A Novel Method for Predicting Oncogenic Types of Human Papillomavirus
Abstract
1. Introduction
2. Related Works
3. Proposed Method
3.1. Proposed Features Detecting Risk Types of HPV
| Algorithm 1 Proposed HPV risk group prediction method |
Input:
|
| Output: Risk classification of testSeq a high risk, probable high risk, or low risk Feature Extraction Step:
|
3.1.1. CpG-Island Feature
3.1.2. TATA-Box Feature
3.1.3. CAAT-Box Feature
3.1.4. K-Mer Feature
3.2. Classification Methods
3.2.1. Random Forest (RF)
3.2.2. Support Vector Machine (SVM)
3.2.3. k-Nearest Neighbor (KNN)
3.2.4. Decision Trees (DT)
3.2.5. Neural Networks (NN)
3.2.6. Adaptive Boosting (AdaBoost)
4. Experimental Results and Discussion
4.1. Experimental Setup
4.2. Human Genome Sequences Dataset
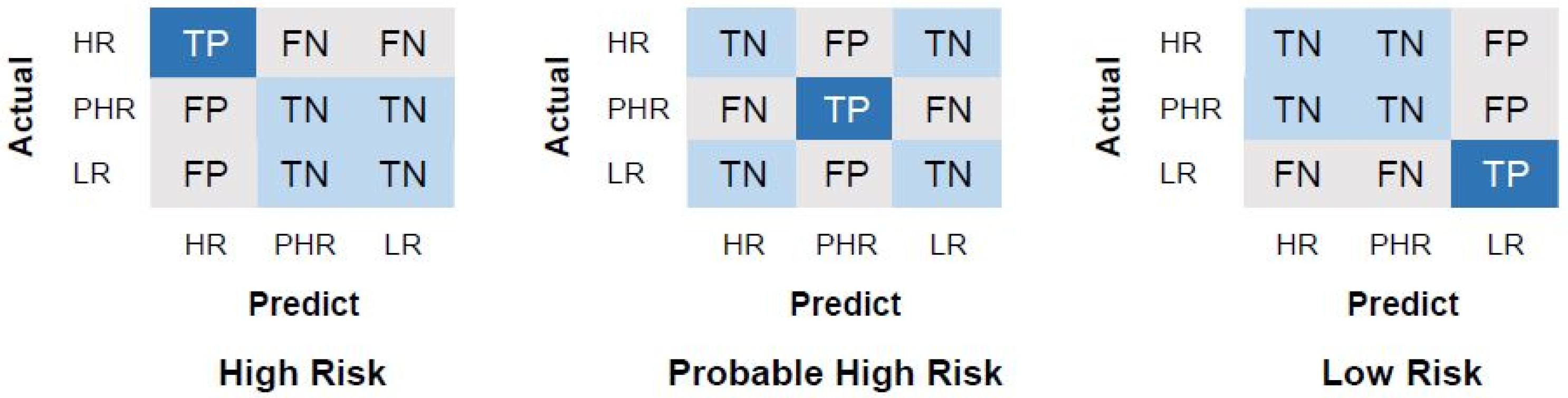
4.3. Performance Evaluation Measurements
4.4. Performance Evaluation of the Proposed CpG Feature for Predicting Risk of the HPV Types
4.5. Performance Evaluation of the Proposed CpG, CAAT-Box and TATA-Box Features for Predicting Risk of the HPV Types
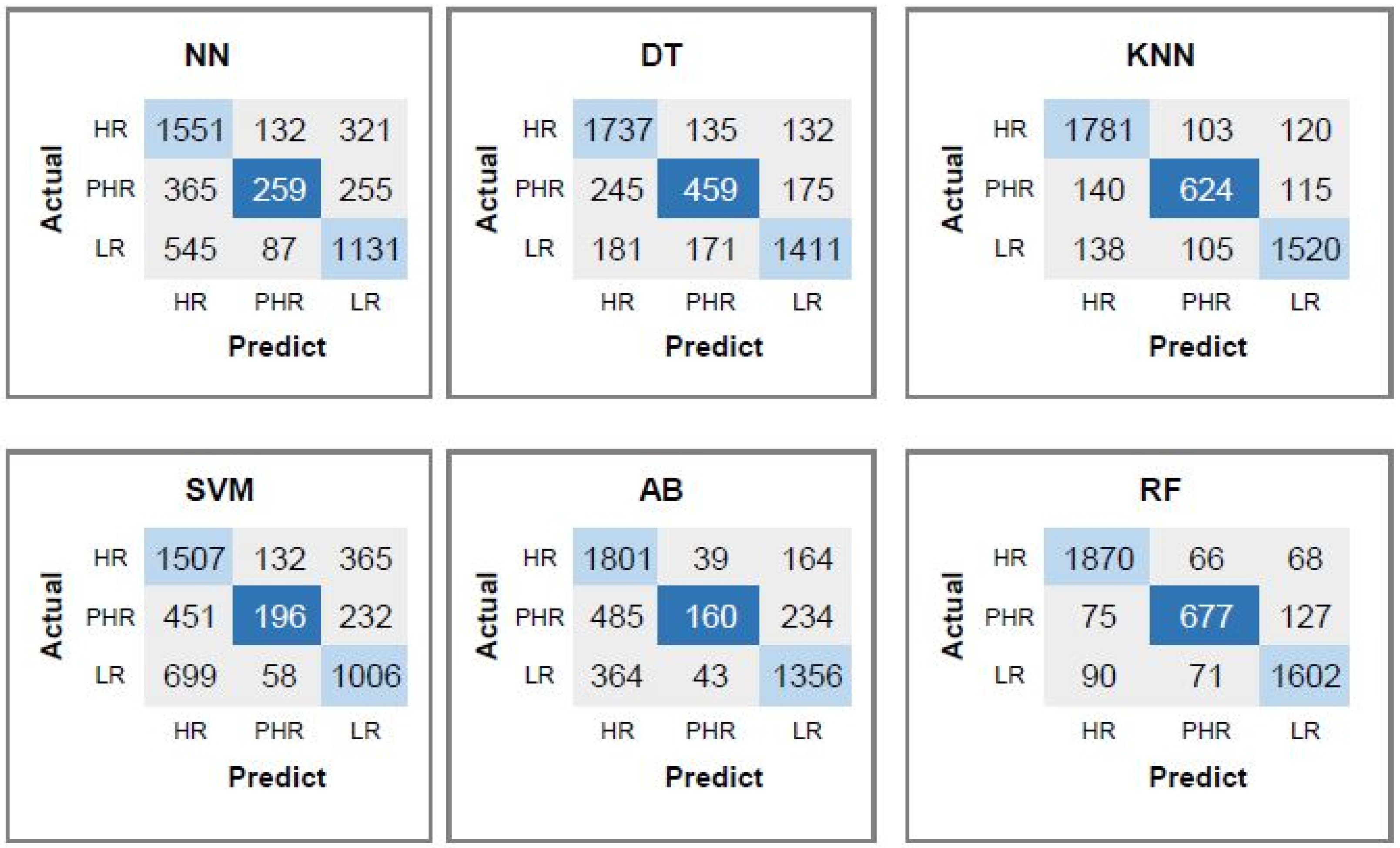
4.6. Performance Evaluation of the Proposed CpG, CAAT-Box, TATA-Box, and Dinucleotide Features for Predicting Risk of the HPV Types
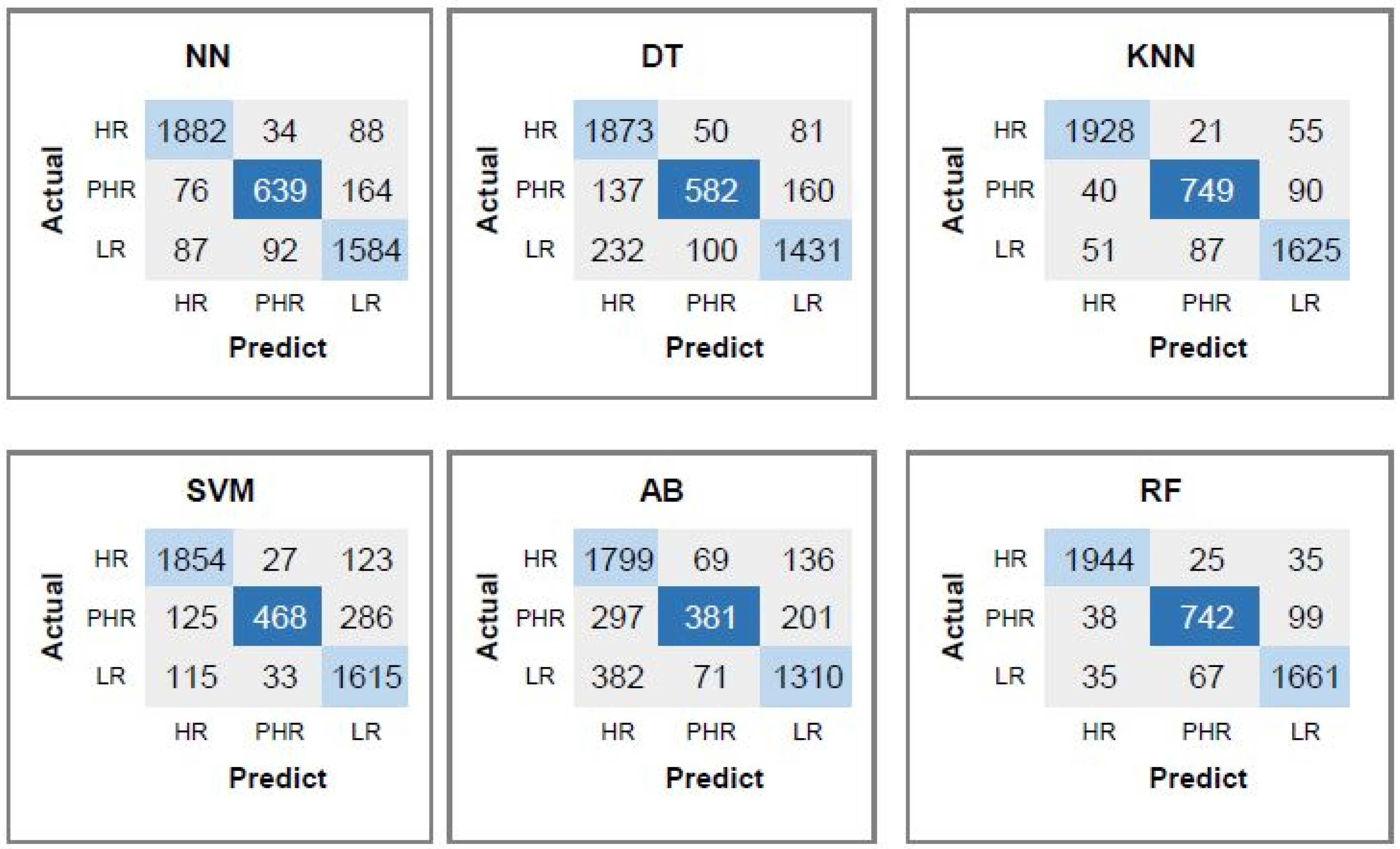
4.7. Performance Evaluation of the Proposed Set of All Features for Predicting Risk of HPV Types
4.8. Explainable AI for Interpreting Feature Importance
4.9. Comparison with Other The-State-of-the-Art Studies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crosbie, E.J.; Einstein, M.H.; Franceschi, S.; Kitchener, H.C. Human papillomavirus and cervical cancer. Lancet 2013, 382, 889–899. [Google Scholar] [CrossRef]
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Collado, J.J.; Gómez, D.; Muñoz, J.; Bosch, F.X.; de Sanjosé, S. Human Papillomavirus and Related Diseases in the World; ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre): Barcelona, Spain, 2023. [Google Scholar]
- Salehiniya, H.; Momenimovahed, Z.; Allahqoli, L.; Momenimovahed, S.; Alkatout, I. Factors related to cervical cancer screening among Asian women. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 6109–6122. [Google Scholar]
- Almeida, A.M.; Queiroz, J.A.; Sousa, F.; Sousa, Â. Cervical cancer and HPV infection: Ongoing therapeutic research to counteract the action of E6 and E7 oncoproteins. Drug Discov. Today 2019, 24, 2044–2057. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, M.; Rizwan, M.; Gregus ml, M.; Abbas, S. Machine Learning Assisted Cervical Cancer Detection. Front. Public Health 2021, 9, 788376. [Google Scholar] [CrossRef]
- Allahqoli, L.; Laganà, A.S.; Mazidimoradi, A.; Salehiniya, H.; Günther, V.; Chiantera, V.; Karimi Goghari, S.; Ghiasvand, M.M.; Rahmani, A.; Momenimovahed, Z.; et al. Diagnosis of Cervical Cancer and Pre-Cancerous Lesions by Artificial Intelligence: A Systematic Review. Diagnostics 2022, 12, 2771. [Google Scholar] [CrossRef] [PubMed]
- Okunade, K.S. Human papillomavirus and cervical cancer. J. Obstet. Gynaecol. 2019, 40, 602–608. [Google Scholar] [CrossRef]
- Liu, J.; Nian, Q.G.; Zhang, Y.; Xu, L.J.; Hu, Y.; Li, J.; Deng, Y.Q.; Zhu, S.Y.; Wu, X.Y.; Qin, E.D.; et al. In Vitro Characterization of Human Adenovirus Type 55 in Comparison with Its Parental Adenoviruses, Types 11 and 14. PLoS ONE 2014, 9, e100665. [Google Scholar] [CrossRef]
- Whitley, R.J.; Roizman, B. Herpes simplex virus infections. Lancet 2001, 357, 1513–1518. [Google Scholar] [CrossRef]
- Sun, H.; Sun, Y.; Pu, J.; Zhang, Y.; Zhu, Q.; Li, J.; Gu, J.; Chang, K.C.; Liu, J. Comparative Virus Replication and Host Innate Responses in Human Cells Infected with Three Prevalent Clades (2.3.4, 2.3.2, and 7) of Highly Pathogenic Avian Influenza H5N1 Viruses. J. Virol. 2014, 88, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Mühr, L.S.A.; Eklund, C.; Dillner, J. Towards quality and order in human papillomavirus research. Virology 2018, 519, 74–76. [Google Scholar] [CrossRef]
