Enhanced Detection of Bacterial Ocular Pathogens: A Comparative Study of Broad-Range Real-Time PCR and Conventional Culture Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Specimens
2.2. Culture
2.3. Nucleic Acid Extraction and PCR
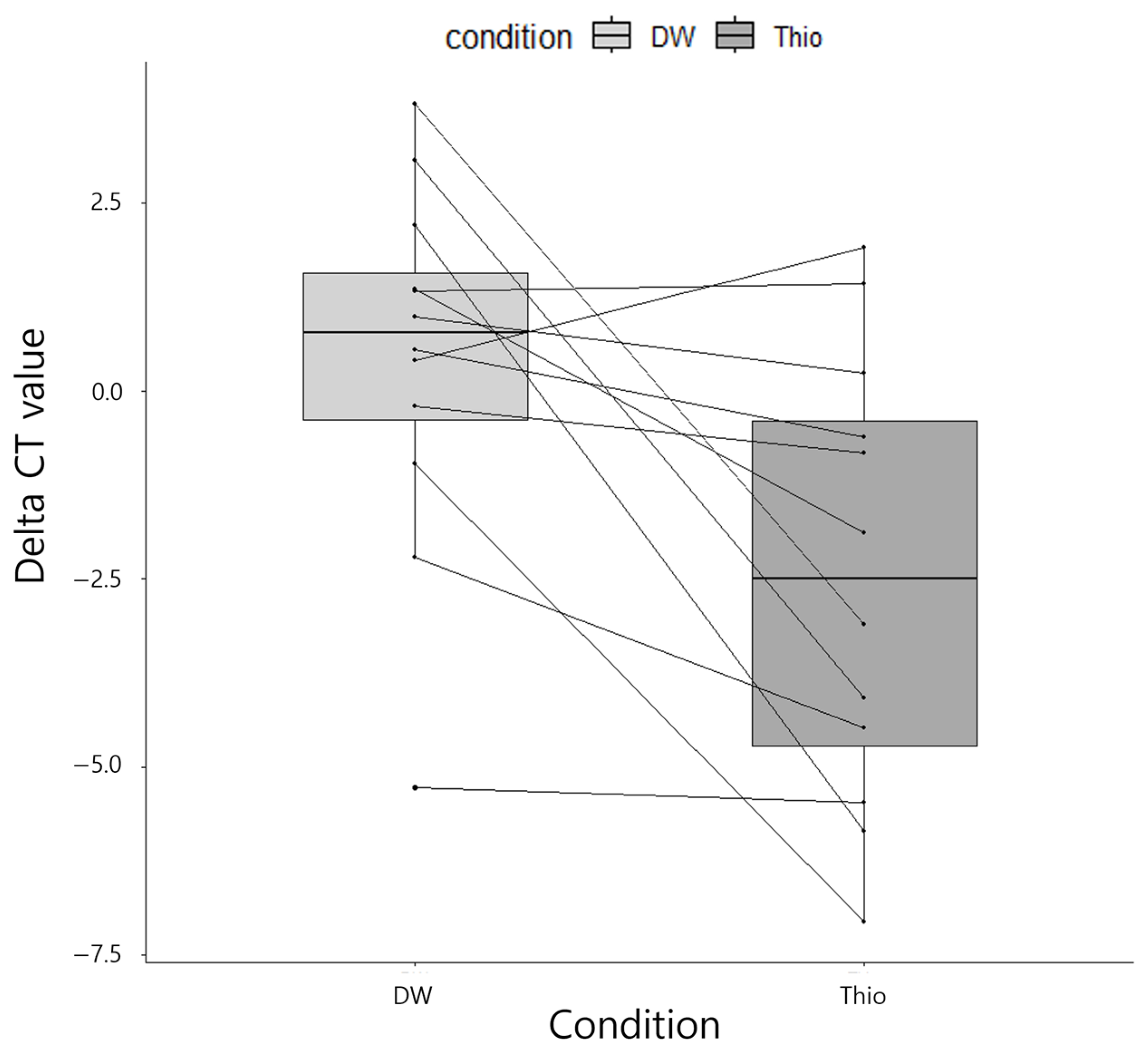
2.4. Validation of an Appropriate Solution for Ocular Specimens in Nucleic Acid Extraction for Identifying Bacteria
2.5. Analysis of the Causes of Discrepancies Between Bacterial Culture Tests and Bacterial Real-Time PCR Results
2.6. 16S rRNA Meta-Taxonomic Analysis
3. Results
3.1. Validation of the Selection of an Appropriate Solution for Ocular Specimens in Nucleic Acid Extraction
3.2. Setting the ΔCT Cutoff Value for Bacterial Real-Time PCR Results Interpretation
3.3. Bacterial Conventional PCR Versus Real-Time PCR
3.4. Analysis of Causes of False-Negative Bacterial Real-Time PCR Results
3.5. 16S rRNA Meta-Taxonomic Analysis
3.6. Broad-Range Fungal PCR and Bacterial Real-Time PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clare, G.; Kempen, J.H.; Pavésio, C. Infectious Eye Disease in the 21st Century—An Overview. Eye 2024, 38, 2014–2027. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, L.J.; Mah, F.S. Clinical Use of Gatifloxacin Ophthalmic Solution for Treatment of Bacterial Conjunctivitis. Clin. Ophthalmol. 2011, 5, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Alkatan, H.M.; Al-Essa, R.S. Challenges in the Diagnosis of Microbial Keratitis: A Detailed Review with Update and General Guidelines. Saudi J. Ophthalmol. 2019, 33, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.R.; Flynn, H.W.; Miller, D.; Forster, R.K.; Alfonso, E.C. Infectious Keratitis Progressing to Endophthalmitis a 15-Year Study of Microbiology, Associated Factors, and Clinical Outcomes. Ophthalmology 2012, 119, 2443–2449. [Google Scholar] [CrossRef]
- Cao, J.; Yang, Y.; Yang, W.; Wu, R.; Xiao, X.; Yuan, J.; Xing, Y.; Tan, X. Prevalence of Infectious Keratitis in Central China. BMC Ophthalmol. 2014, 14, 43. [Google Scholar] [CrossRef]
- Teweldemedhin, M.; Gebreyesus, H.; Atsbaha, A.H.; Asgedom, S.W.; Saravanan, M. Bacterial Profile of Ocular Infections: A Systematic Review. BMC Ophthalmol. 2017, 17, 212. [Google Scholar] [CrossRef]
- Dalmon, C.; Porco, T.C.; Lietman, T.M.; Prajna, N.V.; Prajna, L.; Das, M.R.; Kumar, J.A.; Mascarenhas, J.; Margolis, T.P.; Whitcher, J.P.; et al. The Clinical Differentiation of Bacterial and Fungal Keratitis: A Photographic Survey. Investig. Opthalmol. Vis. Sci. 2012, 53, 1787. [Google Scholar] [CrossRef]
