Optimized Lightweight Architecture for Coronary Artery Disease Classification in Medical Imaging
Abstract
1. Introduction
- Modifying the VGG16 model with ResNet-inspired residual connections, tailored specifically for CAD detection in athletes, proposes a novel lightweight architecture.
- The model achieves superior diagnostic performance compared to state-of-the-art architectures, with significantly reduced computational complexity.
- Comprehensive evaluation using the CADICA dataset demonstrates its effectiveness across key metrics, including accuracy, recall, and AUC-ROC.
- Practical implications for deploying the model in real-time diagnostic workflows in sports medicine are discussed.
2. Related Works
2.1. Traditional Methods for CAD Diagnosis
2.2. Deep Learning in Medical Imaging
2.3. Datasets for CAD Research
2.4. Evaluation Metrics and Benchmarking
2.5. Gaps and Challenges in Current Researches
3. Methodology
3.1. VGG16
3.2. The Proposed Model
4. Experiment and Results
4.1. The CADICA
Strategies for Managing Dataset Limitations and Ensuring Robust Model Evaluation
4.2. Comparison of the Results of the Experiment and Analyses
4.3. Comparison Results with State-of-the-Art Model
| Model | Accuracy | Recall | Precision |
|---|---|---|---|
| ResNet-50 | 0.863 | 0.86 | 0.83 |
| VGG-16 | 0.878 | 0.856 | 0.79 |
| DenseNet121 | 0.871 | 0.853 | 0.821 |
| SqueezeNet | 0.832 | 0.81 | 0.782 |
| EfficientNet-B0 | 0.881 | 0.87 | 0.85 |
| The proposed model | 0.903 | 0.89 | 0.872 |
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Komilovich, E.B.Z. Coronary Artery Disease. Eur. J. Mod. Med. Pract. 2023, 3, 81–87. [Google Scholar]
- Celeski, M.; Di Gioia, G.; Nusca, A.; Segreti, A.; Squeo, M.R.; Lemme, E.; Mango, F.; Ferrera, A.; Ussia, G.P.; Grigioni, F. The Spectrum of Coronary Artery Disease in Elite Endurance Athletes—A Long-Standing Debate: State-of-the-Art Review. J. Clin. Med. 2024, 13, 5144. [Google Scholar] [CrossRef]