- Abreu, A.L.P.; Souza, R.P.; Gimenes, F.; Consolaro, M.E.L. A review of methods for detect human Papillomavirus infection. Virol. J. 2012, 9, 262. [Google Scholar] [CrossRef]
- Faridi, R.; Zahra, A.; Khan, K.; Idrees, M. Oncogenic potential of Human Papillomavirus (HPV) and its relation with cervical cancer. Virol. J. 2011, 8, 269. [Google Scholar] [CrossRef]
- Longworth, M.S.; Laimins, L.A. Pathogenesis of Human Papillomaviruses in Differentiating Epithelia. Microbiol. Mol. Biol. Rev. 2004, 68, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Galati, L.; Chiantore, M.V.; Marinaro, M.; Di Bonito, P. Human Oncogenic Viruses: Characteristics and Prevention Strategies—Lessons Learned from Human Papillomaviruses. Viruses 2024, 16, 416. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Jaffer, N.N.; Khan, M.N.; Rai, M.A.; Shafiq, M.; Ali, A.; Pervez, S.; Khan, N.; Aziz, A.; Ali, S.H. Human papillomavirus subtype 16 is common in Pakistani women with cervical carcinoma. Int. J. Infect. Dis. 2007, 11, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Arroyo Mühr, L.S.; Lagheden, C.; Lei, J.; Eklund, C.; Nordqvist Kleppe, S.; Sparén, P.; Sundström, K.; Dillner, J. Deep sequencing detects human papillomavirus (HPV) in cervical cancers negative for HPV by PCR. Br. J. Cancer 2020, 123, 1790–1795. [Google Scholar] [CrossRef]
- Gravitt, P.E.; Peyton, C.; Wheeler, C.; Apple, R.; Higuchi, R.; Shah, K.V. Reproducibility of HPV 16 and HPV 18 viral load quantitation using TaqMan real-time PCR assays. J. Virol. Methods 2003, 112, 23–33. [Google Scholar] [CrossRef]
- Tucker, R.A.; Unger, E.R.; Holloway, B.P.; Swan, D.C. Real-time PCR-based Fluorescent Assay for Quantitation of Human Papillomavirus Types 6, 11, 16, and 18. Mol. Diagn. 2001, 6, 39–47. [Google Scholar] [CrossRef]
- Kocjan, B.J.; Seme, K.; Poljak, M. Detection and differentiation of human papillomavirus genotypes HPV-6 and HPV-11 by FRET-based real-time PCR. J. Virol. Methods 2008, 153, 245–249. [Google Scholar] [CrossRef]
- Andrews, E.; Seaman, W.T.; Webster-Cyriaque, J. Oropharyngeal carcinoma in non-smokers and non-drinkers: A role for HPV. Oral Oncol. 2009, 45, 486–491. [Google Scholar] [CrossRef]
- Karlsen, F.; Kalantari, M.; Jenkins, A.; Pettersen, E.; Kristensen, G.; Holm, R.; Johansson, B.; Hagmar, B. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J. Clin. Microbiol. 1996, 34, 2095–2100. [Google Scholar] [CrossRef]
- Shin, S.Y.; Lee, I.H.; Cho, Y.M.; Yang, K.A.; Zhang, B.T. EvoOligo: Oligonucleotide probe design with multiobjective evolutionary algorithms. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 2009, 39, 1606–1616. [Google Scholar] [CrossRef]
- Petrou, E.; Chatzipapas, K.; Papadimitroulas, P.; Andrade-Miranda, G.; Katsakiori, P.F.; Papathanasiou, N.D.; Visvikis, D.; Kagadis, G.C. Investigation of Machine and Deep Learning Techniques to Detect HPV Status. J. Pers. Med. 2024, 14, 737. [Google Scholar] [CrossRef]
- de Sanjosé, S.; Perkins, R.B.; Campos, N.; Inturrisi, F.; Egemen, D.; Befano, B.; Rodriguez, A.C.; Jerónimo, J.; Cheung, L.C.; Desai, K.; et al. Design of the HPV-automated visual evaluation (PAVE) study: Validating a novel cervical screening strategy. eLife 2024, 12, RP91469. [Google Scholar] [CrossRef]
- Wenftzensen, N.; Klug, S.J. Cervical Cancer Control in the Era of HPV Vaccination and Novel Biomarkers. Pathobiology 2009, 76, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Forman, D.; de Martel, C.; Lacey, C.J.; Soerjomataram, I.; Lortet-Tieulent, J.; Bruni, L.; Vignat, J.; Ferlay, J.; Bray, F.; Plummer, M.; et al. Global Burden of Human Papillomavirus and Related Diseases. Vaccine 2012, 30, F12–F23. [Google Scholar] [CrossRef]
