Combinatory Flowcytometric Approach in Pediatric Acute Lymphoid Leukemia Identifies Surrogate Minimal Residual Disease Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Inclusion and Study Population
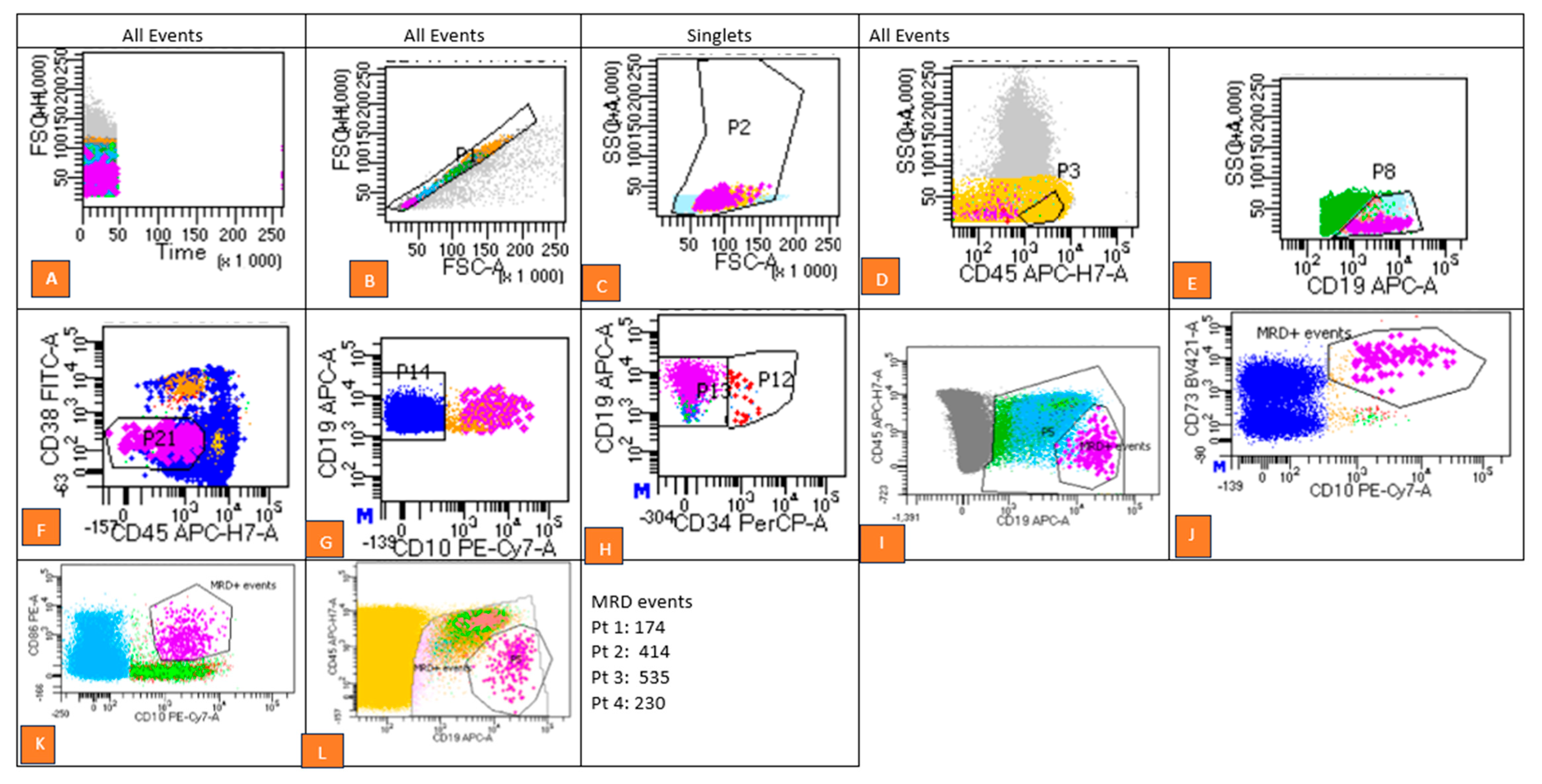
2.2. Flow Cytometry Analysis and Gating Strategy
- Exclusion of doublets using forward scatter-area (FSC-A) versus forward scatter-height (FSC-H) plots.
- Debris removal based on forward scatter (FSC) versus side scatter (SSC) parameters.
- Separation of cell populations using CD45 versus CD19 dot plots to isolate CD19+ CD34+ (immature blasts) from CD19+ CD34− (mature B-cell populations).
