Training State-of-the-Art Deep Learning Algorithms with Visible and Extended Near-Infrared Multispectral Images of Skin Lesions for the Improvement of Skin Cancer Diagnosis
Abstract
1. Introduction
2. Recent Literature
3. Methods
3.1. Multispectral System and Dataset
3.2. Data Preprocessing and Augmentation
3.3. Training Strategy Selection
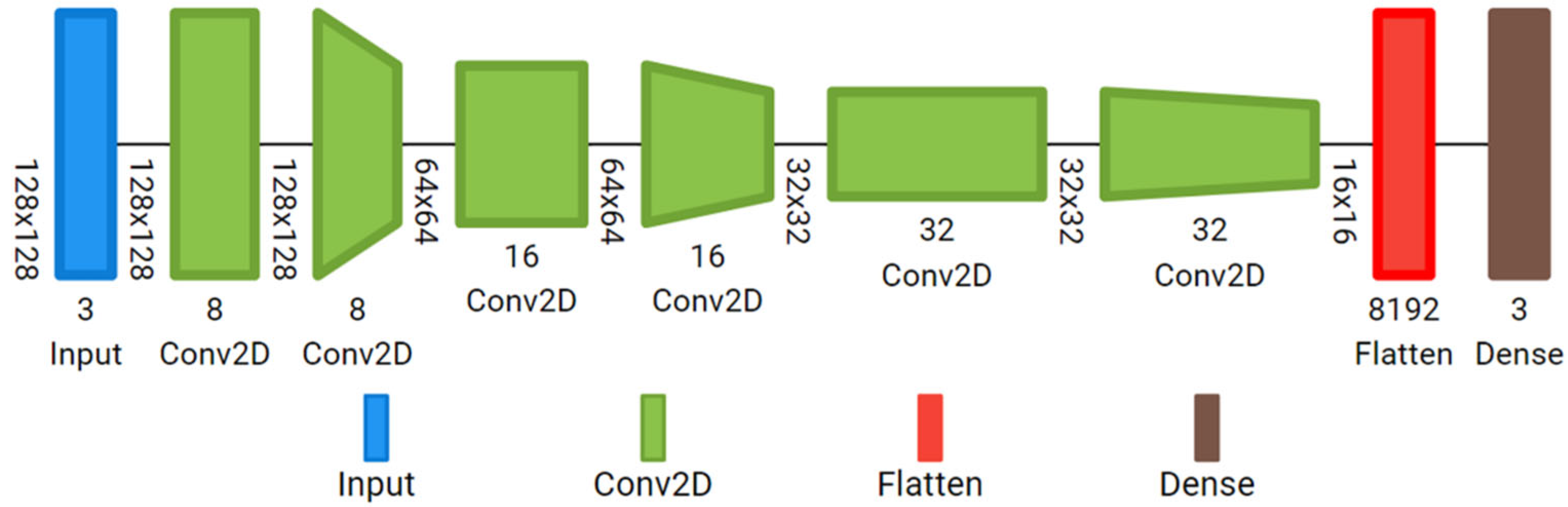
3.4. Deep Learning Architectures
3.5. Performance Metrics
3.6. Implementation
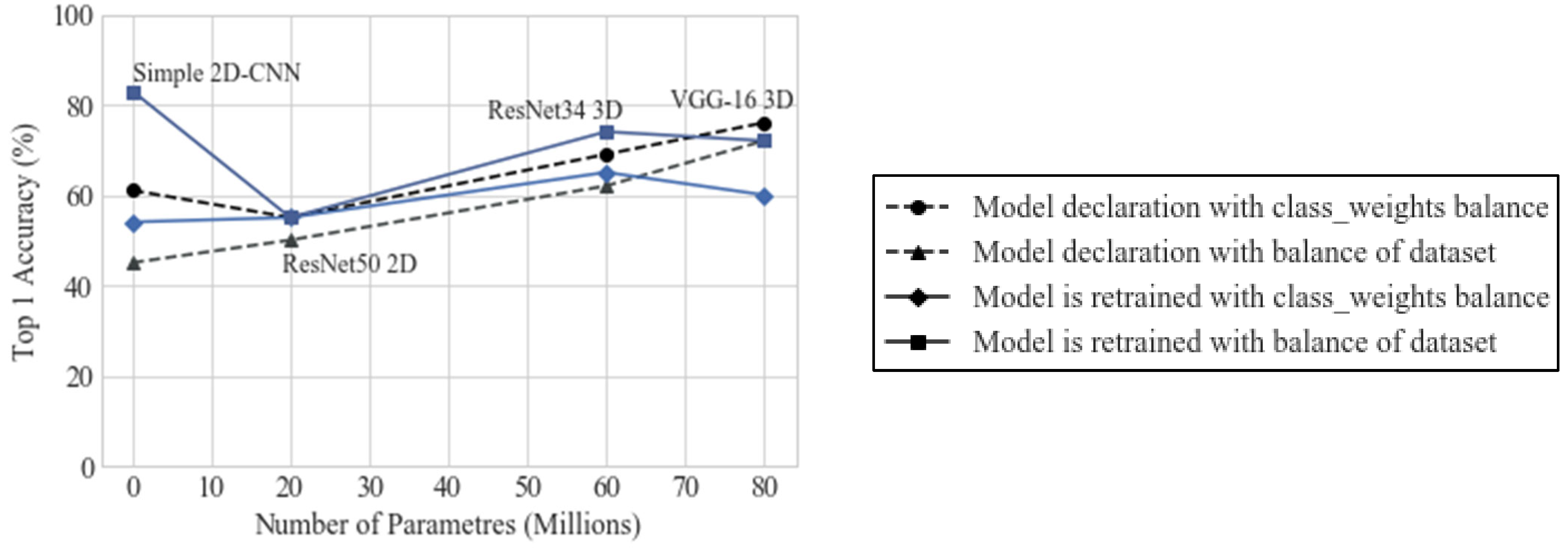
4. Results
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Glossary
| Multispectral Imaging (MSI) | A technique that captures image data at different wavelengths across the electromagnetic spectrum, allowing for the analysis of the spectral information of each pixel. In dermatology, MSI is used to enhance skin lesion analysis by identifying chromophore variations. |
| Convolutional Neural Network (CNN) | A type of Deep Learning model particularly effective for image recognition tasks. CNNs use convolutional layers to extract spatial features, making them suitable for medical image classification, such as distinguishing between types of skin lesions. |
| VGG-16 Model | A specific CNN architecture with 16 layers, often used in image classification. This model processes input images with high spatial resolution, making it effective for detailed medical image analysis. |
| Reflectance Cube | In MSI, a 3D dataset containing images captured at various spectral bands. Each ’slice’ of the cube corresponds to a different wavelength, providing detailed information on how skin reflects light at each band. |
| Data Augmentation | A technique to artificially increase the size of a dataset by applying transformations (such as rotation, flipping, etc.) to existing images. This helps reduce overfitting and improves model generalization, especially in small datasets. |
| Cross-Validation | A statistical method used to evaluate a model’s performance by dividing the dataset into multiple subsets. The model is trained on some subsets and validated on the remaining ones, improving reliability and reducing bias in performance metrics. |
| Spectral Bands | Specific wavelength ranges within the electromagnetic spectrum used in MSI. For skin lesion analysis, spectral bands capture the unique absorption properties of skin chromophores (e.g., melanin, hemoglobin). |
| Attention Maps (Saliency Maps) | Visualizations that show which areas of an image a neural network focuses on during classification. In skin lesion analysis, attention maps highlight lesion regions over the surrounding skin, validating the model’s attention. |
| Binary Classification | A classification task where the model assigns one of two labels to an input. In this context, binary classification refers to distinguishing between benign and malignant lesions. |