- Hou, X.; Shen, G.; Zhou, L.; Li, Y.; Wang, T.; Ma, X. Artificial intelligence in cervical cancer screening and diagnosis. Front. Oncol. 2022, 12, 851367. [Google Scholar] [CrossRef] [PubMed]
- Catarino, R.; Schäfer, S.; Vassilakos, P.; Petignat, P.; Arbyn, M. Accuracy of combinations of visual inspection using acetic acid or lugol iodine to detect cervical precancer: A meta-analysis. BJOG Int. J. Obstet. Gynaecol. 2018, 125, 545–553. [Google Scholar] [CrossRef] [PubMed]
- D’Oria, O.; Corrado, G.; Laganà, A.S.; Chiantera, V.; Vizza, E.; Giannini, A. New Advances in Cervical Cancer: From Bench to Bedside. Int. J. Environ. Res. Public Health 2022, 19, 7094. [Google Scholar] [CrossRef]
- Bao, H.; Zhang, L.; Wang, L.; Zhang, M.; Zhao, Z.; Fang, L.; Cong, S.; Zhou, M.; Wang, L. Significant variations in the cervical cancer screening rate in China by individual-level and geographical measures of socioeconomic status: A multilevel model analysis of a nationally representative survey dataset. Cancer Med. 2018, 7, 2089–2100. [Google Scholar] [CrossRef]
- Bedell, S.L.; Goldstein, L.S.; Goldstein, A.R.; Goldstein, A.T. Cervical Cancer Screening: Past, Present, and Future. Sex. Med. Rev. 2019, 8, 28–37. [Google Scholar] [CrossRef]
- Jia, X.; Yan, H.; Shi, Z.; Gao, C.; Zhai, J.; Ding, H.; Qiao, Y. Systematic training and technological empowerment: Enhancing the quality of cervical and breast cancer screening in low-resource areas of China. Gynecol. Obstet. Clin. Med. 2024, 4, e000107. [Google Scholar] [CrossRef]
- Ye, Y.; Jones, T.E.; Zhao, C. Utility of extended HPV genotyping in cervical cancer screening and clinical management. Gynecol. Obstet. Clin. Med. 2025, 5, e000226. [Google Scholar] [CrossRef]
- Osman, R.; Arslan, H. The Application of AI in Oncology Research in Türkiye: Impact and Future Directions. Gazi Univ. J. Sci. Part A Eng. Innov. 2025, 12, 894–917. [Google Scholar] [CrossRef]
- Seven, İ.; Çalişkan, C.; Köş, F.T.; Arslan, H.; Esen, S.A.; Ceylan, F.; Uncu, D. Predicting survival outcomes in advanced pancreatic cancer using machine learning methods. Medicine 2025, 104, e43904. [Google Scholar] [CrossRef]
- Seven, İ.; Bayram, D.; Arslan, H.; Köş, F.T.; Gümüşlü, K.; Aktürk Esen, S.; Şahin, M.; Şendur, M.A.N.; Uncu, D. Predicting hepatocellular carcinoma survival with artificial intelligence. Sci. Rep. 2025, 15, 6226. [Google Scholar] [CrossRef]
- Sharafi, P.; Arslan, H.; Evans, S.; Varan, A.; Ayter, Ş. A machine learning approach for predicting familial and sporadic disease cases based on clinical symptoms: Introduction of a new dataset (Klinik belirtilere dayalı ailesel ve sporadik hastalık vakalarını tahmin etmek için bir makine öğrenimi yaklaşımı: Yeni bir veri kümesinin tanıtımı). Turk Hij. Deney. Biyol. Derg. 2025, 82, 99–106. [Google Scholar]
- Kos, F.T.; Cecen Kaynak, S.; Aktürk Esen, S.; Arslan, H.; Uncu, D. Comparison of Different Machine Learning Models for Predicting Long-Term Overall Survival in Non-metastatic Colorectal Cancers. Cureus 2024, 16, e75713. [Google Scholar] [CrossRef] [PubMed]
- Esen, İ.; Arslan, H.; Esen, S.A.; Gülşen, M.; Kültekin, N.; Özdemir, O. Early prediction of gallstone disease with a machine learning-based method from bioimpedance and laboratory data. Medicine 2024, 103, e37258. [Google Scholar] [CrossRef]
- Arslan, H. A k-mer based metaheuristic approach for detecting COVID-19 variants. DÜMF Mühendis. Derg. 2023, 14, 17–26. [Google Scholar] [CrossRef]