- Evaluation of investigational marker expression patterns by comparing fluorescence intensities in stained samples with internal control populations. Aberrant expression patterns were identified as MRD-positive.
2.3. Cytogenetic and Molecular Analysis
2.4. Statistical Analysis
3. Results
3.1. Immunophenotypic Aberrancies
3.2. Correlation of MRD Negativity with Cytogenetic Abnormalities
3.3. MRD Expression Data
3.4. Risk Stratification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willemse, M.J.; Seriu, T.; Hettinger, K.; d’Aniello, E.; Hop, W.C.; Panzer-Grümayer, E.R.; Biondi, A.; Schrappe, M.; Kamps, W.A.; Masera, G.; et al. Detection of Minimal Residual Disease Identifies Differences in Treatment Response between T-All and Precursor B-All. Blood 2002, 99, 4386–4393. [Google Scholar] [CrossRef] [PubMed]
- Brisco, M.J.; Condon, J.; Hughes, E.; Neoh, S.H.; Sykes, P.J.; Seshadri, R.; Toogood, I.; Waters, K.; Tauro, G.; Ekert, H.; et al. Outcome prediction in childhood acute lymphoblastic leukaemia by molecular quantification of residual disease at the end of induction. Lancet 1994, 343, 196–200. [Google Scholar] [CrossRef]
- Seth, N.; Mahajan, V.; Kedia, S.; Sutar, A.; Sehgal, K. Minimal Residual Disease (MRD) detection in B- ALL—Experience of a standalone flow cytometry laboratory. Pediatr. Hematol. Oncol. J. 2021, 6, 26–31. [Google Scholar] [CrossRef]
- Dix, C.; Lo, T.H.; Clark, G.; Abadir, E. Measurable Residual Disease in Acute Myeloid Leukemia Using Flow Cytometry: A Review of Where We Are and Where We Are Going. J. Clin. Med. 2020, 9, 1714. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sachdeva, M.U.; Varma, N.; Varma, S.; Marwaha, R.K. Characterization of immunophenotypic aberrancies in adult and childhood acute lymphoblastic leukemia: A study from Northern India. J. Cancer Res. Ther. 2016, 12, 620–626. [Google Scholar] [CrossRef]
- Jalal, S.D.; Al-Allawi, N.A.S.; Al Doski, A.A.S. Immunophenotypic aberrancies in acute lymphoblastic leukemia from 282 Iraqi patients. Int. J. Lab. Hematol. 2017, 39, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Jegalian, A.G.; Wayne, A.S.; Kreitman, R.J.; Mussai, F.J.; Pastan, I.; Yuan, C.M.; Stetler-Stevenson, M. CD22 Expression in Pediatric B-Lineage Acute Lymphoblastic Leukemia. Blood 2009, 114, 4119. [Google Scholar] [CrossRef]
- Panda, S.S.; Radhakrishnan, V.; Ganesan, P.; Rajendranath, R.; Ganesan, T.S.; Rajalekshmy, K.R.; Bhola, R.K.; Das, H.; Sagar, T.G. Flow Cytometry Based MRD and Its Impact on Survival Outcome in Children and Young Adults with ALL: A Prospective Study from a Tertiary Cancer Centre in Southern India. Indian J. Hematol. Blood Transfus. 2020, 36, 300–308. [Google Scholar] [CrossRef]
- Ratti, S.; Lonetti, A.; Follo, M.Y.; Paganelli, F.; Martelli, A.M.; Chiarini, F.; Evangelisti, C. B-ALL Complexity: Is Targeted Therapy Still A Valuable Approach for Pediatric Patients? Cancers 2020, 12, 3498. [Google Scholar] [CrossRef]
- Plesa, A.; Dumezy, F.; Mathis, S.; Lhoumeau, A.-C.; Bardet, V.; Saada, V.; Arnoux, I.; Badaoui, B.; Cornet, E.; Osman, J.; et al. AML MRD By Multiparameter Flow Cytometry Using Laip/Dfn and LSC: Methodological Aspects in a Multicentric Study of the French-Flow MRD AML ALFA Network. Blood 2022, 140, 6279–6281. [Google Scholar] [CrossRef]