- Laterion, W.; Frakes, M.; Cohen, J.; Ender, V.; Wilcox, S.R. Critical Care Transport of Patients with Spontaneous Coronary Artery Dissection: A Case Series. Air Med. J. 2025, in press. [Google Scholar] [CrossRef]
- Azami, P.; Mohammadzadeh, S.; Seirafi, S.; Razeghian-Jahromi, I. A review of cutting-edge biomarkers for diagnosing coronary artery disease. Medicine 2025, 104, e41377. [Google Scholar] [CrossRef] [PubMed]
- Cacciatore, S.; Spadafora, L.; Bernardi, M.; Galli, M.; Betti, M.; Perone, F.; Nicolaio, G.; Marzetti, E.; Martone, A.M.; Landi, F.; et al. Management of coronary artery disease in older adults: Recent advances and gaps in evidence. J. Clin. Med. 2023, 12, 5233. [Google Scholar] [CrossRef] [PubMed]
- Attiq, A.; Afzal, S.; Ahmad, W.; Kandeel, M. Hegemony of inflammation in atherosclerosis and coronary artery disease. Eur. J. Pharmacol. 2024, 966, 176338. [Google Scholar] [CrossRef]
- Nasimov, R.; Nasimova, N.; Mirzakhalilov, S.; Tokdemir, G.; Rizwan, M.; Abdusalomov, A.; Cho, Y.I. GAN-Based Novel Approach for Generating Synthetic Medical Tabular Data. Bioengineering 2024, 11, 1288. [Google Scholar] [CrossRef]
- Forrest, I.S.; Petrazzini, B.O.; Duffy, Á.; Park, J.K.; Marquez-Luna, C.; Jordan, D.M.; Rocheleau, G.; Cho, J.H.; Rosenson, R.S.; Narula, J.; et al. Machine learning-based marker for coronary artery disease: Derivation and validation in two longitudinal cohorts. Lancet 2023, 401, 215–225. [Google Scholar] [CrossRef] [PubMed]
- D’Ancona, G.; Massussi, M.; Savardi, M.; Signoroni, A.; Di Bacco, L.; Farina, D.; Metra, M.; Maroldi, R.; Muneretto, C.; Ince, H.; et al. Deep learning to detect significant coronary artery disease from plain chest radiographs AI4CAD. Int. J. Cardiol. 2023, 370, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.M.; Sharma, V. Applications of AI in cardiovascular disease detection—A review of the specific ways in which AI is being used to detect and diagnose cardiovascular diseases. In AI in Disease Detection: Advancements and Applications; Wiley: Hoboken, NJ, USA, 2025; pp. 123–146. [Google Scholar]