- Arslan, H.; Durmaz, R. A Parallel Algorithm for Designing Primer and Probe for Accurate Detection of Severe Acute Respiratory Syndrome Coronavirus. Black Sea J. Eng. Sci. 2023, 6, 477–485. [Google Scholar] [CrossRef]
- Arslan, H. COVID-19 Hastalarının Mortalitesini Tahmin Etmek için Torbalama ve Arttırma Yöntemleri. DÜMF Mühendis. Derg. 2022, 13, 221–226. [Google Scholar] [CrossRef]
- Abdoh, S.F.; Abo Rizka, M.; Maghraby, F.A. Cervical Cancer Diagnosis Using Random Forest Classifier with SMOTE and Feature Reduction Techniques. IEEE Access 2018, 6, 59475–59485. [Google Scholar] [CrossRef]
- Dong, B.; Lu, Z.; Yang, T.; Wang, J.; Zhang, Y.; Tuo, X.; Wang, J.; Lin, S.; Cai, H.; Cheng, H.; et al. Development, validation, and clinical application of a machine learning model for risk stratification and management of cervical cancer screening based on full-genotyping hrHPV test (SMART-HPV): A modelling study. Lancet Reg. Health–West. Pac. 2025, 55, 101480. [Google Scholar] [CrossRef]
- Tanimu, J.J.; Hamada, M.; Hassan, M.; Kakudi, H.; Abiodun, J.O. A Machine Learning Method for Classification of Cervical Cancer. Electronics 2022, 11, 463. [Google Scholar] [CrossRef]
- Chauhan, R.; Goel, A.; Alankar, B.; Kaur, H. Predictive modeling and web-based tool for cervical cancer risk assessment: A comparative study of machine learning models. MethodsX 2024, 12, 102653. [Google Scholar] [CrossRef] [PubMed]
- Al Mudawi, N.; Alazeb, A. A Model for Predicting Cervical Cancer Using Machine Learning Algorithms. Sensors 2022, 22, 4132. [Google Scholar] [CrossRef]
- Glučina, M.; Lorencin, A.; Anđelić, N.; Lorencin, I. Cervical Cancer Diagnostics Using Machine Learning Algorithms and Class Balancing Techniques. Appl. Sci. 2023, 13, 1061. [Google Scholar] [CrossRef]
- Alsmariy, R.; Healy, G.; Abdelhafez, H. Predicting Cervical Cancer using Machine Learning Methods. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 173–184. [Google Scholar] [CrossRef]
- Nithya, B.; Ilango, V. Evaluation of machine learning based optimized feature selection approaches and classification methods for cervical cancer prediction. SN Appl. Sci. 2019, 1, 641. [Google Scholar] [CrossRef]
- Vidya, R.; Nasira, G. Prediction of cervical cancer using hybrid induction technique: A solution for human hereditary disease patterns. Indian J. Sci. Technol. 2016, 9, 1–10. [Google Scholar] [CrossRef]
- Sahay, A.; Gopakumar, G.; Gokulan, S.; Subham, D.; Thakur, A. Applying Machine Learning Algorithms to Investigate Cervical Cancer. In Proceedings of the 2024 International Conference on Intelligent and Innovative Technologies in Computing, Electrical and Electronics (IITCEE), Bangalore, India, 24–25 January 2024; pp. 1–5. [Google Scholar]
- Harden, M.E.; Munger, K. Human papillomavirus molecular biology. Mutat. Res./Rev. Mutat. Res. 2017, 772, 3–12. [Google Scholar] [CrossRef]
- Park, S.B.; Hwang, S.; Zhang, B.T. Classification of the Risk Types of Human Papillomavirus by Decision Trees. In Proceedings of the Intelligent Data Engineering and Automated Learning; Liu, J., Cheung, Y.m., Yin, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 540–544. [Google Scholar]
- Muñoz, N.; Bosch, F.X.; de Sanjosé, S.; Herrero, R.; Castellsagué, X.; Shah, K.V.; Snijders, P.J.; Meijer, C.J. Epidemiologic Classification of Human Papillomavirus Types Associated with Cervical Cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Senel, B. Harnessing machine learning in HPV diagnostics: Model performance, explainability, and clinical integration. Int. J. Health Serv. Res. Policy 2025, 10, 175–190. [Google Scholar] [CrossRef]
- Nowicki, M.; Mroczek, M.; Mukhedkar, D.; Bała, P.; Nikolai Pimenoff, V.; Arroyo Mühr, L.S. HPV-KITE: Sequence analysis software for rapid HPV genotype detection. Briefings Bioinform. 2025, 26, bbaf155. [Google Scholar] [CrossRef]