| Categorical Cross-Entropy (CE) | A loss function used in classification tasks with multiple classes, measuring the difference between predicted and actual class probabilities. Lower CE indicates a better-performing model. |
| Accuracy | A metric that represents the proportion of correctly classified instances out of the total cases. It is a primary metric in evaluating model performance for lesion classification. |
| Sensitivity (SE) | Also known as recall, it measures the model’s ability to correctly identify true positive cases (e.g., malignant lesions). High sensitivity is crucial in medical diagnostics to avoid missing critical cases. |
| Specificity (SP) | The ability of a model to correctly identify true negatives (e.g., benign lesions). High specificity reduces the number of false positives, which is important to prevent unnecessary biopsies. |
| Precision (P) | A measure of how many of the model’s positive predictions are actually correct. High precision indicates fewer false positives, which is valuable for accurate diagnosis. |
| F1 Score | The harmonic mean of precision and sensitivity, providing a balanced measure of a model’s accuracy in binary classification tasks, particularly in imbalanced datasets. |
| Crossed Polarizers | Optical filters used in MSI to reduce specular reflections from skin, enhancing the quality of the captured images by minimizing the glare from sweat or skin oils. |
| Random Grid Search | A hyperparameter tuning method where values are randomly selected within a specified range to optimize model performance without an exhaustive search. |
| Transfer Learning | A Deep Learning technique where a pre-trained model is fine-tuned on a new, smaller dataset. It is generally avoided in MSI with skin images due to the unique spectral characteristics of these data. |
| Learning Rate (LR) | A hyperparameter in Deep Learning that controls the step size at each iteration of optimization. The learning rate is crucial for model convergence and stability. |
References
- Goodson, A.G.; Grossman, D. Strategies for early melanoma detection: Approaches to the patient with nevi. J. Am. Acad. Dermatol. 2009, 60, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Guy, G.P.; Ekwueme, D.U.; Tangka, F.K.; Richardson, L.C. Melanoma Treatment Costs. Am. J. Prev. Med. 2012, 43, 537–545. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: https://www.who.int/en/ (accessed on 19 October 2020).
- Wolff, K.; Allen, J.R. Fitzpatrick’s Color Atlas and Synopsis of Clinical Dermatology; McGraw-Hill Professional: New York, NY, USA, 2009; pp. 154–196. [Google Scholar]
- American Cancer Society. Available online: https://www.cancer.org (accessed on 19 October 2020).