- Subathra, R.; Sumathy, V. A smart CardioSenseNet framework with advanced data processing models for precise heart disease detection. Comput. Biol. Med. 2025, 185, 109473. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, H.; Liu, X.; Chen, C.; Shu, M. Cardiovascular disease detection based on deep learning and multi-modal data fusion. Biomed. Signal Process. Control 2025, 99, 106882. [Google Scholar] [CrossRef]
- Rathore, S.; Gautam, A.; Raghav, P.; Subramaniam, V.; Gupta, V.; Rathore, M.; Rathore, A.; Rathore, S.; Iyengar, S. Fully automated coronary artery calcium score and risk categorization from chest CT using deep learning and multiorgan segmentation: A validation study from National Lung Screening Trial (NLST). IJC Heart Vasc. 2025, 56, 101593. [Google Scholar] [CrossRef] [PubMed]
- Abdusalomov, A.; Mirzakhalilov, S.; Umirzakova, S.; Ismailov, O.; Sultanov, D.; Nasimov, R.; Cho, Y.I. Lightweight Deep Learning Framework for Accurate Detection of Sports-Related Bone Fractures. Diagnostics 2025, 15, 271. [Google Scholar] [CrossRef]
- Uddin, K.M.M.; Ripa, R.; Yeasmin, N.; Biswas, N.; Dey, S.K. Machine learning-based approach to the diagnosis of cardiovascular vascular disease using a combined dataset. Intell. Based Med. 2023, 7, 100100. [Google Scholar] [CrossRef]
- Shukur, B.S.; Mijwil, M.M. Involving machine learning techniques in heart disease diagnosis: A performance analysis. Int. J. Electr. Comput. Eng. 2023, 13, 2177. [Google Scholar]
- Alizadehsani, R.; Abdar, M.; Roshanzamir, M.; Khosravi, A.; Kebria, P.M.; Khozeimeh, F.; Nahavandi, S.; Sarrafzadegan, N.; Acharya, U.R. Machine learning-based coronary artery disease diagnosis: A comprehensive review. Comput. Biol. Med. 2019, 111, 103346. [Google Scholar] [CrossRef]
- Hassannataj Joloudari, J.; Azizi, F.; Nematollahi, M.A.; Alizadehsani, R.; Hassannatajjeloudari, E.; Nodehi, I.; Mosavi, A. GSVMA: A genetic support vector machine ANOVA method for CAD diagnosis. Front. Cardiovasc. Med. 2022, 8, 760178. [Google Scholar] [CrossRef] [PubMed]