- Ai, W.; Wu, C.; Jia, L.; Xiao, X.; Xu, X.; Ren, M.; Xue, T.; Zhou, X.; Wang, Y.; Gao, C. Deep Sequencing of HPV16 E6 Region Reveals Unique Mutation Pattern of HPV16 and Predicts Cervical Cancer. Microbiol. Spectr. 2022, 10, e01401-22. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Hwang, K.; Ann, J.; Kim, J.; Nam, J. Next-generation sequencing for typing human papillomaviruses and predicting multi-infections and their clinical symptoms. Microbiol. Immunol. 2021, 65, 273–278. [Google Scholar] [CrossRef]
- Remita, M.A.; Halioui, A.; Malick Diouara, A.A.; Daigle, B.; Kiani, G.; Diallo, A.B. A machine learning approach for viral genome classification. BMC Bioinform. 2017, 18, 208. [Google Scholar] [CrossRef]
- Asensio-Puig, L.; Alemany, L.; Pavón, M.A. A Straightforward HPV16 Lineage Classification Based on Machine Learning. Front. Artif. Intell. 2022, 5, 851841. [Google Scholar] [CrossRef]
- Hao, L.; Jiang, Y.; Zhang, C.; Han, P. Genome composition-based deep learning predicts oncogenic potential of HPVs. Front. Cell. Infect. Microbiol. 2024, 14, 1430424. [Google Scholar] [CrossRef]
- Tiwary, B.K. CarcinoHPVPred: An ensemble of machine learning models for HPV carcinogenicity prediction using genomic data. Carcinogenesis 2022, 43, 1083–1091. [Google Scholar]
- Kim, S.; Eom, J.H. Prediction of the human Papillomavirus risk types using gap-spectrum kernels. In Advances in Neural Networks—ISNN 2006; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2006; pp. 710–715. [Google Scholar]
- Wang, C.; Hai, Y.; Liu, X.; Liu, N.; Yao, Y.; He, P.; Dai, Q. Prediction of High-Risk Types of Human Papillomaviruses Using Statistical Model of Protein “Sequence Space”. Comput. Math. Methods Med. 2015, 2015, 756345. [Google Scholar] [CrossRef]
- Joung, J.G.; O, S.J.; Zhang, B.T. Prediction of the Risk Types of Human Papillomaviruses by Support Vector Machines. In Proceedings of the PRICAI 2004: Trends in Artificial Intelligence; Zhang, C., W. Guesgen, H., Yeap, W.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 723–731. [Google Scholar]
- Kim, S.; Kim, J.; Zhang, B.T. Ensembled support vector machines for human papillomavirus risk type prediction from protein secondary structures. Comput. Biol. Med. 2009, 39, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Shetty, A.; Shah, V. Survey of cervical cancer prediction using machine learning: A comparative approach. In Proceedings of the 2018 9th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Bengaluru, India, 10–12 July 2018; pp. 1–6. [Google Scholar]
- Rahimi, M.; Akbari, A.; Asadi, F.; Emami, H. Cervical cancer survival prediction by machine learning algorithms: A systematic review. BMC Cancer 2023, 23, 341. [Google Scholar] [CrossRef]
- Simón, D.; Cristina, J.; Musto, H. Nucleotide composition and Codon usage across viruses and their respective hosts. Front. Microbiol. 2021, 12, 646300. [Google Scholar] [CrossRef]
- Abeel, T.; Saeys, Y.; Rouzé, P.; Van de Peer, Y. ProSOM: Core promoter prediction based on unsupervised clustering of DNA physical profiles. Bioinformatics 2008, 24, i24–i31. [Google Scholar] [CrossRef] [PubMed]
- Arslan, H.; Arslan, H. A new COVID-19 detection method from human genome sequences using CpGisland features and KNN classifier. Eng. Sci. Technol. Int. J. 2021, 24, 839–847. [Google Scholar] [CrossRef]
- Ponger, L.; Mouchiroud, D. CpGProD: Identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics 2002, 18, 631–633. [Google Scholar] [CrossRef]