- Das, N.; Gupta, R.; Gupta, S.K.; Bakhshi, S.; Seth, R.; Kumar, C.; Rai, S.; Singh, S.; Prajapati, V.K.; Gogia, A.; et al. Critical evaluation of the utility of pre- and post-therapy immunophenotypes in assessment of measurable residual disease in B-ALL. Ann. Hematol. 2021, 100, 2487–2500. [Google Scholar] [CrossRef]
- Al-Mudallal, S.S. Expression of CD10, CD13, CD19, CD20, CD22, CD33, CD34 and CD45 on B Lymphoblasts in Bone Marrow Aspirate of Adult Patients with Newly Diagnosed Acute Lymphoblasticleukemia Using Multicolor Flow Cytometry. Iraqi J. Hematol. 2014, 3, 47–50. [Google Scholar] [CrossRef]
- Belurkar, S.; Mantravadi, H.; Manohar, C.; Kurien, A. Correlation of morphologic and cytochemical diagnosis with flowcytometric analysis in acute leukemia. J. Cancer Res. Ther. 2013, 9, 71–79. [Google Scholar] [CrossRef]
- Olga, C.; Lyudmila Yuryevna, G.; Alexander, P.; Nikolay Nikolayevich, T. B-Cell Precursors: Immunophenotypic Features in the Detection of Minimal Residual Disease in Acute Leukemia. In Normal and Malignant B-Cell; Mourad, A., Ed.; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar] [CrossRef]
- Tembhare, P.R.; Chatterjee, G.; Khanka, T.; Ghogale, S.; Badrinath, Y.; Deshpande, N.; Panda, D.; Patkar, N.V.; Narula, G.; Girase, K.; et al. Eleven-marker 10-color flow cytometric assessment of measurable residual disease for T-cell acute lymphoblastic leukemia using an approach of exclusion. Cytom. Part B Clin. Cytom. 2021, 100, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Griesinger, F.; Pirò-Noack, M.; Kaib, N.; Falk, M.; Renziehausen, A.; Troff, C.; Grove, D.; Schnittger, S.; Büchner, T.; Ritter, J.; et al. Leukaemia-associated immunophenotypes (LAIP) are observed in 90% of adult and childhood acute lymphoblastic leukaemia: Detection in remission marrow predicts outcome. Br. J. Haematol. 1999, 105, 241–255. [Google Scholar]
- Gaipa, G.; Basso, G.; Biondi, A.; Campana, D. Detection of minimal residual disease in pediatric acute lymphoblastic leukemia. Cytom. Part B Clin. Cytom. 2013, 84, 359–369. [Google Scholar] [CrossRef]
- Coustan-Smith, E.; Song, G.; Clark, C.; Key, L.; Liu, P.; Mehrpooya, M.; Stow, P.; Su, X.; Shurtleff, S.; Pui, C.-H.; et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood 2011, 117, 6267–6276. [Google Scholar] [CrossRef]
- Słota, Ł.; Sędek, Ł.; Kulis, J.; Perkowski, B.; Malinowska, I.; Zawitkowska, J.; Kazanowska, B.; Derwich, K.; Niedźwiecki, M.; Mizia-Malarz, A.; et al. Expression of CD73 on leukemic blasts increases during follow-up—A promising candidate marker for minimal residual disease detection in pediatric B-cell precursor acute lymphoblastic leukemia. Cent.-Eur. J. Immunol. 2022, 47, 84–91. [Google Scholar] [CrossRef]
- Tembhare, P.R.; Ghogale, S.; Ghatwai, N.; Badrinath, Y.; Kunder, N.; Patkar, N.V.; Bibi, A.R.; Chatterjee, G.; Arora, B.; Narula, G.; et al. Evaluation of new markers for minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia: CD73 and CD86 are the most relevant new markers to increase the efficacy of MRD 2016; 00B: 000-000. Cytom. Part B Clin. Cytom. 2018, 94, 100–111. [Google Scholar] [CrossRef]