- Joloudari, J.H.; Hassannataj Joloudari, E.; Saadatfar, H.; Ghasemigol, M.; Razavi, S.M.; Mosavi, A.; Nabipour, N.; Shamshirband, S.; Nadai, L. Coronary artery disease diagnosis; ranking the significant features using a random trees model. Int. J. Environ. Res. Public Health 2020, 17, 731. [Google Scholar] [CrossRef]
- Kolukisa, B.; Bakir-Gungor, B. Ensemble feature selection and classification methods for machine learning-based coronary artery disease diagnosis. Comput. Stand. Interfaces 2023, 84, 103706. [Google Scholar] [CrossRef]
- Ainiwaer, A.; Hou, W.Q.; Kadier, K.; Rehemuding, R.; Liu, P.F.; Maimaiti, H.; Qin, L.; Ma, X.; Dai, J.G. A Machine Learning Framework for Diagnosing and Predicting the Severity of Coronary Artery Disease. Rev. Cardiovasc. Med. 2023, 24, 168. [Google Scholar] [CrossRef]
- Özbilgin, F.; Kurnaz, Ç.; Aydın, E. Prediction of coronary artery disease using machine learning techniques with iris analysis. Diagnostics 2023, 13, 1081. [Google Scholar] [CrossRef] [PubMed]
- Abd-Ellah, M.K.; Khalaf, A.A.; Gharieb, R.R.; Hassanin, D.A. Automatic diagnosis of common carotid artery disease using different machine learning techniques. J. Ambient. Intell. Humaniz. Comput. 2023, 14, 113–129. [Google Scholar] [CrossRef]
- Arun Kumar, S.; Sasikala, S. Review on deep learning-based CAD systems for breast cancer diagnosis. Technol. Cancer Res. Treat. 2023, 22, 15330338231177977. [Google Scholar] [CrossRef] [PubMed]
- Kaba, Ş.; Haci, H.; Isin, A.; Ilhan, A.; Conkbayir, C. The application of deep learning for the segmentation and classification of coronary arteries. Diagnostics 2023, 13, 2274. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.Y.; Yang, C.C. Investigating the feasibility of using DenseNet to improve coronary calcification detection in CT. Acad. Radiol. 2023, 30, 1600–1613. [Google Scholar] [CrossRef]