- Kottaridi, C.; Kyrgiou, M.; Pouliakis, A.; Magkana, M.; Aga, E.; Spathis, A.; Mitra, A.; Makris, G.; Chrelias, C.; Mpakou, V.; et al. Quantitative measurement of L1 human papillomavirus type 16 methylation for the prediction of preinvasive and invasive cervical disease. J. Infect. Dis. 2017, 215, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.L.A.; Tsang, T.Y.; Yau, P.L.; Kwok, T.T. Human papillomavirus type 16 E6 suppresses microRNA-23b expression in human cervical cancer cells through DNA methylation of the host gene C9orf3. Oncotarget 2017, 8, 12158. [Google Scholar] [CrossRef]
- Galván, S.C.; Martínez-Salazar, M.; Galván, V.M.; Méndez, R.; Díaz-Contreras, G.T.; Alvarado-Hermida, M.; Alcántara-Silva, R.; García-Carrancá, A. Analysis of CpG methylation sites and CGI among human papillomavirus DNA genomes. BMC Genom. 2011, 12, 580. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.K.; El-Osta, A. The rise of DNA methylation and the importance of chromatin on multidrug resistance in cancer. Exp. Cell Res. 2003, 290, 177–194. [Google Scholar] [CrossRef]
- Vo ngoc, L.; Huang, C.Y.; Cassidy, C.J.; Medrano, C.; Kadonaga, J.T. Identification of the human DPR core promoter element using machine learning. Nature 2020, 585, 459–463. [Google Scholar] [CrossRef]
- Smits, P.H.M.; Smits, H.L.; Minnaar, R.P.; ter Schegget, J. Regulation of human papillomavirus type 16 (HPV-16) transcription by loci on the short arm of chromosome 11 is mediated by the TATAAAA motif of the HPV-16 promoter. J. Gen. Virol. 1993, 74, 121–124. [Google Scholar] [CrossRef]
- Demeret, C.; Desaintes, C.; Yaniv, M.; Thierry, F. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J. Virol. 1997, 71, 9343–9349. [Google Scholar] [CrossRef]
- Massimi, P.; Thomas, M.; Bouvard, V.; Ruberto, I.; Campo, M.S.; Tommasino, M.; Banks, L. Comparative transforming potential of different human papillomaviruses associated with non-melanoma skin cancer. Virology 2008, 371, 374–379. [Google Scholar] [CrossRef]
- Gharelo, R.Z.; Bandehagh, A. Analysis of the Promoter Region of the Gene Encoding Sodium/Hydrogen Exchanger 1 Protein. J. Mol. Genet. Med. 2017, 11, 1747-0862. [Google Scholar] [CrossRef]
- Hirt, L.; Hirsch-Behnam, A.; de Villiers, E.M. Nucleotide sequence of human papillomavirus (HPV) type 41: An unusual HPV type without a typical E2 binding site consensus sequence. Virus Res. 1991, 18, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Karlen, S.; Offord, E.A.; Beard, P. Herpes simplex virions interfere with the expression of human papillomavirus type 18 genes. J. Gen. Virol. 1993, 74, 965–973. [Google Scholar] [CrossRef]
- Desaintes, C.; Hallez, S.; Van Alphen, P.; Burny, A. Transcriptional activation of several heterologous promoters by the E6 protein of human papillomavirus type 16. J. Virol. 1992, 66, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Bauknecht, T.; Shi, Y. Overexpression of C/EBPβ represses human papillomavirus type 18 upstream regulatory region activity in HeLa cells by interfering with the binding of TATA-binding protein. J. Virol. 1998, 72, 2113–2124. [Google Scholar] [CrossRef]
- Alvarez, J.; Gagnon, D.; Coutlée, F.; Archambault, J. Characterization of an HPV33 natural variant with enhanced transcriptional activity suggests a role for C/EBPβ in the regulation of the viral early promoter. Sci. Rep. 2019, 9, 5113. [Google Scholar] [CrossRef]
- Dlamini, G.S.; Müller, S.J.; Meraba, R.L.; Young, R.A.; Mashiyane, J.; Chiwewe, T.; Mapiye, D.S. Classification of COVID-19 and other pathogenic sequences: A dinucleotide frequency and machine learning approach. IEEE Access 2020, 8, 195263–195273. [Google Scholar] [CrossRef]
- Ren, J.; Ahlgren, N.A.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. VirFinder: A novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 2017, 5, 69. [Google Scholar] [CrossRef]