- Özenci, V.; Rossolini, G.M. Rapid Microbial Identification and Antimicrobial Susceptibility Testing to Drive Better Patient Care: An Evolving Scenario. J. Antimicrob. Chemother. 2019, 74, i2–i5. [Google Scholar] [CrossRef]
- Somerville, T.F.; Corless, C.E.; Sueke, H.; Neal, T.; Kaye, S.B. 16S Ribosomal RNA PCR Versus Conventional Diagnostic Culture in the Investigation of Suspected Bacterial Keratitis. Transl. Vis. Sci. Technol. 2020, 9, 2. [Google Scholar] [CrossRef]
- Kang, M.C.; Lim, D.H.; Huh, H.J.; Yoo, I.Y.; Chung, T.-Y. Diagnostic Utility of Polymerase Chain Reaction for Acanthamoeba in Contact Lens-Related Keratitis with Epithelial Defects. J. Korean Ophthalmol. Soc. 2019, 60, 1312–1317. [Google Scholar] [CrossRef]
- Ogawa, M.; Sugita, S.; Shimizu, N.; Watanabe, K.; Nakagawa, I.; Mochizuki, M. Broad-Range Real-Time PCR Assay for Detection of Bacterial DNA in Ocular Samples from Infectious Endophthalmitis. Jpn. J. Ophthalmol. 2012, 56, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Sugita, S.; Watanabe, K.; Shimizu, N.; Mochizuki, M. Novel Diagnosis of Fungal Endophthalmitis by Broad-Range Real-Time PCR Detection of Fungal 28S Ribosomal DNA. Graefe’s Arch. Clin. Exp. Ophthalmol. 2012, 250, 1877–1883. [Google Scholar] [CrossRef]
- Eleinen, K.G.A.; Mohalhal, A.A.; Elmekawy, H.E.; Abdulbaki, A.M.; Sherif, A.M.; El-Sherif, R.H.; Rahman, E.M.A. Polymerase Chain Reaction-Guided Diagnosis of Infective Keratitis—A Hospital Based Study. Curr. Eye Res. 2012, 37, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Sagerfors, S.; Edslev, S.; Lindblad, B.E.; Lilje, B.; Stegger, M.; Söderquist, B. In the Eye of the Ophthalmologist: The Corneal Microbiome in Microbial Keratitis. Graefe’s Arch. Clin. Exp. Ophthalmol. 2024, 262, 1579–1589. [Google Scholar] [CrossRef]
- Gallon, P.; Parekh, M.; Ferrari, S.; Fasolo, A.; Ponzin, D.; Borroni, D. Metagenomics in Ophthalmology: Hypothesis or Real Prospective? Biotechnol. Rep. 2019, 23, e00355. [Google Scholar] [CrossRef]
- Parekh, M.; Romano, V.; Franch, A.; Leon, P.; Birattari, F.; Borroni, D.; Kaye, S.B.; Ponzin, D.; Ahmad, S.; Ferrari, S. Shotgun Sequencing to Determine Corneal Infection. Am. J. Ophthalmol. Case Rep. 2020, 19, 100737. [Google Scholar] [CrossRef]
- Borroni, D.; Bonzano, C.; Sánchez-González, J.-M.; Rachwani-Anil, R.; Zamorano-Martín, F.; Pereza-Nieves, J.; Traverso, C.E.; Lorente, M.G.; Rodríguez-Calvo-de-Mora, M.; Esposito, A.; et al. Shotgun Metagenomic Sequencing in Culture Negative Microbial Keratitis. Eur. J. Ophthalmol. 2022, 33, 1589–1595. [Google Scholar] [CrossRef]