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Li, Y.; Shen, L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Ellinger, I.; Ecker, R.; Pitiot, A.; Wang, C. Fusing fine-tuned deep features for skin lesion classification. Comput. Med. Imaging Graph. 2019, 71, 19–29. [Google Scholar] [CrossRef]
- Harangi, B. Skin lesion classification with ensembles of deep convolutional neural networks. J. Biomed. Inform. 2018, 86, 25–32. [Google Scholar] [CrossRef]
- Serte, S.; Demirel, H. Wavelet-based deep learning for skin lesion classification. IET Image Process. 2019, 14, 720–726. [Google Scholar] [CrossRef]
- Dorj, U.O.; Lee, K.K.; Choi, J.Y.; Lee, M. The skin cancer classification using deep convolutional neural network. Multimed. Tools Appl. 2018, 77, 9909–9924. [Google Scholar] [CrossRef]
- Mandache, D.; Dalimier, E.; Durkin, J.R.; Boceara, C.; Olivo-Marin, J.-C.; Meas-Yedid, V. Basal cell carcinoma detection in full field OCT images using convolutional neural networks. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 784–787. [Google Scholar] [CrossRef]
- Milton, M.A.A. Automated Skin Lesion Classification Using Ensemble of Deep Neural Networks in ISIC 2018: Skin Lesion Analysis Towards Melanoma Detection Challenge. arXiv 2019, arXiv:1901.10802. [Google Scholar] [CrossRef]
- Mendes, D.B.; da Silva, N.C. Skin Lesions Classification Using Convolutional Neural Networks in Clinical Images. arXiv 2018, arXiv:1812.02316. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Saba, T.; Khan, M.A.; Rehman, A.; Marie-Sainte, S.L. Region Extraction and Classification of Skin Cancer: A Heterogeneous framework of Deep CNN Features Fusion and Reduction. J. Med. Syst. 2019, 43, 289. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Chao, M.; Lo, Y.-C. Deep Fully Convolutional Networks with Jaccard Distance. IEEE Trans. Med. Imaging 2017, 36, 1876–1886. [Google Scholar] [CrossRef] [PubMed]
- Refianti, R.; Mutiara, A.B.; Priyandini, R.P. Classification of melanoma skin cancer using convolutional neural network. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 409–417. [Google Scholar] [CrossRef]
- Maiti, A.; Chatterjee, B. Improving detection of Melanoma and Naevus with deep neural networks. Multimed. Tools Appl. 2020, 79, 15635–15654. [Google Scholar] [CrossRef]
- Premaladha, J.; Ravichandran, K.S. Novel Approaches for Diagnosing Melanoma Skin Lesions Through Supervised and Deep Learning Algorithms. J. Med. Syst. 2016, 40, 96. [Google Scholar] [CrossRef]
- Rodrigues, D.A.; Ivo, R.F.; Satapathy, S.C.; Wang, S.; Hemanth, J.; Filho, P.P.R. A new approach for classification skin lesion based on transfer learning, deep learning, and IoT system. Pattern Recognit. Lett. 2020, 136, 8–15. [Google Scholar] [CrossRef]
- Sagar, A.; Jacob, D. Convolutional Neural Networks for Classifying Melanoma Images. bioRxiv 2020. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Codella, N.; Cai, J.; Abedini, M.; Garnavi, R.; Halpern, A.; Smith, J.R. Deep Learning, Sparse Coding, and SVM for Melanoma Recognition in Dermoscopy Images. In International Workshop on Machine Learning in Medical Imaging; Springer International Publishing: Cham, Switzerland, 2015; pp. 118–126. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.-A. Automated Melanoma Recognition in Dermoscopy Images via Very Deep Residual Networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef]
- Adegun, A.A.; Viriri, S. Deep learning-based system for automatic melanoma detection. IEEE Access 2020, 8, 7160–7172. [Google Scholar] [CrossRef]
- The International Skin Imaging Collaboration (ISIC). Available online: https://www.isic-archive.com (accessed on 25 July 2022).
- Jones, O.T.; Matin, R.N.; van der Schaar, M.; Bhayankaram, K.P.; Ranmuthu CK, I.; Islam, M.S.; Behiyat, D.; Boscott, R.; Calanzani, N.; Emery, J.; et al. Artificial intelligence and machine learning algorithms for early detection of skin cancer in community and primary care settings: A systematic review. Lancet Digit. Health 2022, 4, e466–e476. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Jain, S.; Singhania, U.; Tripathy, B.; Nasr, E.A.; Aboudaif, M.K.; Kamrani, A.K. Deep learning-based transfer learning for classification of skin cancer. Sensors 2021, 21, 8142. [Google Scholar] [CrossRef]
- Maniraj, S.; Maran, P.S. A hybrid deep learning approach for skin cancer diagnosis using subband fusion of 3D wavelets. J. Supercomput. 2022, 78, 12394–12409. [Google Scholar] [CrossRef]
- Abbas, Q.; Gul, A. Detection and Classification of Malignant Melanoma Using Deep Features of NASNet. SN Comput. Sci. 2022, 4, 21. [Google Scholar] [CrossRef]
- Ho, C.; Calderon-Delgado, M.; Chan, C.; Lin, M.; Tjiu, J.; Huang, S.; Chen, H.H. Detecting mouse squamous cell carcinoma from submicron full-field optical coherence tomography images by deep learning. J. Biophotonics 2021, 14, e202000271. [Google Scholar] [CrossRef]
- Jojoa-Acosta, M.F.; Tovar LY, C.; Garcia-Zapirain, M.B.; Percybrooks, W.S. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med. Imaging 2021, 21, 6. [Google Scholar] [CrossRef]