- Rhein, P.; Mitlohner, R.; Basso, G.; Gaipa, G.; Dworzak, M.N.; Kirschner-Schwabe, R.; Hagemeier, C.; Stanulla, M.; Schrappe, M.; Ludwig, W.D.; et al. CD11b is a therapy resistance- and minimal residual disease-specific marker in precursor B-cell acute lymphoblastic leukemia. Blood 2010, 115, 3763–3771. [Google Scholar] [CrossRef]
- Guillaume, N.; Penther, D.; Vaida, I.; Gruson, B.; Harrivel, V.; Claisse, J.F.; Capiod, J.C.; Lefrere, J.J.; Damaj, G. CD66c expression in B-cell acute lymphoblastic leukemia: Strength and weakness. Int. J. Lab. Hematol. 2011, 33, 92–96. [Google Scholar] [CrossRef]
- McGinnis, E.; Yang, D.; Au, N.; Morrison, D.; Chipperfield, K.M.; Setiadi, A.F.; Liu, L.; Tsang, A.; Vercauteren, S.M. Clinical and laboratory features associated with myeloperoxidase expression in pediatric B-lymphoblastic leukemia. Cytom. Part B Clin. Cytom. 2021, 100, 446–453. [Google Scholar] [CrossRef]
- Thabet, M.; Hassan, E.; ELsalakawy, W.; Abd-Allah, N.E.-H.; Abdelrahman, R. Prognostic significance of tetraspanin (CD81) expression in patients with acute lymphoblastic leukemia. Egypt. J. Hematol. Bone Marrow Transplant. 2024, 11, 7–17. [Google Scholar] [CrossRef]
- Veltroni, M.; De Zen, L.; Sanzari, M.C.; Maglia, O.; Dworzak, M.N.; Ratei, R.; Biondi, A.; Basso, G.; Gaipa, G. Expression of CD58 in normal, regenerating and leukemic bone marrow B cells: Implications for the detection of minimal residual disease in acute lymphocytic leukemia. Haematologica 2003, 88, 1245–1252. [Google Scholar]
- Kansal, R. Diagnosis and Molecular Pathology of Lymphoblastic Leukemias and Lymphomas in the Era of Genomics and Precision Medicine: Historical Evolution and Current Concepts—Part 1: Lymphoid Neoplasms. Lymphatics 2023, 1, 55–76. [Google Scholar] [CrossRef]
- Vaghela, N.; Anand, I.S.; Trivedi, D.H.; Jani, M. Prognostic value of peripheral blood blast percentage on day 8 in long term cure in patients with ALL. World J. Pharm. Pharm. Sci. 2014, 3, 1839–1847. [Google Scholar]
- Rogers, S.L.; Zhao, Y.; Jiang, X.; Eaves, C.J.; Mager, D.L.; Rouhi, A. Expression of the leukemic prognostic marker CD7 is linked to epigenetic modifications in chronic myeloid leukemia. Mol. Cancer 2010, 9, 41. [Google Scholar] [CrossRef]
- Loganathan, A.; Bharadwaj, R.; Srinivasan, A.; Scott, J.X. Cytogenetics and Molecular Genetics in Pediatric Acute Lymphoblastic Leukemia (ALL) and Its Correlation with Induction Outcomes. South Asian J. Cancer 2022, 11, 353–360. [Google Scholar] [CrossRef]
- Bowman, W.P.; Larsen, E.L.; Devidas, M.; Linda, S.B.; Blach, L.; Carroll, A.J.; Carroll, W.L.; Pullen, D.J.; Shuster, J.; Willman, C.L.; et al. Augmented therapy improves outcome for pediatric high risk acute lymphocytic leukemia: Results of Children’s Oncology Group trial P9906. Pediatr. Blood Cancer 2011, 57, 569–577. [Google Scholar] [CrossRef]
- Verma, S.; Dhanda, H.; Singh, A.; Rishi, B.; Tanwar, P.; Chaudhry, S.; Siraj, F.; Misra, A. Systematic review of epigenetic targets in acute myeloid leukemia. Am. J. Blood Res. 2021, 11, 458–471. [Google Scholar]