- Liu, C.M.; Kachur, S.; Dwan, M.G.; Abraham, A.G.; Aziz, M.; Hsueh, P.-R.; Huang, Y.-T.; Busch, J.D.; Lamit, L.J.; Gehring, C.A.; et al. FungiQuant: A Broad-Coverage Fungal Quantitative Real-Time PCR Assay. BMC Microbiol. 2012, 12, 255. [Google Scholar] [CrossRef]
- Zucol, F.; Ammann, R.A.; Berger, C.; Aebi, C.; Altwegg, M.; Niggli, F.K.; Nadal, D. Real-Time Quantitative Broad-Range PCR Assay for Detection of the 16S RRNA Gene Followed by Sequencing for Species Identification. J. Clin. Microbiol. 2006, 44, 2750–2759. [Google Scholar] [CrossRef]
- Goldenberger, D.; Künzli, A.; Vogt, P.; Zbinden, R.; Altwegg, M. Molecular Diagnosis of Bacterial Endocarditis by Broad-Range PCR Amplification and Direct Sequencing. J. Clin. Microbiol. 1997, 35, 2733–2739. [Google Scholar] [CrossRef]
- Ung, L.; Bispo, P.J.M.; Shanbhag, S.S.; Gilmore, M.S.; Chodosh, J. The Persistent Dilemma of Microbial Keratitis: Global Burden, Diagnosis, and Antimicrobial Resistance. Surv. Ophthalmol. 2019, 64, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.D.P.; Lira, R.P.C.; Arieta, C.E.L.; de Castro, R.S.; Bonon, S.H.A. The Prevalence of Adenoviral Conjunctivitis at the Clinical Hospital of the State University of Campinas, Brazil. Clinics 2015, 70, 748–750. [Google Scholar] [CrossRef] [PubMed]
- Lam, D.S.C.; Houang, E.; Fan, D.S.P.; Lyon, D.; Seal, D.; Wong, E.; on behalf of the Hong Kong Microbial Keratitis Study Group. Incidence and Risk Factors for Microbial Keratitis in Hong Kong: Comparison with Europe and North America. Eye 2002, 16, 608–618. [Google Scholar] [CrossRef]
- Cho, E.Y.; Lee, S.B. Gram-Negative Bacterial Keratitis: A 15-Year Review of Clinical Aspects. J. Korean Ophthalmol. Soc. 2015, 56, 1479–1488. [Google Scholar] [CrossRef][Green Version]
- Ahmadikia, K.; Gharehbolagh, S.A.; Fallah, B.; Eshkaleti, M.N.; Malekifar, P.; Rahsepar, S.; Getso, M.I.; Sharma, S.; Mahmoudi, S. Distribution, Prevalence, and Causative Agents of Fungal Keratitis: A Systematic Review and Meta-Analysis (1990 to 2020). Front. Cell. Infect. Microbiol. 2021, 11, 698780. [Google Scholar] [CrossRef]
- Mohd-Tahir, F.; Norhayati, A.; Siti-Raihan, I.; Ibrahim, M. A 5-Year Retrospective Review of Fungal Keratitis at Hospital Universiti Sains Malaysia. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 851563. [Google Scholar] [CrossRef]
- Fernandes, M.; Vira, D.; Dey, M.; Tanzin, T.; Kumar, N.; Sharma, S. Comparison Between Polymicrobial and Fungal Keratitis: Clinical Features, Risk Factors, and Outcome. Am. J. Ophthalmol. 2015, 160, 873–881.e2. [Google Scholar] [CrossRef]