- Young, F.; Rogers, S.; Robertson, D.L. Predicting host taxonomic information from viral genomes: A comparison of feature representations. PLoS Comput. Biol. 2020, 16, e1007894. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Burges, C.J. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, L.; Zhao, D.; Zhou, J.; Zhang, W. Emotion recognition from Chinese speech for smart affective services using a combination of SVM and DBN. Sensors 2017, 17, 1694. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Fang, S.C.; Nuttle, H.L. A neural network model with bounded-weights for pattern classification. Comput. Oper. Res. 2004, 31, 1411–1426. [Google Scholar] [CrossRef]
- Keerthi, S.S.; Lin, C.J. Asymptotic behaviors of support vector machines with Gaussian kernel. Neural Comput. 2003, 15, 1667–1689. [Google Scholar] [CrossRef]
- Brown, I.; Mues, C. An experimental comparison of classification algorithms for imbalanced credit scoring data sets. Expert Syst. Appl. 2012, 39, 3446–3453. [Google Scholar] [CrossRef]
- Deng, Z.; Zhu, X.; Cheng, D.; Zong, M.; Zhang, S. Efficient kNN classification algorithm for big data. Neurocomputing 2016, 195, 143–148. [Google Scholar] [CrossRef]
- Abu Alfeilat, H.A.; Hassanat, A.B.; Lasassmeh, O.; Tarawneh, A.S.; Alhasanat, M.B.; Eyal Salman, H.S.; Prasath, V.S. Effects of distance measure choice on k-nearest neighbor classifier performance: A review. Big Data 2019, 7, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006. [Google Scholar]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef]
- Aha, D.W.; Kibler, D.; Albert, M.K. Instance-based learning algorithms. Mach. Learn. 1991, 6, 37–66. [Google Scholar] [CrossRef]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. NCBI Virus Variation Sequence Search Interface (VSSI). 2025. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/ (accessed on 4 January 2025).
- Goutte, C.; Gaussier, E. A Probabilistic Interpretation of Precision, Recall and F-Score, with Implication for Evaluation. In Proceedings of the Advances in Information Retrieval; Losada, D.E., Fernández-Luna, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 345–359. [Google Scholar]
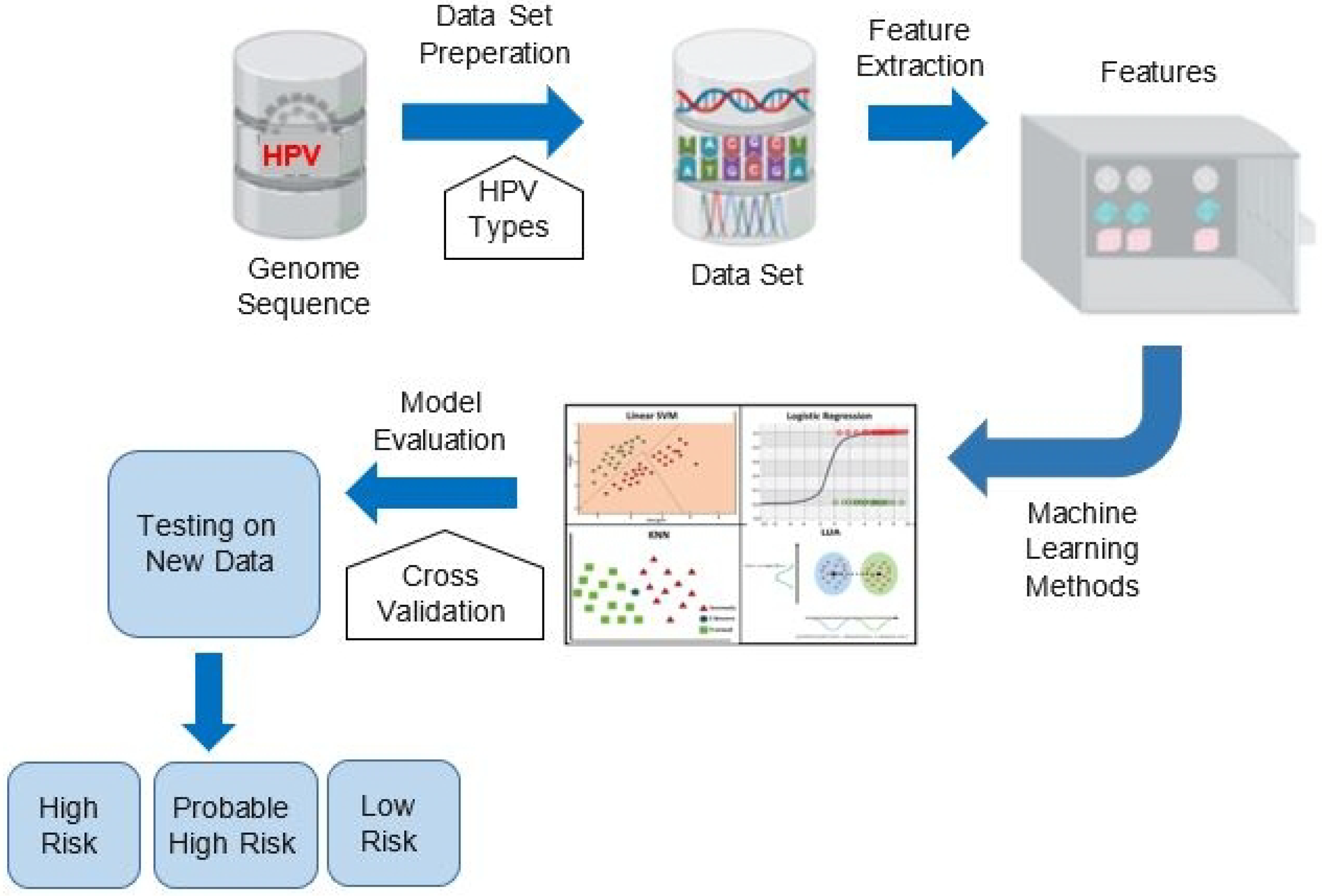
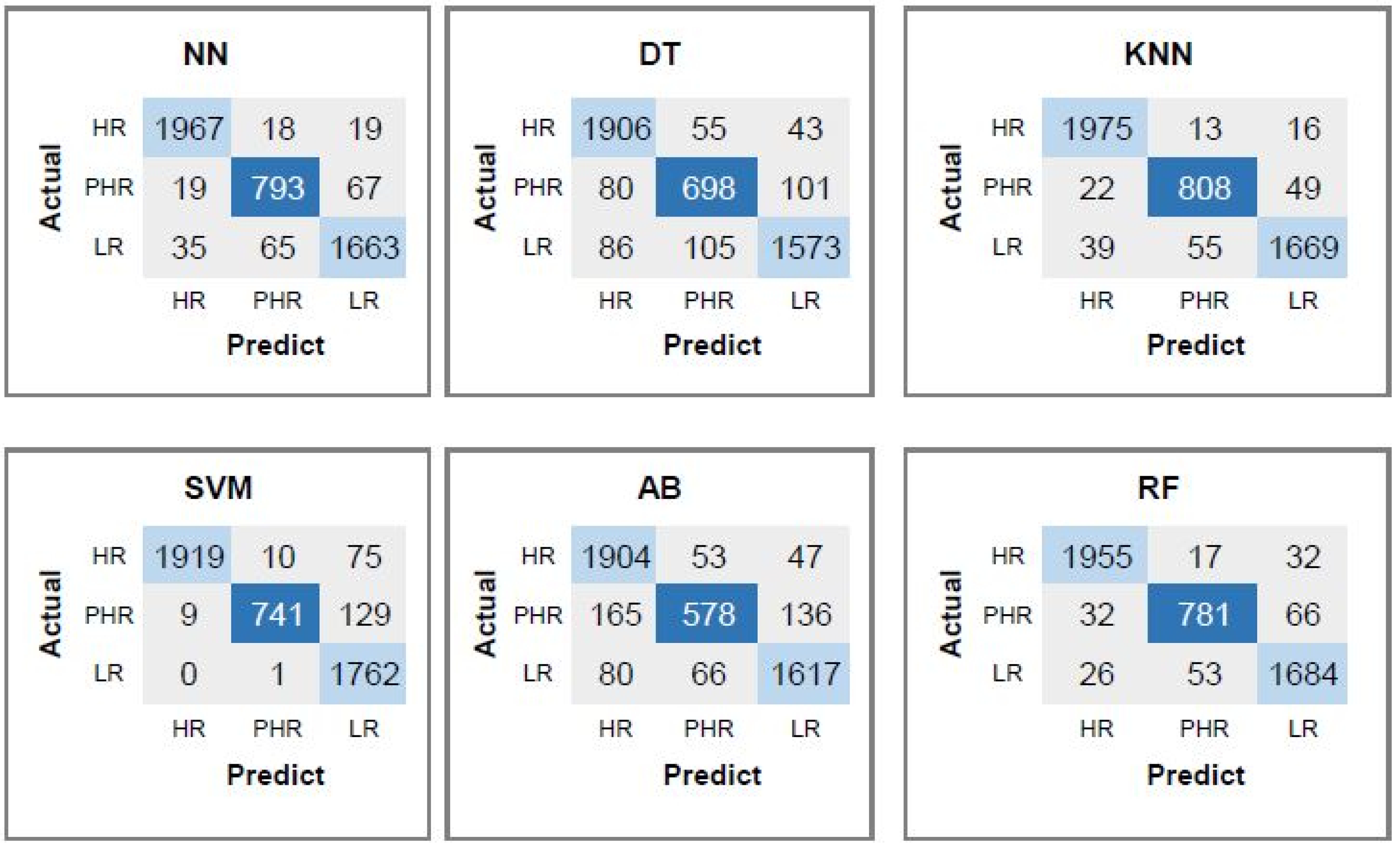

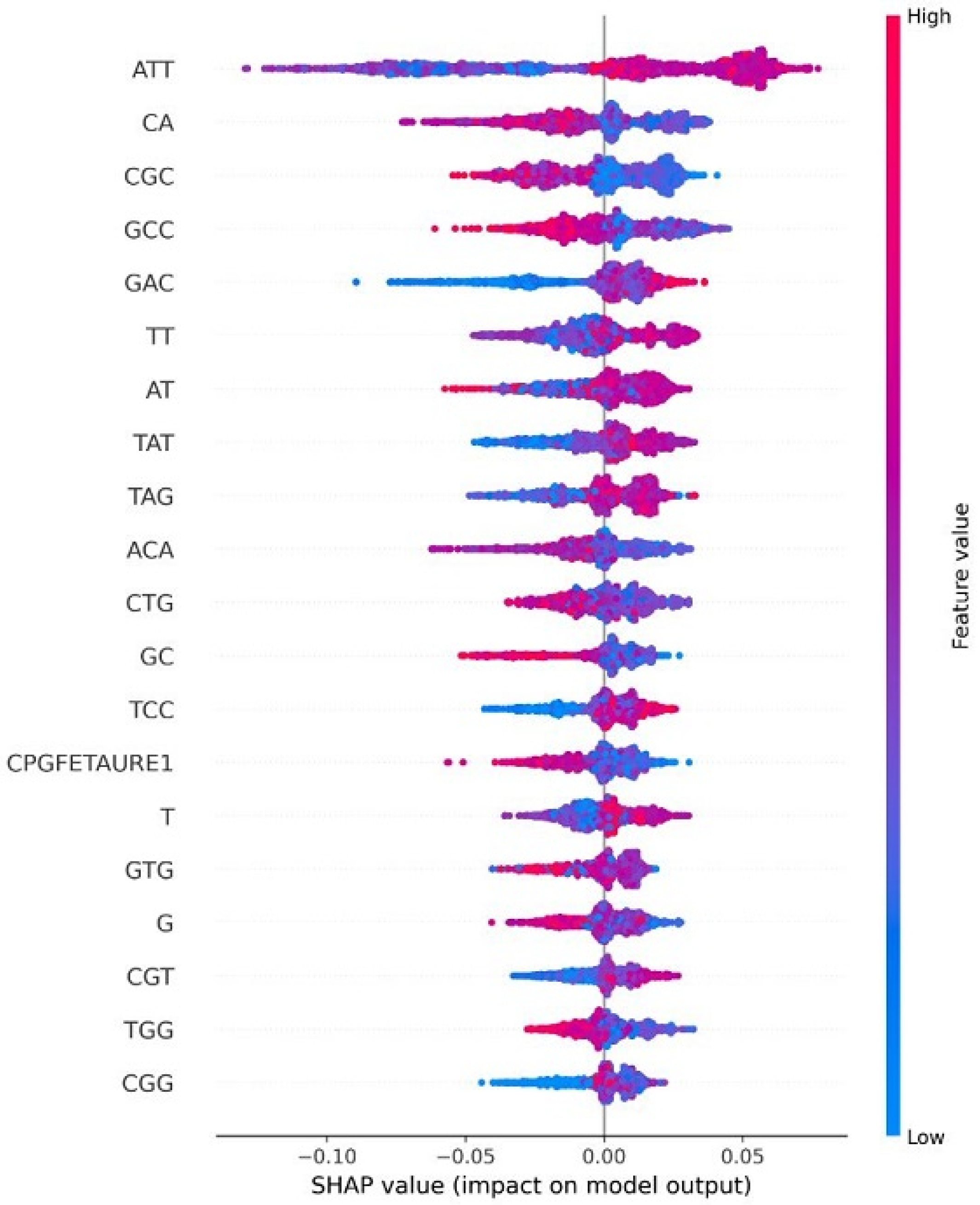
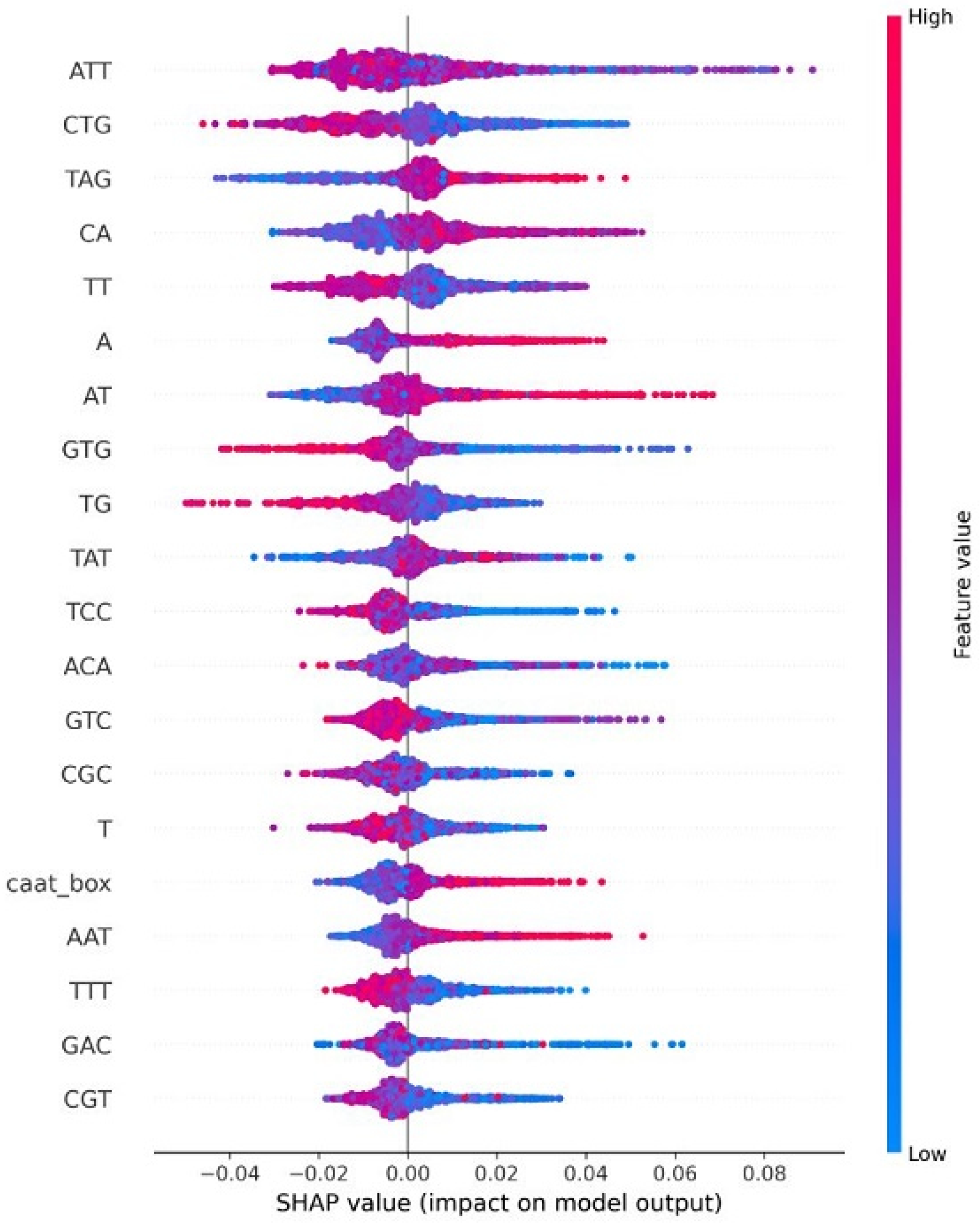
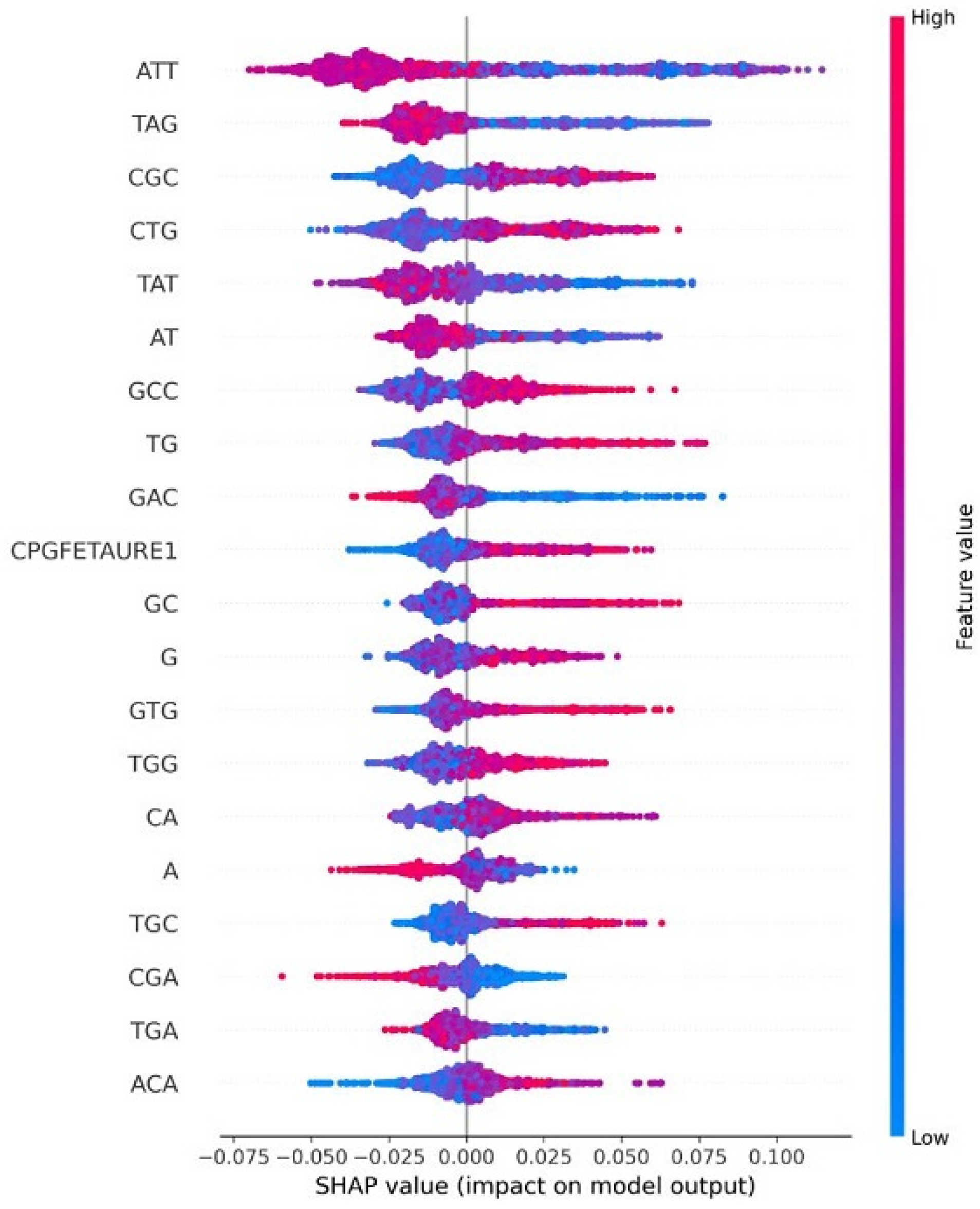
| HPV Risk Type | HPV Types |
|---|---|
| High Risk | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 |
| Probable High Risk | 26, 30, 34, 53, 66, 67, 68, 69, 70, 73, 82, 85, 97 |
| Low Risk | 6, 11, 40, 42, 43, 44, 54, 55, 57, 61, 62, 64, 71, 72, |
| 74, 81, 83, 84, 86, 87, 89, 90, 91 |
| High Risk HPV Types | Total Sequences in NCBI | Sequences Used in This Study |
|---|---|---|
| HPV16 | 12,856 | 167 |
| HPV18 | 1609 | 167 |
| HPV31 | 3118 | 167 |
| HPV33 | 733 | 167 |
| HPV35 | 1276 | 167 |
| HPV39 | 325 | 167 |
| HPV45 | 388 | 167 |
| HPV51 | 476 | 167 |
| HPV52 | 1978 | 167 |
| HPV56 | 372 | 167 |
| HPV58 | 2538 | 167 |
| HPV59 | 392 | 167 |
| Probable High Risk HPV Types | Total Sequences in NCBI | Sequences Used in This Study |
|---|---|---|
| HPV30 | 60 | 60 |
| HPV66 | 379 | 379 |
| HPV67 | 66 | 66 |
| HPV68 | 229 | 229 |
| HPV69 | 31 | 31 |
| HPV73 | 45 | 45 |
| HPV82 | 51 | 51 |
| HPV85 | 11 | 11 |
| HPV97 | 7 | 7 |
| Low Risk HPV Types | Total Sequences in NCBI | Sequences Used in This Study |
|---|---|---|
| HPV11 | 794 | 794 |
| HPV40 | 85 | 85 |
| HPV42 | 266 | 266 |
| HPV43 | 67 | 67 |
| HPV44 | 107 | 107 |
| HPV57 | 24 | 24 |
| HPV62 | 40 | 40 |
| HPV71 | 43 | 43 |
| HPV72 | 16 | 16 |
| HPV74 | 40 | 40 |
| HPV81 | 143 | 143 |
| HPV83 | 26 | 26 |
| HPV84 | 22 | 22 |
| HPV87 | 26 | 26 |
| HPV86 | 16 | 16 |
| HPV89 | 27 | 27 |
| HPV91 | 21 | 21 |
| Performance Metric | Formula for Each Class c | Average Metric |
|---|---|---|
| Precision (Pre) | ||
| Recall (Rec) | ||
