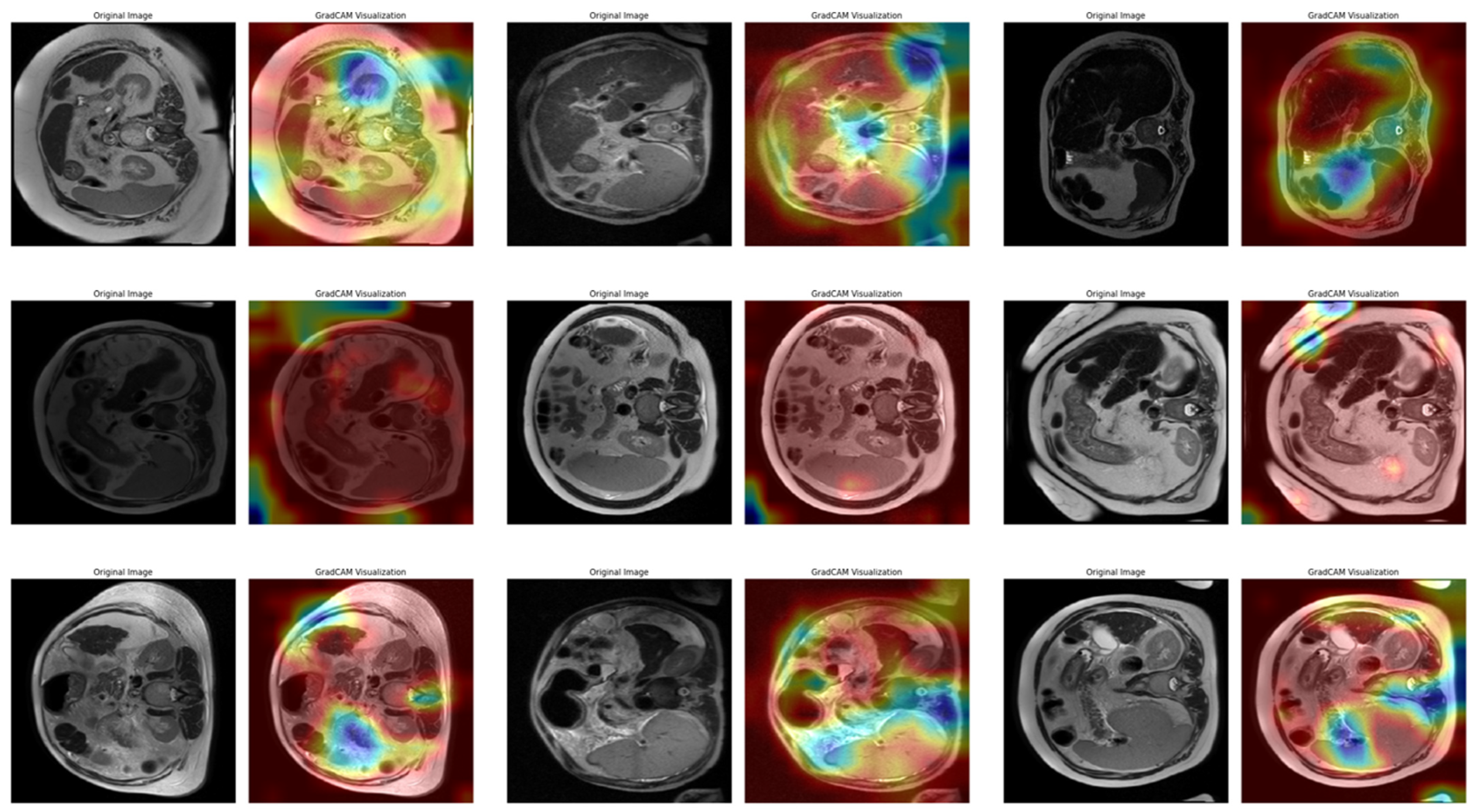
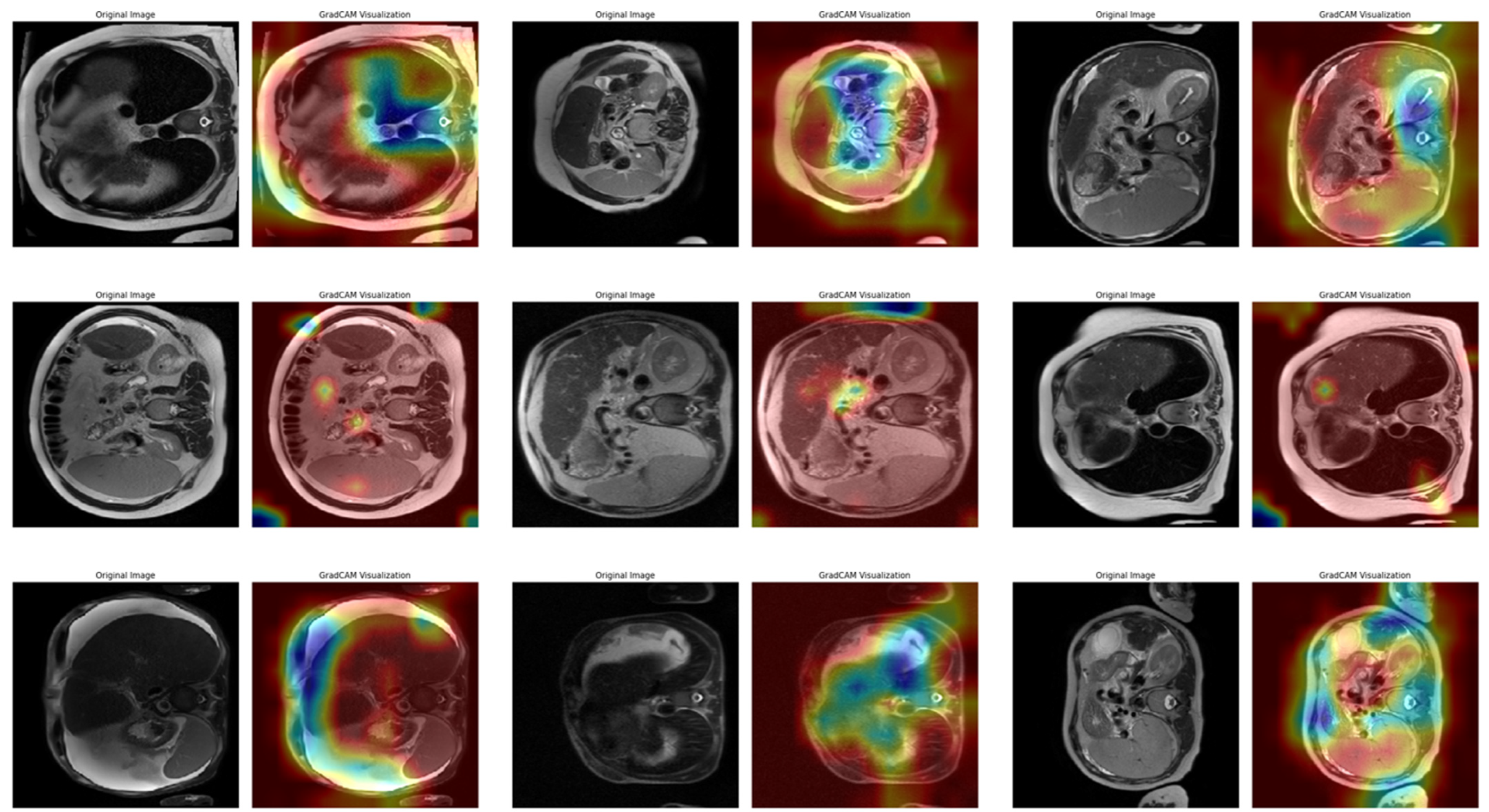
In Task 4 (the last task), the study applied XAI. The Grad-CAM method, developed to better understand the decision mechanisms of DL models, allows for visualizing which image regions the model focuses on when making a particular prediction [
35]. This method is particularly used to explain the decisions of DL-based models, such as CNNs, and understand which features the model considers in the classification process. Grad-CAM uses the activation maps in the final convolution layer of the model to identify the regions that contribute the most to the prediction of the class of interest. First, the derivatives of the model output for a given class are taken to calculate the importance of the feature maps in the final convolution layer. Then, using these importance ratings, the feature maps are weighted, and a class-specific heatmap is obtained. Finally, this heatmap is overlaid on the original image to visualize which regions the model considers when making predictions.