- Rey-Barroso, L.; Peña-Gutiérrez, S.; Yáñez, C.; Burgos-Fernández, F.J.; Vilaseca, M.; Royo, S. Optical technologies for the improvement of skin cancer diagnosis: A review. Sensors 2021, 21, 252. [Google Scholar] [CrossRef]
- Campanella, G.; Navarrete-Dechent, C.; Liopyris, K.; Monnier, J.; Aleissa, S.; Minhas, B.; Scope, A.; Longo, C.; Guitera, P.; Pellacani, G.; et al. Deep Learning for Basal Cell Carcinoma Detection for Reflectance Confocal Microscopy. J. Investig. Dermatol. 2022, 142, 97–103. [Google Scholar] [CrossRef]
- Lihacova, I.; Bondarenko, A.; Chizhov, Y.; Uteshev, D.; Bliznuks, D.; Kiss, N.; Lihachev, A. Multi-Class CNN for Classification of Multispectral and Autofluorescence Skin Lesion Clinical Images. J. Clin. Med. 2022, 11, 2833. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Hsiao, Y.-P.; Mukundan, A.; Tsao, Y.-M.; Chang, W.-Y.; Wang, H.-C. Classification of Skin Cancer Using Novel Hyperspectral Imaging Engineering via YOLOv5. J. Clin. Med. 2023, 12, 1134. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.L.; Lu, C.T.; Karmakar, R.; Nampalley, K.; Mukundan, A.; Hsiao, Y.P.; Hsieh, S.C.; Wang, H.C. Assessing the efficacy of the Spectrum-Aided Vision Enhancer (SAVE) to detect acral lentiginous melanoma, melanoma in situ, nodular melanoma, and superficial spreading melanoma. Diagnostics 2024, 14, 1672. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Ding, Y.; Jiang, H.; Thiers, B.H.; Wang, J.Z. Automatic diagnosis of melanoma using machine learning methods on a spectroscopic system. BMC Med. Imaging 2014, 14, 36. [Google Scholar] [CrossRef]
- Godoy, S.E.; Ramirez, D.A.; Myers, S.A.; von Winckel, G.; Krishna, S.; Berwick, M.; Padilla, R.S.; Sen, P.; Krishna, S. Dynamic infrared imaging for skin cancer screening. Infrared Phys. Technol. 2015, 70, 147–152. [Google Scholar] [CrossRef]
- Kim, S.; Cho, D.; Kim, J.; Kim, M.; Youn, S.; Jang, J.E.; Je, M.; Lee, D.H.; Lee, B.; Farkas, D.L.; et al. Smartphone-based multi-spectral imaging: System development and potential for mobile skin diagnosis. Biomed. Opt. Express 2016, 7, 5294. [Google Scholar] [CrossRef]
- Delpueyo, X.; Vilaseca, M.; Royo, S.; Ares, M.; Rey-Barroso, L.; Sanabria, F.; Puig, S.; Malvehy, J.; Pellacani, G.; Noguero, F.; et al. Multispectral imaging system based on light-emitting diodes for the detection of melanomas and basal cell carcinomas: A pilot study. J. Biomed. Opt. 2017, 22, 065006. [Google Scholar] [CrossRef]
- Rey-Barroso, L.; Burgos-Fernández, F.; Delpueyo, X.; Ares, M.; Royo, S.; Malvehy, J.; Puig, S.; Vilaseca, M. Visible and Extended Near-Infrared Multispectral Imaging for Skin Cancer Diagnosis. Sensors 2018, 18, 1441. [Google Scholar] [CrossRef]
- Romero Lopez, A.; Giro-i-Nieto, X.; Burdick, J.; Marques, O. Skin lesion classification from dermoscopic images using deep learning techniques. In Proceedings of the 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20–21 February 2017; pp. 49–54. [Google Scholar] [CrossRef]
- De Lucena, D.V.; da Silva Soares, A.; Coelho, C.J.; Wastowski, I.J.; Filho, A.R.G. Detection of Tumoral Epithelial Lesions Using Hyperspectral Imaging and Deep Learning. In Proceedings of the Computational Science–ICCS 2020: 20th International Conference, Amsterdam, The Netherlands, 3–5 June 2020; Springer International Publishing: Cham, Switzerland, 2020; pp. 599–612. [Google Scholar] [CrossRef]
- La Salvia, M.; Torti, E.; Leon, R.; Fabelo, H.; Ortega, S.; Balea-Fernandez, F.; Martinez-Vega, B.; Castaño, I.; Almeida, P.; Carretero, G.; et al. Neural Networks-Based On-Site Dermatologic Diagnosis through Hyperspectral Epidermal Images. Sensors 2022, 22, 7139. [Google Scholar] [CrossRef]
- Igareta, A. Method to Generate Class Weights Given a Multi-Class or Multi-Label Set of Classes Using Python, Supporting One-Hot-Encoded Labels. 2021. Available online: https://gist.github.com/angeligareta/83d9024c5e72ac9ebc34c9f0b073c64c (accessed on 10 July 2022).
- Keras API Reference: Keras Applications ResNet and ResNetV2. Available online: https://keras.io/api/applications/resnet/ (accessed on 20 July 2022).
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Neptune.ai. Data Augmentation in Python: Everything You Need to Know by Vladimir Lyashenko. Available online: https://neptune.ai/blog/data-augmentation-in-python (accessed on 4 November 2023).
- Perez, F.; Vasconcelos, C.; Avila, S.; Valle, E. Data Augmentation for Skin Lesion Analysis. In Proceedings of the 2018 Context-Aware Operating Theaters 2018, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis, Proceedings of the CARE CLIP OR 2.0 ISIC 2018, Granada, Spain, 16–20 September 2018; Springer: Cham, Switzerland, 2018; Volume 11041. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Kaggle. Kaggle: Your Machine Learning and Data Science Community. Topic: Why No Augmentation Applied to Test or Validation Data and Only to Train Data? Available online: https://www.kaggle.com/questions-and-answers/291581 (accessed on 5 November 2023).