- Shimizu, D.; Miyazaki, D.; Ehara, F.; Shimizu, Y.; Uotani, R.; Inata, K.; Sasaki, S.; Inoue, Y. Effectiveness of 16S Ribosomal DNA Real-Time PCR and Sequencing for Diagnosing Bacterial Keratitis. Graefe’s Arch. Clin. Exp. Ophthalmol. 2020, 258, 157–166. [Google Scholar] [CrossRef]



| Patients | Variable | Number | % | Specimens | Variable | Number | % |
|---|---|---|---|---|---|---|---|
| Total | 111 | 100 | Total | 160 | 100 | ||
| Sex | M | 64 | 57.66 | Type | Cornea | 78 | 48.75 |
| F | 47 | 42.34 | Anterior chamber | 31 | 19.38 | ||
| DM | Yes | 32 | 28.83 | Vitreous humor | 23 | 14.38 | |
| No | 79 | 71.17 | Contact lens | 14 | 8.75 | ||
| Clinical diagnosis | Keratitis | 66 | 59.46 | Discharge | 4 | 2.50 | |
| Endophthalmitis | 32 | 28.83 | Balanced salt solution | 3 | 1.88 | ||
| Uveitis | 4 | 3.60 | Therapeutic contact lens | 2 | 1.25 | ||
| Orbital/eyelid mass | 2 | 1.80 | Intraocular lens | 2 | 1.25 | ||
| Orbital cellulitis | 2 | 1.80 | Contact lens solution | 1 | 0.63 | ||
| Retinal vasculitis | 1 | 0.90 | Foreign body | 1 | 0.63 | ||
| Scleritis | 1 | 0.90 | Conjunctiva | 1 | 0.63 | ||
| Eyeball rupture | 1 | 0.90 | Culture (Septic, 101) | No growth | 51 | 50.5 | |
| Intraorbital foreign body | 1 | 0.90 | Corynebacterium macginleyi | 9 | 8.91 | ||
| Conjunctivitis | 1 | 0.90 | Staphylococcus epidermidis | 7 | 6.93 | ||
| Staphylococcus aureus | 6 | 5.94 | |||||
| Serratia marcescens | 4 | 3.96 | |||||
| Pseudomonas aeruginosa | 3 | 2.97 | |||||
| Others | 21 | 20.79 | |||||
| Culture (Aseptic, 59) | No growth | 46 | 77.97 | ||||
| Pseudomonas aeruginosa | 5 | 8.47 | |||||
| Streptococcus mitis/oralis | 3 | 5.08 | |||||
| Corynebacterium striatum | 2 | 3.39 | |||||
| Streptococcus dysgalactiae | 2 | 3.39 | |||||
| Staphylococcus epidermidis | 1 | 1.69 | |||||
| Conventional PCR [13] | Real-Time PCR [18,19] | |
|---|---|---|
| Target | 16SrRNA | 16SrRNA |
| Forward | 5′-CAGGCCTAACAGATGCAAGTC-3′ | 5′-AGTTTGATCMTGGCTCAG-3′ |
| Reverse | 5′-GGGCGGWGTGTACAAGGC-3′ | 5′-GGACTACHAGGGTATCTAAT-3′ |
| Probe | None | 5′(HEX)-CGTATTACCGCGGCTGCTGGCAC-(BHQ1)3′ |
| PCR conditions | Initial activation: 95 × 15 min Total cycles: 30 cycles Denaturation: 95 × 1 min Annealing: 55 × 1 min Extension: 72 × 1.5 min | Initial activation: 95 × 10 min Total cycles: 45 cycles Denaturation: 95 × 1 min Annealing: 50 × 1 min Extension: 72 × 1 min |
| Target | D1/D2 region | 18s rRNA |