- Yang, W.; Chen, C.; Yang, Y.; Chen, L.; Yang, C.; Gong, L.; Wang, J.; Shi, F.; Wu, D.; Yan, F. Diagnostic performance of deep learning-based vessel extraction and stenosis detection on coronary computed tomography angiography for coronary artery disease: A multi-reader multi-case study. La Radiol. Med. 2023, 128, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, S.; Zhang, Y.; Chang, Q.; Zhang, Y.; Li, D.; Qiu, J.; Hu, L.; Peng, X.; Du, Y.; et al. Congenital heart disease detection by pediatric electrocardiogram based deep learning integrated with human concepts. Nat. Commun. 2024, 15, 976. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, X.; Tian, C.; Chen, S.; Deng, Y.; Liao, X.; Wang, Q.; Si, W. PlaqueNet: Deep learning enabled coronary artery plaque segmentation from coronary computed tomography angiography. Vis. Comput. Ind. Biomed. Art 2024, 7, 6. [Google Scholar] [CrossRef]
- Shahrbabak, S.M.; Kim, S.; Youn, B.D.; Cheng, H.M.; Chen, C.H.; Mukkamala, R.; Hahn, J.O. Peripheral artery disease diagnosis based on deep learning-enabled analysis of non-invasive arterial pulse waveforms. Comput. Biol. Med. 2024, 168, 107813. [Google Scholar] [CrossRef]
- Jiménez-Partinen, A.; Molina-Cabello, M.A.; Thurnhofer-Hemsi, K.; Palomo, E.J.; Rodríguez-Capitán, J.; Molina-Ramos, A.I.; Jiménez-Navarro, M. CADICA: A new dataset for coronary artery disease detection by using invasive coronary angiography. Expert Syst. 2024, 41, e13708. [Google Scholar] [CrossRef]
- Ramesh, B.; Lakshmanna, K. A Novel Early Detection and Prevention of Coronary Heart Disease Framework Using Hybrid Deep Learning Model and Neural Fuzzy Inference System. IEEE Access 2024, 12, 26683–26695. [Google Scholar] [CrossRef]
- Duan, H.; Yi, S.; Ren, Y. DCA-YOLOv8: A Novel Framework Combined with AICI Loss Function for Coronary Artery Stenosis Detection. Sensors 2024, 24, 8134. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, H.; Su, F.; Qiu, S.; Tong, H.; Huang, M.; Yao, J. Identification of middle cerebral artery stenosis in transcranial Doppler using a modified VGG-16. Front. Neurol. 2024, 15, 1394435. [Google Scholar] [CrossRef] [PubMed]