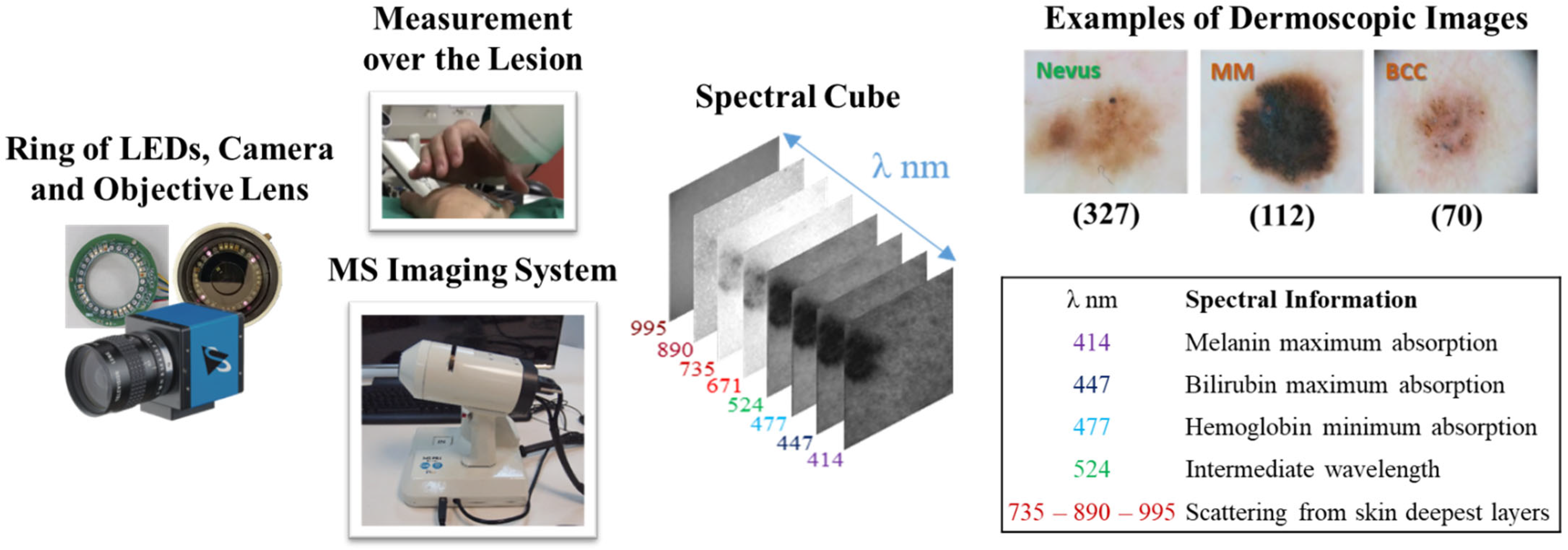
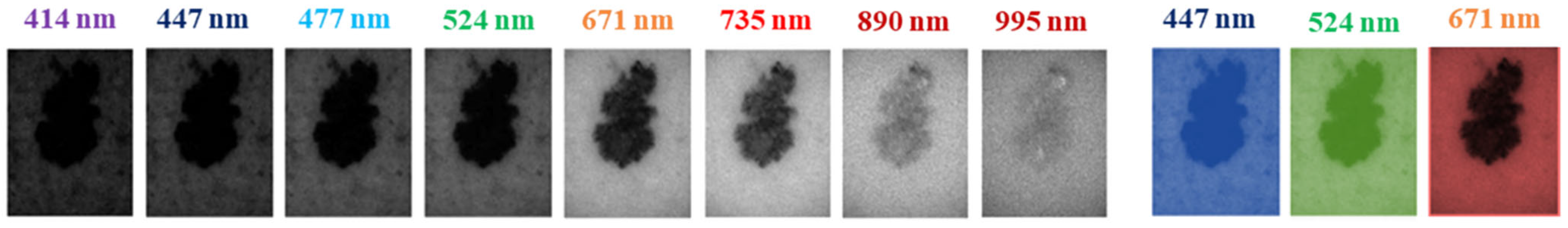
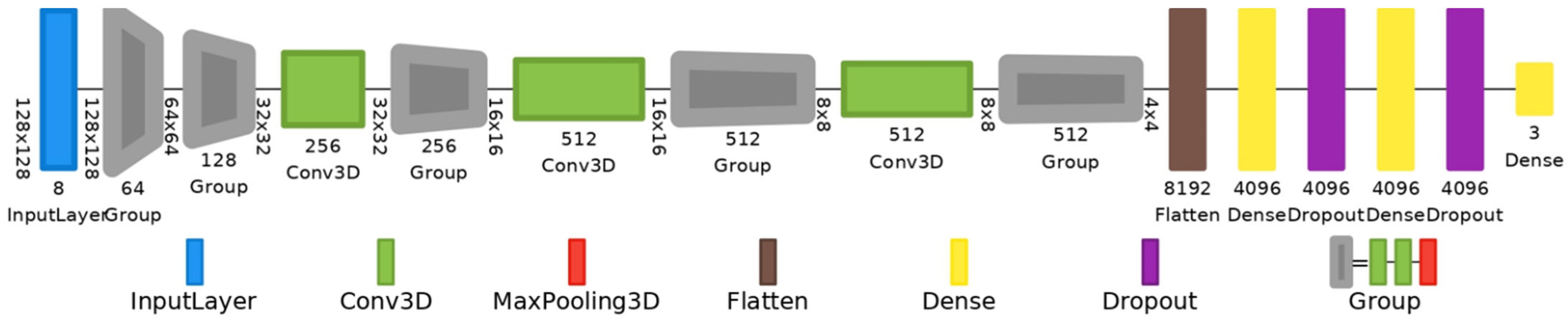
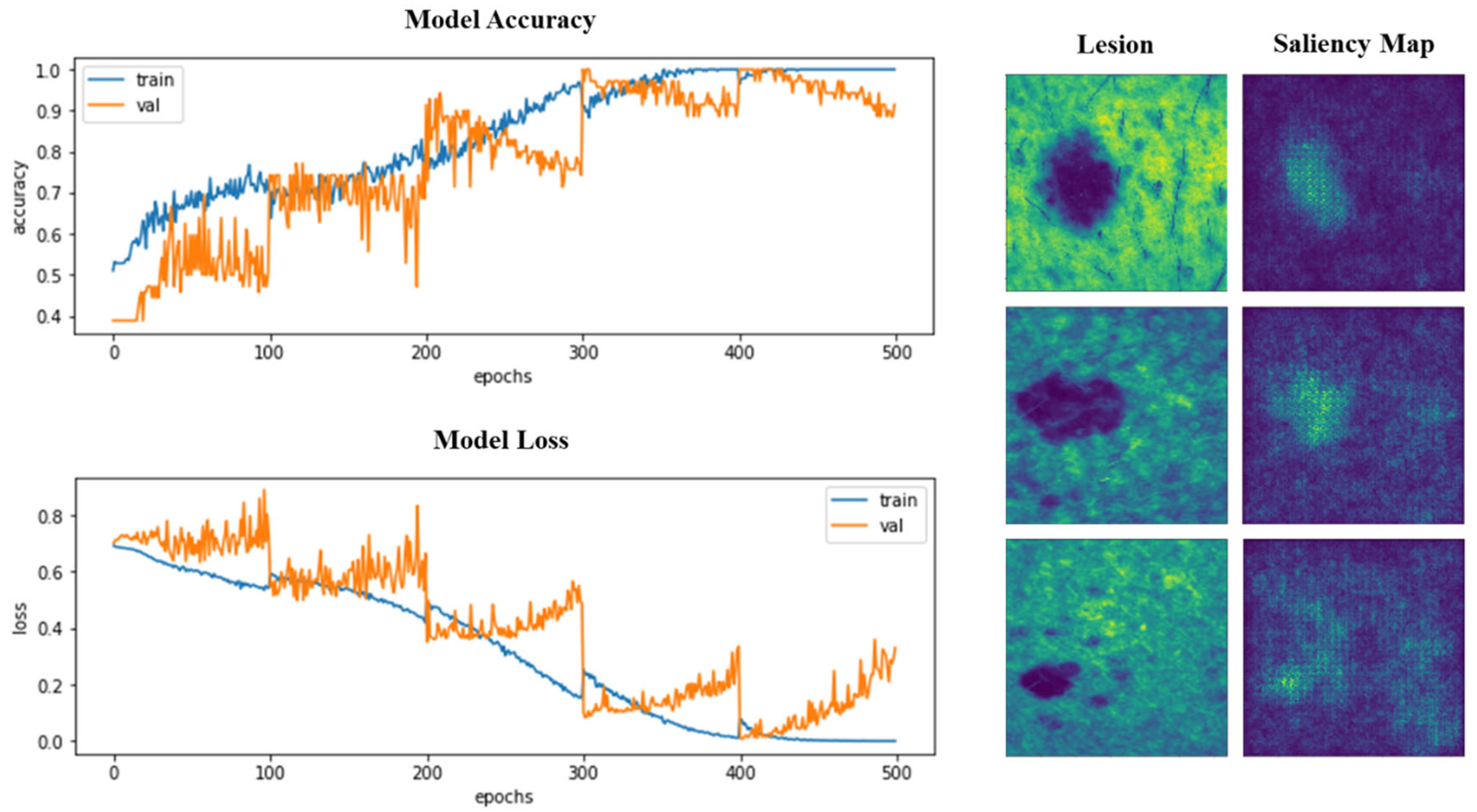
| Dataset Samples | Model | Approach | Data Splitting (%) | Accuracy | AUC | SE | SP | |
|---|---|---|---|---|---|---|---|---|
| Harangi et al., 2018 [9] | ISIC 2017 (2600): MM (491), Nevi (1765), SK (344). | Ensembled CNN out of AlexNet, VGGNet, GoogLeNet. | 1. Aggregation of robust CNNs. 2. Feature extraction. | Training, validation (80%). Test (20%). | 0.85 MM, 0.88 SK | 0.85 MM, 0.93 SK | 0.40 MM, 0.71 SK | 0.72 MM, 0.85 SK |
| Mahbod et al., 2018 [8] | ISIC 2016 + 2017 (2787): MM (518), SK (396), B. lesions (2389). | Pre-trained ensembled CNNs. | 1. Color normalization + Mean RGB value subtraction + Data augmentation. 2. Ensembled CNNs for feature extraction and SVM as classifier. | Training, validation (80%). Test (20%). | - | 0.73 MM, 0.93 SK (for pre-trained) | 0.87 MM, 0.96 SK (for fine-tuned) | - | - |
| Mandache et al., 2018 [12] | FF-OCT images (40–108,082 patches): BCC (48,970), H. skin (59,112). | VGG-16 and InceptionV3 pre-trained on ImageNet. CNN of 10 layers (trained from scratch). | Feature extraction and classification. | Training, validation (80%): Test (20%). | 0.96 (proposed CNN)-0.89 (VGG-16)-0.91 (InceptionV3) | - | - | - |
| Refianti et al., 2019 [18] | ISIC 2017 (198): MM (99), non-MMC (99). | LeNet-5. | 1. Data augmentation. 2. Feature extraction with CNN. | Training, validation (80%): Test (20%) | 0.95 | - | 0.91 | - |
| Saba et al., 2019 [16] | ISIC 2016 + 2017 + PH2 (4229): B. lesions, Nevi, MM. | InceptionV3. | 1. Contrast enhancement + HSV color transformation + lesion boundary extraction (segmentation). 2. Feature extraction with Inception V3 | Cross-validation kfold = 20. Training, validation (70%): Test (30%). | 0.98 (averaged-PH2), 0.95 (ISIC 2016), 0.95 (ISIC 2017-best) | 0.98 (ISIC 2017-best) | 0.95 (ISIC 2017-best) | 0.98 (ISIC 2017-best) |
| Serte et al., 2019 [10] | ISIC 2017 (2750): MM, SK, M. Nevi. | Pre-trained ensemble of ResNet-18 and ResNet50. | 1. Gray-scale transformation. 2. Implementation of wavelet transform (WT = FT) + Data augmentation of MM and SK. 3. Fine-tuning with WT and images + Model fusing. | Training and validation (80%): Test (20%). | 0.84 MM, 0.79 SK | - | 0.96 MM, 0.81 SK | - |
| Adegun et al., 2020 [28] | ISIC 2017 + PH2 (2860): MM, non-MMC. | Encoder-decoder network. | 1. Remove of noise (hair, artifacts) + Zero mean unit variance normalization + Data augmentation. 2. Multi-stage and multi-scale pixel-wise classification of lesions. | Training, validation (80%): Test (20%): 600 ISIC + 60 PH2. | 0.92 ISIC, 0.93 PH2 | - | - | - |