| Forward | 5′-GCATATCAATAAGCGGAGGAAAAG-3′ | 5′-GGRAAACTCACCAGGTCCAG-3′ |
| Reverse | 5′-GGTCCGTGTTTCAAGACG-3′ | 5′-GSWCTATCCCCAKCACGA-3′ |
| Probe | None | 5′(FAM)-TGGTGCATGGCCGTT-(BHQ1)3′ |
| PCR conditions | Initial activation: 95 × 15 min Total cycles: 35 cycles Denaturation: 95 × 30 s Annealing: 50 × 30 s Extension: 72 × 30 s | Initial activation: 95 × 10 min Total cycles: 50 cycles Denaturation: 95 × 15 s Annealing & extension: 65 × 1 min |
| Specimen Type (No.) | Growth over Occasional | Growth Under Occasional or No Growth | Sensitivity (95% CI) | Specificity (95% CI) | ||
|---|---|---|---|---|---|---|
| No. of Positive Real-Time PCR (%) | No. of Negative Real-Time PCR (%) | No. of Positive Real-Time PCR (%) | No. of Negative Real-Time PCR (%) | |||
| Septic (101) | 18 (48.6) | 10 (27.0) | 12 (9.8) | 61 (49.6) | 0.643 (0.441–0.814) | 0.836 (0.73–0.912) |
| Aseptic (59) | 8 (21.6) | 1 (2.7) | 6 (4.9) | 44 (35.8) | 0.889 (0.518–0.997) | 0.88 (0.757–0.955) |
| Specimen type (No.) | Growth over occasional | Growth under occasional or no growth | Sensitivity (95% CI) | Specificity (95% CI) | ||
| No. of positives with conventional PCR (%) | No. of negatives with conventional PCR (%) | No. of positives with conventional PCR (%) | No. of negatives with conventional PCR (%) | |||
| Septic (101) | 12 (32.4) | 16 (43.2) | 15 (12.2) | 58 (47.2) | 0.429 (0.245–0.628) | 0.795 (0.684–0.88) |
| Aseptic (59) | 3 (8.1) | 6 (16.2) | 2 (1.6) | 48 (39.0) | 0.333 (0.075–0.701) | 0.96 (0.863–0.995) |
| Specimen Type (No.) | Positive for Real-Time PCR | Negative for Real-Time PCR | Agreement (95% CI) | ||
|---|---|---|---|---|---|
| No. of Positives with Conventional PCR (%) | No. of Negatives with Conventional PCR (%) | No. of Positives with Conventional PCR (%) | No. of Negatives with Conventional PCR (%) | ||
| Septic (101) | 14 (43.8) | 13 (40.6) | 16 (12.5) | 58 (45.3) | 0.713 (0.618–0.792) |
| Aseptic (59) | 2 (6.3) | 3 (9.4) | 12 (9.4) | 42 (32.8) | 0.746 (0.622–0.839) |
| Variable | Growth on Culture | p-Value | ||
|---|---|---|---|---|
| Negatives with Real-Time PCR (N = 29) | Positives with Real-Time PCR (N = 11) | |||
| Sex | Female | 18 (62.1%) | 5 (45.5%) | 0.477 |
| Male | 11 (37.9%) | 6 (54.5%) | ||
| Age | 47 ± 21.5 | 43.3 ± 21.1 | 0.629 | |
| DM | No | 17 (58.6%) | 9 (81.8%) | 0.27 |
| Yes | 12 (41.4%) | 2 (18.2%) | ||
| Contact lens | No | 20 (69.0%) | 7 (63.6%) | 1 |
| Yes | 9 (31.0%) | 4 (36.4%) | ||
| Trauma | No | 29 (100.0%) | 11 (100.0%) | NA |
| Operation history | No | 20 (69.0%) | 6 (54.5%) | 0.469 |