- Mrózek, K.; Harper, D.P.; Aplan, P.D. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Hematol. /Oncol. Clin. N. Am. 2009, 23, 991–1010. [Google Scholar] [CrossRef] [PubMed]
- Al-Anazi, W.; Dridi, W.; Al-Anazi, K. Cytogenetics, Molecular Genetics and Epigenetics and Their Impact on The Management of Acute Lymphoblastic Leukemia. J. Mol. Genet. Med. 2017, 11, 1747-0862. [Google Scholar]
- Kruse, A.; Abdel-Azim, N.; Kim, H.N.; Ruan, Y.; Phan, V.; Ogana, H.; Wang, W.; Lee, R.; Gang, E.J.; Khazal, S.; et al. Minimal Residual Disease Detection in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2020, 21, 1054. [Google Scholar] [CrossRef] [PubMed]
- Liu-Dumlao, T.; Kantarjian, H.; Thomas, D.A.; O’Brien, S.; Ravandi, F. Philadelphia-positive acute lymphoblastic leukemia: Current treatment options. Curr. Oncol. Rep. 2012, 14, 387–394. [Google Scholar] [CrossRef]
- Forero, R.M.; Hernández, M.; Hernández-Rivas, J.M. Genetics of Acute Lymphoblastic Leukemia. In Leukemia; Margarita, G., Gueorgui, B., Eds.; IntechOpen: Rijeka, Croatia, 2013. [Google Scholar] [CrossRef][Green Version]
| Parameter | Total (n (%)) |
|---|---|
| Total Patients | 218 |
| Age Range | 9 months–15 years |
| Median | 7.5 years (MRD-positive) 6.5 years (MRD-negative) |
| Gender | Male 130 (59.6%), Female 88 (40.4%) |
| No. of Patients under different risks | High Risk 130 (59.6%), Intermediate Risk 58 (26.6%), Standard Risk 30 (13.8%) |
| No. of patients with different cytogenetic abnormalities | Hyper-ploidy 26 (11.9%), Hypoploidy 4 (1.8%), Others 20 (9.2%) |
| Treatment Response (MRD) | Positive 80 (36.7%), Negative 138 (63.3%) |
| Markers | Incidence | MRD Positive | Incidence Cited by Other Studies Refs % | ||
|---|---|---|---|---|---|
| No. of: Positive | Positive % | No. of Cases Out of 208 | Positive % | ||
| CD 10 | 190 | 93.6 | 35 | 83 | ~70–83 [12,13] |
| CD 19 | 194 | 95.6 | 33 | 79 | ~90–95 [12,13] |
| CD 79a | 127 | 62.6 | 17 | 40 | ~40–50 [14] |
| HLA-DR | 180 | 88.7 | 35 | 83 | ~15–25 [15] |
| CD 58 | 19 | 9.4 | 8 | 19 | ~10–20 [16,17] |
| TdT | 18 | 8.9 | 5 | 12 | ~20–70 [14] |
| CD 38 | 88 | 43.3 | 17 | 40 | ~20–40 [18] |
| CD 34 | 142 | 70.0 | 25 | 60 | ~50–70 [11] |
| CD 22 | 37 | 18.2 | 6 | 14 | ~10–20 [7] |
| CD 73 | 10 | 4.9 | 8 | 19 | <5 [19] |
| CD 86 | 9 | 4.4 | 7 | 17 | ~50 [20] |
| Recurrent Genetic Abnormality | Prognosis | Total 65 Patients | MRD-Positive | MRD-Negative | Alive | Expired | Relapse |
|---|---|---|---|---|---|---|---|
| Hyper ploidy: >50 chromosomes | Good | 26 | 22 | 4 | 20 | 3 | 3 |
| Hypoploidy: <45 chromosomes | Poor | 4 | 1 | 3 | 3 | 1 | 0 |
| t (9;22)(q34;q11) | Intermediate | 12 | 1 | 11 | 10 | 2 | 0 |
| t (12;22)(p13;q22) | Good | 13 | 1 | 12 | 10 | 2 | 1 |
| t (1;19)(q23;p13) | Intermediate | 10 | 3 | 7 | 7 | 1 | 1 |
| Markers | Incidence | MRD-Positive | Incidence Cited by Other Studies Refs % | ||
|---|---|---|---|---|---|
| No. of Positive | Positive % | No. of Cases Out of 208 | Positive % | ||
| CD 13 | 14 | 6.9 | 2 | 5 | ~30–50 [4,16] |
| CD33 | 20 | 9.9 | 5 | 12 | ~10–25 [4,16] |
| CD 11b | 3 | 1.5 | 2 | 5 | >1 [21] |
| CD 66c | 5 | 2.5 | 1 | 2 | ~40 [14,22] |
| CD 14 | 2 | 1.0 | 1 | 2 | ~10 [13] |
| MPO | 1 | 0.5 | 0 | 0 | ~2 [23] |