| Maiti et al., 2020 [19] | ISIC 2017 + MED NODE-(2170): MM (1070), Nevi (1100). | AlexNet, VGG custom CNN. | 1. Contrast enhancement + Segmentation. 2. Feature extraction with CNNs. | Training and validation (100%) No test set. | 0.72 (AlexNet), 0.68 (VGGNet), 0.97 (custom CNN-best) | - | - | - |
| Rodrigues et al., 2020 [21] | ISIC 2017 + PH2-(1100): MM (174), Nevi (726), MM (40), C. Nevi (80), A. Nevi (80). | Pre-trained VGG, Inception, ResNet, Inception-ResNet, Xception, MobileNet, DenseNet, NASNet. | 1. Fine-tuning for feature extraction. 2. Use of classic classifiers: Bayes, MLP, SVM, KNN, and RF. 3. IoT system. | 5 instances for each combination of CNN and classifier. Training, validation (90%): Test (10%). | 0.97 ISIC, 0.93 PH2 (DenseNet20 and KNN-best) | - | 0.97 ISIC, 0.93 PH2 | - |
| Ho et al., 2021 [35] | FF-OCT tomograms-(297–130,383): SCC (43,900), Dysplasia (42,583), H. skin (43,900). | ResNet-18 | 1. Model training. 2. Heat map extraction. | 10-fold cross-validation. Training, validation (85%). Test (15%). | 0.81 | - | - | - |
| Jojoa-Acosta et al., 2021 [36] | ISIC 2017-(2742): B. lesions (2220), M. lesions (522). | ResNet152 | 1. ROI extraction using the Mask and Region-based CNN. 2. Data augmentation balancing lesion ratio. 3. Fine-tuning of ResNet152 for feature extraction and classification. | Training, validation (90%). Test (10%). | 0.91 | . | 0.87 (over MM) | - |
| Mendes et al., 2021 [14] | MED-NODE + Edinburgh + Atlas-(3816): 12 lesions classes including MM, Nevi, BCC and SCC. | Pre-trained ResNet-152 | 1. Data augmentation over training and validation. 2. Fine-tuning of the model. | Training, validation (80%). Test (20%). | 0.78 | - | - | - |
| Abbas and Gul, 2022 [34] | ISIC 2020 (30,000+ images, subset of 4000 images used: Melanoma: 584, Nevus: 2000, Others: 1416). | NASNet (Modified) with global average pooling, fine-tuned. | 1. Transfer learning from NASNet pre-trained on ImageNet. 2. Label-preserving augmentations applied (rotation, flipping). 3. Data pre-processing includes ROI cropping and artifact removal. | Training: 75%, Testing: 25% | 0.98 | - | 0.98 | 0.98 |
| Authors | Dataset Samples | MS Sensitivity | Model | Approach | Data Splitting (%) | Accuracy | AUC | SE | SP |
|---|---|---|---|---|---|---|---|---|---|
| de Lucena et al., 2020 [48] | SWIR spectroscopy images (35): MM (12 samples, 34 parts), D. nevi (72 parts), H. skin (17 parts). | IR: 900 nm–2500 nm (256 spectral bands). | RetinaNet (Resnet50 model on backbone). | 1. Reduction in the spectral dimension of each SWIR. 2. Feature extraction and classification with RetinaNet. | Training, validation and test. | 0.688 MM, 0.725 Nevi | - | - | - |