- Satyasai, B.; Sharma, R. CM-VGG16: ConvMixer-Enhanced VGG16 Model for Automatic Detection of Heart Valve Diseases from Phonocardiogram Signals. IEEE Sens. J. 2024, 25, 3998–4005. [Google Scholar] [CrossRef]
- Staal, J.; Abràmoff, M.D.; Niemeijer, M.; Viergever, M.A.; Van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Hoover, A.D.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Armato, S.G., III; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
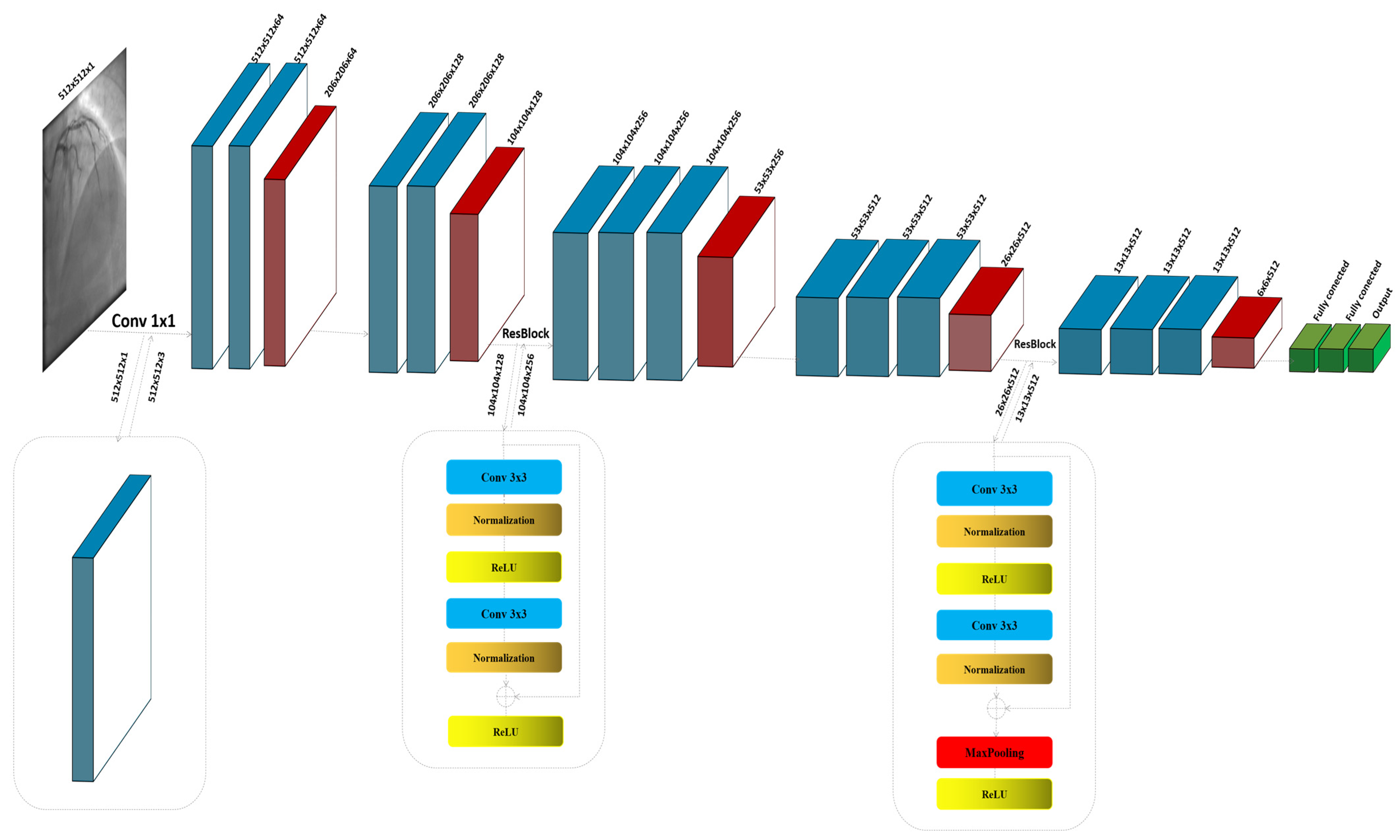
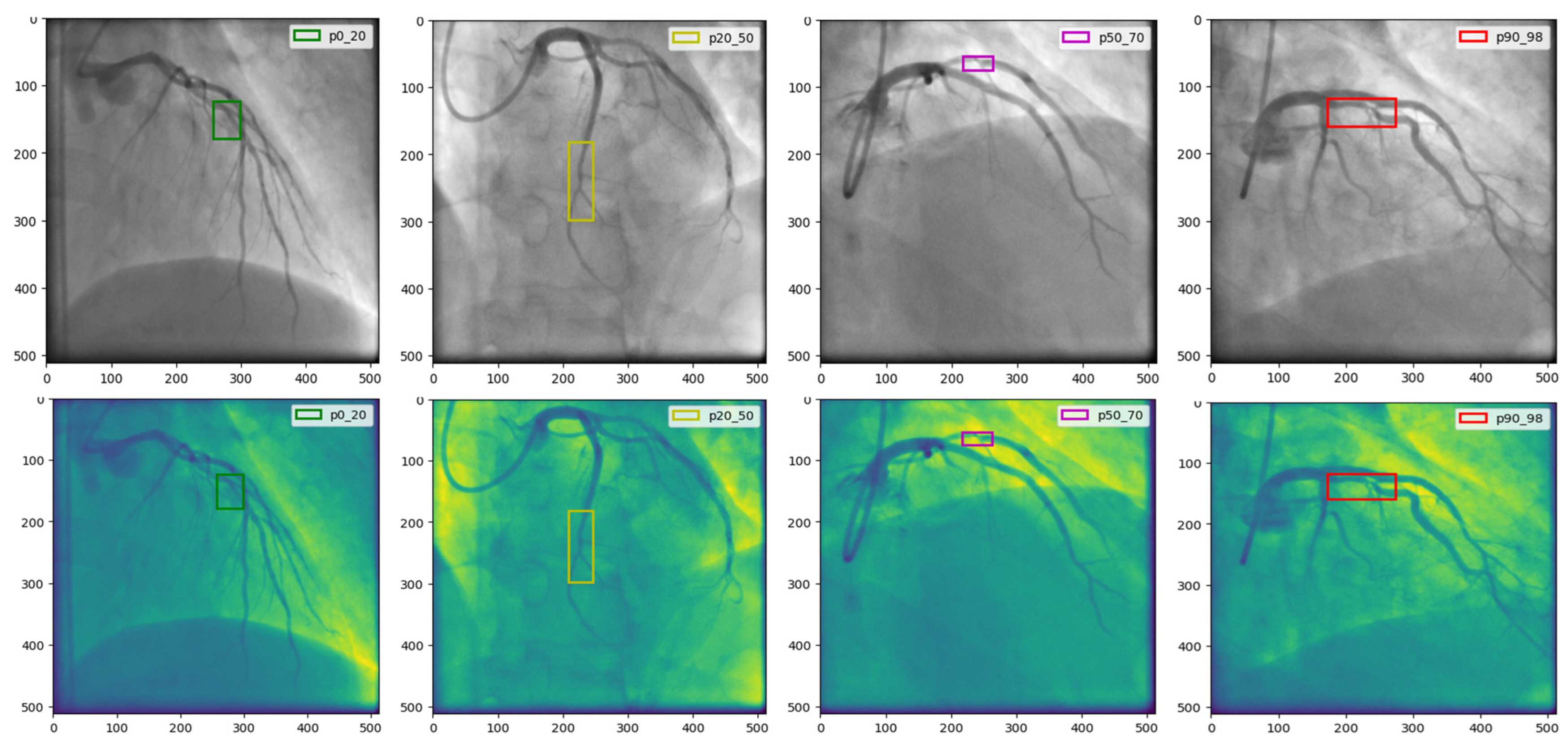
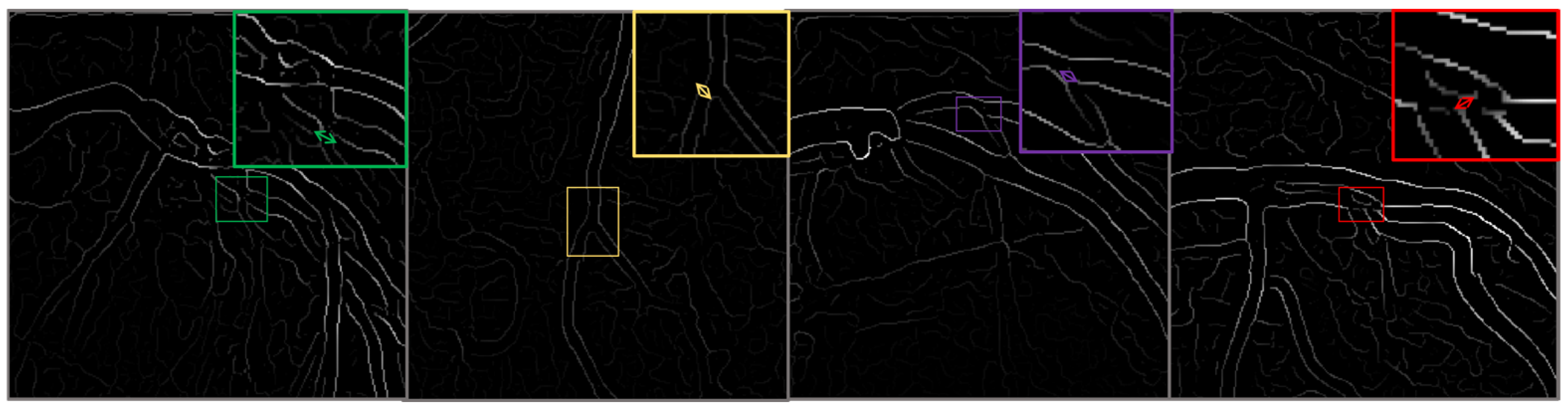

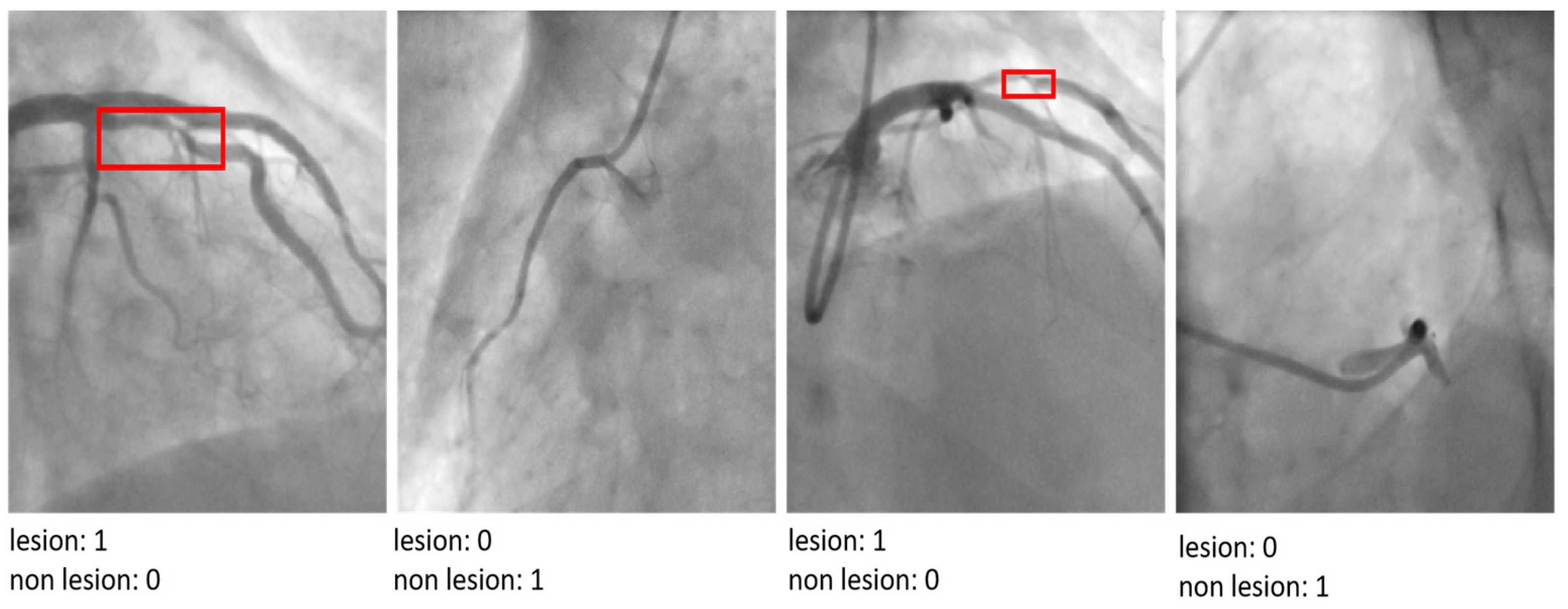
| Parameter | Value/Setting | Assumptions/Rationale |
|---|---|---|
| Input Image Size | 512 × 512 pixels | Large images ensure detailed feature extraction of coronary lesions while preserving diagnostic information. |
| Learning Rate | 0.001 | A low learning rate stabilizes training and prevents overshooting the optimal solution. |
| Batch Size | 16 | Selected to balance memory efficiency and gradient estimation stability. |
| Optimizer | Adam | Chosen for its ability to adapt learning rates dynamically for faster convergence. |
| Loss Function | Binary Cross-Entropy | Suitable for binary classification tasks like lesion detection (lesion vs. non-lesion). |
| Epochs | 50 | Early stopping was applied to terminate training if validation performance plateaued. |
| Weight Initialization | He Initialization | Optimized for deep networks with ReLU activation functions to address vanishing gradients. |
| Data Augmentation | Rotation, Scaling, Flipping, Elastic Deformations | Enhances model robustness to real-world variations in imaging conditions. |
| Cross-Validation | 5-fold Patient-Level Split | Ensures robust evaluation by preventing information leakage and simulating real-world scenarios. |
| Number of Residual Blocks | 2 | Balances model depth and computational efficiency to maintain lightweight architecture. |
| Metric | Formulation Description |
|---|---|
| Accuracy | |
| Sensitivity (Recall) | |
| Specificity | |