In this study, the Grad-CAM method was used to describe the predictions of the VGG16-based model. After each k-fold CV step of the model’s training process, heat maps obtained by applying Grad-CAM on a sample from the validation set were recorded. Thus, it was analyzed which image regions the model makes decisions by focusing on. The modeling process was based on the VGG16 architecture previously trained on ImageNet, with the last convolutional layers fixed and only the upper layers reconstructed. The final layers of the customized model consist of a dense layer with smoothing, dropout (0.5), and Softmax activation. The model is compiled with a categorical cross-entropy loss function and the Adam optimization algorithm (LR = ) for a three-class problem. The dataset consists of labeled liver images containing three stages of cirrhosis. The images were resized to pixels to fit the model and normalized using ImageDataGenerator (pixel value scaling: ). To assess the generalizability of the model, a 10-fold CV was applied in this task, too. In each fold, smart data splitting (flow_from_dataframe) was used for training and validation, and the training process was conducted by monitoring accuracy and loss metrics.
The Grad-CAM heatmap (
) for class
was obtained by weighting each feature map (
) in the relevant layer by the importance coefficients (
), representing its contribution to the class score and emphasizing positive contributions (Equation (3)) [
36]:
where
is calculated as the spatial average of the gradients of the score of class
concerning the
-th feature map (Equation (4)) [
36]:
Thanks to this formulation, it is possible to visualize which spatial regions are more effective when the model predicts the class through their positive gradient contributions. The ReLU function ensures that only regions that contribute positively to the class score are considered. In this way, Grad-CAM allows inferences to be made about the model’s internal learning processes and validates the model’s decisions, especially in critical areas such as medical image analysis.
2.3. Evaluation Metrics
The correct selection and application of evaluation metrics used in medical research is crucial to objectively assess the performance of models. In this context, not only basic metrics such as accuracy rate but also measures based on loss function, precision, recall/sensitivity, and F1-score were considered. In addition, to comprehensively analyze the classification performance of the model, area under the ROC curve (AUC) values were calculated, and performance comparisons were made within the framework of ensemble learning methods. The evaluation process is based on the true positive (TP), true negative (TN), false positive (FP), and false negative (FN) values for each class.
The accuracy metric used to measure the overall success of the model is calculated as the ratio of correctly predicted instances to the total number of instances, as in Equation (5). A high accuracy rate indicates that the model is generally successful in its predictions for all classes:
Precision refers to the probability that samples predicted as positive are positive and is calculated as shown in Equation (6). It is a critical metric for avoiding false positive predictions and is used to assess the selectivity of the model, especially in imbalanced datasets. A high precision value indicates that the model minimizes false positive errors and is more reliable in its positive classifications:
Recall (sensitivity) indicates the model’s ability to correctly recognize instances belonging to the positive class. It measures how many true positives in the dataset are correctly predicted. Recall is important in preventing false negatives and is used to determine the effectiveness of the model, especially in critical applications such as medical diagnostics. A high recall value indicates that the model is successful in identifying positive examples. This metric is also called the True Positive Rate (TPR) and is calculated, as in Equation (7):
A metric that balances precision and recall is called the F1-score, which is calculated as the harmonic mean of these two metrics (Equation (8)). It is used to more reliably assess the performance of the model in imbalanced datasets. A high F1-score indicates that the model correctly identifies all instances belonging to the positive class while minimizing false positives. Therefore, it is often a preferred metric to measure the overall classification success of the model:
The AUC—ROC Curve is a method used to evaluate the classification success of the model at different thresholds. The ROC curve shows the relationship between the TPR and False Positive Rate (FPR) of the model, while the AUC value represents the discriminative power of the model. As the AUC value approaches
, the classification success of the model increases, and as it approaches
, it is understood that the model makes random predictions. AUC is the calculation of the area under the ROC curve (Equations (9) and (10)):
Evaluating these metrics together allows the strengths and weaknesses of the model to be identified and provides a more comprehensive analysis of model performance.