| CD 81 | 6 | 3.0 | 1 | 2 | ~90–95 [24] |
| CD 123 | 14 | 6.9 | 5 | 12 | ~8.2 [25] |
| Markers | Incidence | MRD Status | Incidence Cited by Other Studies Refs % | ||
|---|---|---|---|---|---|
| No. of Positive | Positive % | No. of Cases Out 208 | Positive | ||
| CD 2 | 2 | 1 | 1 | 2.4 | ~3.6 [11] |
| CD 3 | 2 | 1 | 1 | 2.4 | ~25 [14,15] |
| CyCD 3 | 2 | 1 | 1 | 2.4 | ~5 [26,27] |
| SurCD 3 | 4 | 2 | 1 | 2.4 | ~20 [26] |
| CD 5 | 2 | 1 | 0 | 0.0 | ~3 [11] |
| CD 7 | 4 | 2 | 2 | 4.8 | ~20 [14,15] |
| CD 8 | 2 | 1 | 0 | 0.0 | ~3 [11] |
| CD 56 | 3 | 1 | 1 | 2.4 | ~4.3–6 [26] |
| CD Markers | Total ((n) = 208) | MRD-Positive (%) | MRD-Negative (%) | p-Value |
|---|---|---|---|---|
| CD10 | 186 | 83.3 | 16.7 | 0.109 |
| CD20 | 81 | 85.2 | 14.8 | 0.365 |
| CD22 | 36 | 86.1 | 13.9 | 0.501 |
| CD7 | 122 | 88.5 | 11.5 | 0.003 * |
| CD73 | 9 | 22.2 | 77.8 | 0.00 * |
| CD86 | 7 | 28.6 | 71.4 | 0.002 * |
| CD64 | 2 | 100 | 0 | 1.0 |
| CD2 | 1 | 0 | 100 | 0.178 |
| CD3 | 1 | 0 | 100 | 0.178 |
| CyCD3 | 1 | 0 | 100 | 0.178 |
| CD5 | 1 | 100 | 0 | 1.0 |
| CD8 | 1 | 100 | 0 | 1.0 |
| CD 56 | 1 | 100 | 0 | 1.0 |
| CD 38 | 85 | 17.7 | 82.35 | 0.970 |
| CD 34 | 136 | 15.4 | 84.6 | 0.228 |
| HLA-DR | 175 | 17.7 | 82.3 | 1.0 |
| TdT | 18 | 22.2 | 11.8 | 0.533 |
| CD13 | 13 | 7.7 | 92.3 | 0.471 |
| CD 33 | 19 | 21.1 | 82.6 | 0.752 |
| CD 14 | 1 | 100 | 0 | 0.178 |
| CD 11b | 1 | 100 | 0 | 0.178 |
| CD 81 | 5 | 20 | 8 | 1.0 |
| CD 58 | 15 | 33.3 | 66.7 | 0.150 |
| CD 66c | 3 | 0 | 100 | 1.0 |
| CD 123 | 13 | 30.8 | 69.2 | 0.253 |
| Risk Status | Total no.: 184 | MRD-Positive | MRD-Positive | |||
|---|---|---|---|---|---|---|
| Expired | Alive | Survivors | Defaulter | |||
| HR | 127 | 24 | 7 | 13 | 2 | 1 |
| IR | 27 | 2 | 0 | 2 | 0 | 0 |
| SR | 30 | 3 | 1 | 2 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
George, N.G.; Rishi, B.; Ray, S.; Kaur, M.; Kamal, R.; Garg, S.; Mehndiratta, S.; Chopra, N.; Zaman, S.; Singh, A.; et al. Combinatory Flowcytometric Approach in Pediatric Acute Lymphoid Leukemia Identifies Surrogate Minimal Residual Disease Markers. Diagnostics 2025, 15, 658. https://doi.org/10.3390/diagnostics15060658
George NG, Rishi B, Ray S, Kaur M, Kamal R, Garg S, Mehndiratta S, Chopra N, Zaman S, Singh A, et al. Combinatory Flowcytometric Approach in Pediatric Acute Lymphoid Leukemia Identifies Surrogate Minimal Residual Disease Markers. Diagnostics. 2025; 15(6):658. https://doi.org/10.3390/diagnostics15060658
Chicago/Turabian StyleGeorge, Noreen Grace, Bhavika Rishi, Sanghmitra Ray, Manpreet Kaur, Raj Kamal, Shikha Garg, Sumit Mehndiratta, Nidhi Chopra, Shamsuz Zaman, Amitabh Singh, and et al. 2025. "Combinatory Flowcytometric Approach in Pediatric Acute Lymphoid Leukemia Identifies Surrogate Minimal Residual Disease Markers" Diagnostics 15, no. 6: 658. https://doi.org/10.3390/diagnostics15060658
APA StyleGeorge, N. G., Rishi, B., Ray, S., Kaur, M., Kamal, R., Garg, S., Mehndiratta, S., Chopra, N., Zaman, S., Singh, A., & Misra, A. (2025). Combinatory Flowcytometric Approach in Pediatric Acute Lymphoid Leukemia Identifies Surrogate Minimal Residual Disease Markers. Diagnostics, 15(6), 658. https://doi.org/10.3390/diagnostics15060658