| Precision | |
| F1-Score | |
| Area Under the ROC Curve (AUC-ROC) | Area under the curve of the Receiver Operating Characteristic; higher values indicate better performance. |
| Number of parameters | The number of parameters in a neural network indicates the total count of trainable weights and biases in the model. |
| Floating Point Operations Per Second FLOPs | FLOPs are calculated to understand the computational complexity of processing a single input through the network. |
| Challenge | Solution | Description |
|---|---|---|
| Risk of Information Leakage | Patient-level data split | The dataset was split by patients, not images, ensuring no overlap of patient data across training, validation, and test sets. This approach eliminates the possibility of the model learning patient-specific features that could bias results. |
| Overfitting | Extensive data augmentation | Applied spatial (rotation, flipping, scaling), color (brightness, contrast), and elastic deformation augmentations to increase diversity in the training set while maintaining biological plausibility. |
| Dataset Size and Diversity | Cross-validation at patient level | Performed k-fold cross-validation with patient-level splits, ensuring that each fold used distinct patient data. This robust evaluation method provided insights into model generalization across different patient subsets. |
| Model Complexity | Regularization techniques (dropout, weight decay) | Integrated dropout layers and weight decay to constrain the model and reduce the risk of overfitting on the small dataset. |
| Training Efficiency | Early stopping | Monitored validation loss and implemented early stopping to terminate training once the model’s performance began to degrade, preventing over-optimization. |
| Evaluation Reliability | Independent test set | Reserved a separate subset of patient data exclusively for final testing. This ensured that the test set remained unseen during training and validation, providing an unbiased evaluation of the model’s diagnostic performance. |