| Yes | 9 (31.0%) | 5 (45.4%) | ||
| Admission | No | 16 (55.2%) | 4 (36.4%) | 0.479 |
| Yes | 13 (44.8%) | 7 (63.6%) | ||
| Gram stain | Negative | 16 (55.2%) | 2 (18.2%) | 0.073 |
| Positive | 13 (44.8%) | 9 (81.8%) | ||
| Sample to extraction (day) | 10 (0–113) | 1 (0–15) | 0.003 | |
| No. | Clinical Diagnosis | Specimen | ΔCT | DNA Concentration (ng/µL) | Culture Results | Microbiome Results |
|---|---|---|---|---|---|---|
| 17 | Endophthalmitis | Intraocular lens | −10.41 | 37.45 | Pseudomonas aeruginosa, occasional | Pseudomonas aeruginosa |
| 117 | Keratitis | Contact lens | −8.16 | 11.2 | Serratia marcescens, many Bacillus cereus, occasional | Serratia marcescens |
| 15 | Endophthalmitis | Cornea | −5.15 | 19.95 | Pseudomonas aeruginosa, moderate | Pseudomonas aeruginosa |
| 20 | Keratitis | Contact lens | −7.27 | 46 | Aeromonas hydrophila, many | Aeromonas hydrophila |
| No. | Clinical diagnosis | Specimen | ΔCT | DNA concentration (ng/µL) | Culture results | Microbiome results |
| 17 | Endophthalmitis | Intraocular lens | −10.41 | 37.45 | Pseudomonas aeruginosa, occasional | Pseudomonas aeruginosa |
| 117 | Keratitis | Contact lens | −8.16 | 11.2 | Serratia marcescens, many Bacillus cereus, occasional | Serratia marcescens |
| 15 | Endophthalmitis | Cornea | −5.15 | 19.95 | Pseudomonas aeruginosa, moderate | Pseudomonas aeruginosa |
| 20 | Keratitis | Contact lens | −7.27 | 46 | Aeromonas hydrophila, many | Aeromonas hydrophila |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Kim, K.; Lee, Y.; Ryoo, N. Enhanced Detection of Bacterial Ocular Pathogens: A Comparative Study of Broad-Range Real-Time PCR and Conventional Culture Methods. Diagnostics 2025, 15, 966. https://doi.org/10.3390/diagnostics15080966
Park S, Kim K, Lee Y, Ryoo N. Enhanced Detection of Bacterial Ocular Pathogens: A Comparative Study of Broad-Range Real-Time PCR and Conventional Culture Methods. Diagnostics. 2025; 15(8):966. https://doi.org/10.3390/diagnostics15080966
Chicago/Turabian StylePark, Sunggyun, Kyoungbo Kim, Youhyun Lee, and Namhee Ryoo. 2025. "Enhanced Detection of Bacterial Ocular Pathogens: A Comparative Study of Broad-Range Real-Time PCR and Conventional Culture Methods" Diagnostics 15, no. 8: 966. https://doi.org/10.3390/diagnostics15080966
APA StylePark, S., Kim, K., Lee, Y., & Ryoo, N. (2025). Enhanced Detection of Bacterial Ocular Pathogens: A Comparative Study of Broad-Range Real-Time PCR and Conventional Culture Methods. Diagnostics, 15(8), 966. https://doi.org/10.3390/diagnostics15080966