| La Salvia et al., 2022 [49] | HS images (76–125 bands): MM-like lesions, ME lesions, BM lesions, BE lesions. | VIS-exNIR: 450 nm–950 nm (76 spectral bands). | Pre-trained ResNet-18, ResNet-50, ResNet-101, and a ResNet-50 variant, which exploits 3D convolutions. | 1. Segmentation with U-Net, U-Net++, and two other networks. 2. The best results are fed to ResNets for feature extraction and classification + Data augmentation. | Both binary classification (B. vs. M. lesions) and multiclass. 10-fold cross-validation. Results are calculated over validation folds and averaged. | - | 0.46 MM, 0.16 ME, 0.46 BM, 0.35 BE | 0.91 (binary | 0.50 MM, 0.88 ME, 0.79 BM, 0.75 BE | 0.88 (binary) | 0.98 MM, 0.83 ME, 0.90 BM, 0.93 BE | 0.89 (binary) |
| Lihacova et al., 2022 [39] | MS images (1304–4 images): MM-like lesions (74), PB lesions (405), HK lesions (323), non-MMC (172), other B. lesions (330). | VIS-exNIR: 526 nm, 663 nm and 994 nm (3 spectral bands) + AF image under 405 nm. | Pre-trained InceptionV3, VGG-16 and ResNet-50. DARTS custom model trained from scratch. | 1. Data augmentation. 2. Fine-tuning pre-trained models and training from scratch DARTS architecture. | 5-fold cross-validation Results are calculated over validation folds and averaged. | - | - | 0.72 MM, 0.83 PB, 0.61 HK, 0.57 non-MMC, 0.84 other benign (DARTS-best) | 0.97 MM, 0.90 PB, 0.91 HK, 0.95 non-MMC, 0.93 other benign (DARTS-best) |
| Lin et al., 2024 [41] | 878 images: Acral Lentiginous Melanoma (342), Superficial Spreading Melanoma (253), Nodular Melanoma (100), Melanoma in situ (183). | SAVE: HSI synthesized from RGB. Band selection of 415 nm, 540 nm, 600 nm, 700 nm, and 780 nm; | YOLO (v5, v8, v9), SAVE algorithm integration. | 1. RGB to HSI conversion using SAVE. 2. Training on augmented dataset (7:2:1 split). 3. Comparison across YOLO versions with metrics like precision, recall, mAP, and F1-score. | Training: 70%, Validation: 20%, Testing: 10%. | - | - | YOLO v8-SAVE: Precision > 90%, Recall 71%, mAP 0.801; YOLO v8-RGB: Precision > 84%, Recall 76%, Map 0.81 Superficial Spreading Melanoma: Precision decreases 7% in YOLO v5-SAVE, increases 1% in YOLO v8-SAVE. |
| Reflectance Cubes | ID | Diagnostic | Benign/Malignant | Reflectance Images |
|---|---|---|---|---|
| 112 | 1 | Melanoma | M | 8 |
| 1 | MIS (melanoma in situ) | M | ||
| 1 | Lentigo | M | ||
| 327 | 2 | Dermic Nevus | B | 8 |
| 2 | Dysplastic Nevus | B | ||
| 2 | Blue Nevus | B | ||
| 70 | 3 | BCC | M | 8 |
| Model | Top 1 Acc. | SE | SP | P | F1 |
|---|---|---|---|---|---|
| 2D-CNN | |||||
| Nevus | 0.83 | 0.86 | 0.88 | 0.80 | 0.83 |
| Melanoma | 0.79 | 0.96 | 0.92 | 0.85 | |
| BCC | 0.86 | 0.88 | 0.86 | 0.86 | |
| Malignant vs. Benign | 0.88 | 0.89 | 0.86 | 0.93 | 0.91 |
| 3D VGG-16 | |||||
| Nevus | 0.71 | 0.71 | 0.83 | 0.71 | 0.66 |
| Melanoma | 0.64 | 0.81 | 0.64 | 0.57 | |
| BCC | 0.79 | 0.86 | 0.79 | 0.75 | |
| Malignant vs. Benign | 0.81 | 0.71 | 0.86 | 0.85 | 0.77 |
| 3D ResNet34 | |||||
| Nevus | 0.74 | 0.79 | 0.77 | 0.65 | 0.71 |
| Melanoma | 0.50 | 1.00 | 1.00 | 0.67 | |