| Feature Robustness | Explainable AI tools as Grad-CAM | Used interpretability tools to confirm that the model relied on relevant image features and not spurious patterns or artifacts, ensuring the model learned clinically meaningful representations. |
| Model | Accuracy | Recall | Precision | F1-Score | AUC-ROC | Parameters (M) | FLOPs (G) |
|---|---|---|---|---|---|---|---|
| Ref. [27] | 0.87 | 0.87 | 0.82 | 0.85 | 0.88 | 23.6 | 4.1 |
| Ref. [28] | 0.87 | 0.86 | 0.80 | 0.83 | 0.89 | 138.4 | 15.3 |
| Ref. [29] | 0.87 | 0.86 | 0.83 | 0.84 | 0.89 | 7.98 | 2.8 |
| Ref. [32] | 0.84 | 0.82 | 0.81 | 0.80 | 0.87 | 1.3 | 0.8 |
| Ref. [33] | 0.88 | 0.88 | 0.86 | 0.86 | 0.91 | 5.3 | 0.4 |
| Proposed Model | 0.903 | 0.890 | 0.90 | 0.90 | 0.912 | 1.2 | 3.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdusalomov, A.; Mirzakhalilov, S.; Umirzakova, S.; Kalandarov, I.; Mirzaaxmedov, D.; Meliboev, A.; Cho, Y.I. Optimized Lightweight Architecture for Coronary Artery Disease Classification in Medical Imaging. Diagnostics 2025, 15, 446. https://doi.org/10.3390/diagnostics15040446
Abdusalomov A, Mirzakhalilov S, Umirzakova S, Kalandarov I, Mirzaaxmedov D, Meliboev A, Cho YI. Optimized Lightweight Architecture for Coronary Artery Disease Classification in Medical Imaging. Diagnostics. 2025; 15(4):446. https://doi.org/10.3390/diagnostics15040446
Chicago/Turabian StyleAbdusalomov, Akmalbek, Sanjar Mirzakhalilov, Sabina Umirzakova, Ilyos Kalandarov, Dilmurod Mirzaaxmedov, Azizjon Meliboev, and Young Im Cho. 2025. "Optimized Lightweight Architecture for Coronary Artery Disease Classification in Medical Imaging" Diagnostics 15, no. 4: 446. https://doi.org/10.3390/diagnostics15040446
APA StyleAbdusalomov, A., Mirzakhalilov, S., Umirzakova, S., Kalandarov, I., Mirzaaxmedov, D., Meliboev, A., & Cho, Y. I. (2025). Optimized Lightweight Architecture for Coronary Artery Disease Classification in Medical Imaging. Diagnostics, 15(4), 446. https://doi.org/10.3390/diagnostics15040446