| BCC | 0.93 | 0.78 | 0.72 | 0.81 | |
| Malignant vs. Benign | 0.79 | 0.79 | 0.79 | 0.88 | 0.83 |
| Keras ResNet50 | |||||
| Nevus | 0.55 | 0.64 | 0.67 | 0.56 | 0.60 |
| Melanoma | 0.79 | 0.50 | 0.48 | 0.59 | |
| BCC | 0.21 | 1.00 | 1.00 | 0.35 | |
| Malignant vs. Benign | 0.71 | 0.75 | 0.64 | 0.80 | 0.77 |
| Model | Top 1 Acc. | SE | SP | P | F1 |
|---|---|---|---|---|---|
| 2D-CNN (Reflectance Cube) | |||||
| Nevus | 0.83 | 0.86 | 0.88 | 0.80 | 0.83 |
| Melanoma | 0.79 | 0.96 | 0.92 | 0.85 | |
| BCC | 0.86 | 0.88 | 0.86 | 0.86 | |
| Malignant vs. Benign | 0.88 | 0.89 | 0.86 | 0.93 | 0.91 |
| 2D-CNN (RGB bands) | |||||
| Nevus | 0.36 | 0.07 | 0.89 | 0.25 | 0.11 |
| Melanoma | 0.93 | 0.18 | 0.36 | 0.52 | |
| BCC | 0.07 | 0.96 | 0.50 | 0.13 | |
| Malignant vs. Benign | 0.62 | 0.07 | 0.89 | 0.25 | 0.11 |
| 3D VGG-16 (Reflectance Cube) | |||||
| Nevus | 0.71 | 0.71 | 0.83 | 0.71 | 0.66 |
| Melanoma | 0.64 | 0.81 | 0.64 | 0.57 | |
| BCC | 0.79 | 0.86 | 0.79 | 0.75 | |
| Malignant vs. Benign | 0.81 | 0.71 | 0.86 | 0.85 | 0.77 |
| 3D VGG-16 (RGB bands) | |||||
| Nevus | 0.45 | 0.43 | 0.62 | 0.43 | 0.43 |
| Melanoma | 0.43 | 0.87 | 0.75 | 0.55 | |
| BCC | 0.50 | 0.48 | 0.35 | 0.41 | |
| Malignant vs. Benign | 0.62 | 0.43 | 0.71 | 0.43 | 0.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rey-Barroso, L.; Vilaseca, M.; Royo, S.; Díaz-Doutón, F.; Lihacova, I.; Bondarenko, A.; Burgos-Fernández, F.J. Training State-of-the-Art Deep Learning Algorithms with Visible and Extended Near-Infrared Multispectral Images of Skin Lesions for the Improvement of Skin Cancer Diagnosis. Diagnostics 2025, 15, 355. https://doi.org/10.3390/diagnostics15030355
Rey-Barroso L, Vilaseca M, Royo S, Díaz-Doutón F, Lihacova I, Bondarenko A, Burgos-Fernández FJ. Training State-of-the-Art Deep Learning Algorithms with Visible and Extended Near-Infrared Multispectral Images of Skin Lesions for the Improvement of Skin Cancer Diagnosis. Diagnostics. 2025; 15(3):355. https://doi.org/10.3390/diagnostics15030355
Chicago/Turabian StyleRey-Barroso, Laura, Meritxell Vilaseca, Santiago Royo, Fernando Díaz-Doutón, Ilze Lihacova, Andrey Bondarenko, and Francisco J. Burgos-Fernández. 2025. "Training State-of-the-Art Deep Learning Algorithms with Visible and Extended Near-Infrared Multispectral Images of Skin Lesions for the Improvement of Skin Cancer Diagnosis" Diagnostics 15, no. 3: 355. https://doi.org/10.3390/diagnostics15030355
APA StyleRey-Barroso, L., Vilaseca, M., Royo, S., Díaz-Doutón, F., Lihacova, I., Bondarenko, A., & Burgos-Fernández, F. J. (2025). Training State-of-the-Art Deep Learning Algorithms with Visible and Extended Near-Infrared Multispectral Images of Skin Lesions for the Improvement of Skin Cancer Diagnosis. Diagnostics, 15(3), 355. https://doi.org/10.3390/diagnostics15030355