| F1-score (F1) | 2 × | |
| Accuracy (Acc) |
| HPV | Type Based-Results | Average Results | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Method | Type | Pre | Rec | F1 | Acc | Pre | Rec | F1 | Acc (%) |
| HR | 0.8 | 0.87 | 0.83 | 0.85 | |||||
| DT | PHR | 0.6 | 0.52 | 0.56 | 0.84 | 0.74 | 0.73 | 0.73 | 85.09 |
| LR | 0.82 | 0.8 | 0.81 | 0.86 | |||||
| HR | 0.86 | 0.89 | 0.88 | 0.89 | |||||
| KNN | PHR | 0.75 | 0.71 | 0.73 | 0.9 | 0.83 | 0.82 | 0.82 | 89.65 |
| LR | 0.87 | 0.86 | 0.86 | 0.9 | |||||
| HR | 0.57 | 0.75 | 0.65 | 0.65 | |||||
| SVM | PHR | 0.51 | 0.22 | 0.31 | 0.81 | 0.57 | 0.52 | 0.52 | 72.21 |
| LR | 0.63 | 0.57 | 0.6 | 0.71 | |||||
| HR | 0.68 | 0.9 | 0.77 | 0.77 | |||||
| AB | PHR | 0.66 | 0.18 | 0.29 | 0.83 | 0.71 | 0.62 | 0.61 | 80.93 |
| LR | 0.77 | 0.77 | 0.77 | 0.83 | |||||
| HR | 0.92 | 0.93 | 0.93 | 0.94 | |||||
| RF | PHR | 0.83 | 0.77 | 0.8 | 0.93 | 0.88 | 0.87 | 0.88 | 92.87 |
| LR | 0.89 | 0.91 | 0.9 | 0.92 | |||||
| HR | 0.63 | 0.77 | 0.69 | 0.71 | |||||
| NN | PHR | 0.54 | 0.29 | 0.38 | 0.82 | 0.61 | 0.57 | 0.58 | 75.53 |
| LR | 0.66 | 0.64 | 0.65 | 0.74 | |||||
| HPV | Type Based Results | Average Results | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Method | Type | Pre | Rec | F1 | Acc | Pre | Rec | F1 | Acc (%) |
| HR | 0.84 | 0.93 | 0.88 | 0.89 | |||||
| DT | PHR | 0.8 | 0.66 | 0.72 | 0.9 | 0.83 | 0.8 | 0.81 | 89.09 |
| LR | 0.86 | 0.81 | 0.83 | 0.88 | |||||
| HR | 0.95 | 0.96 | 0.96 | 0.96 | |||||
| KNN | PHR | 0.87 | 0.85 | 0.86 | 0.95 | 0.92 | 0.91 | 0.91 | 95.06 |
| LR | 0.92 | 0.92 | 0.92 | 0.94 | |||||
| HR | 0.89 | 0.93 | 0.9 | 0.92 | |||||
| SVM | PHR | 0.89 | 0.53 | 0.67 | 0.9 | 0.86 | 0.79 | 0.81 | 89.83 |
| LR | 0.8 | 0.92 | 0.85 | 0.88 | |||||
| HR | 0.73 | 0.9 | 0.8 | 0.81 | |||||
| AB | PHR | 0.73 | 0.43 | 0.54 | 0.86 | 0.75 | 0.69 | 0.71 | 83.41 |
| LR | 0.8 | 0.74 | 0.77 | 0.83 | |||||
| HR | 0.96 | 0.97 | 0.97 | 0.97 | |||||
| RF | PHR | 0.96 | 0.93 | 0.95 | 0.96 | 0.96 | 0.95 | 0.96 | 96.32 |
| LR | 0.96 | 0.95 | 0.96 | 0.96 | |||||
| HR | 0.92 | 0.94 | 0.93 | 0.94 | |||||
| NN | PHR | 0.84 | 0.73 | 0.78 | 0.92 | 0.87 | 0.85 | 0.86 | 92.24 |
| LR | 0.86 | 0.9 | 0.88 | 0.91 | |||||
| HPV | Type Based Results | Average Results | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Method | Type | Pre | Rec | F1 | Acc | Pre | Rec | F1 | Acc (%) |
| HR | 0.92 | 0.95 | 0.94 | 94.32 | |||||
| DT | PHR | 0.81 | 0.79 | 0.80 | 92.66 | 0.88 | 0.88 | 0.88 | 93.26 |
| LR | 0.92 | 0.89 | 0.90 | 92.79 | |||||
| HR | 0.97 | 0.99 | 0.98 | 98.06 | |||||
| KNN | PHR | 0.92 | 0.92 | 0.92 | 97.01 | 0.95 | 0.95 | 0.956 | 97.22 |
| LR | 0.96 | 0.95 | 0.95 | 96.58 | |||||
| HR | 1.00 | 0.96 | 0.98 | 97.98 | |||||
| SVM | PHR | 0.99 | 0.84 | 0.91 | 96.79 | 0.96 | 0.93 | 0.94 | 96.79 |
| LR | 0.90 | 1.00 | 0.95 | 95.59 | |||||
| HR | 0.89 | 0.95 | 0.92 | 92.57 | |||||
| AB | PHR | 0.83 | 0.66 | 0.73 | 90.96 | 0.87 | 0.84 | 0.85 | 92.15 |
| LR | 0.90 | 0.92 | 0.91 | 92.92 | |||||
| HR | 0.97 | 0.98 | 0.97 | 97.70 | |||||
| RF | PHR | 0.92 | 0.89 | 0.90 | 96.38 | 0.94 | 0.94 | 0.94 | 96.76 |
| LR | 0.95 | 0.96 | 0.95 | 96.19 | |||||
| HR | 0.97 | 0.98 | 0.98 | 98.04 | |||||
| NN | PHR | 0.91 | 0.90 | 0.90 | 96.36 | 0.94 | 0.94 | 0.94 | 96.80 |
| LR | 0.95 | 0.94 | 0.95 | 96.00 | |||||
| HPV | Type Based Results | Average Results | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Method | Type | Pre | Rec | F1 | Acc | Pre | Rec | F1 | Acc (%) |
| HR | 0.94 | 0.97 | 0.95 | 95.98 | |||||
| DT | PHR | 0.88 | 0.84 | 0.86 | 94.90 | 0.92 | 0.91 | 0.91 | 95.05 |
| LR | 0.93 | 0.92 | 0.92 | 94.27 | |||||
| HR | 0.97 | 0.99 | 0.98 | 98.30 | |||||
| KNN | PHR | 0.92 | 0.92 | 0.92 | 97.14 | 0.95 | 0.95 | 0.95 | 97.46 |
| LR | 0.97 | 0.95 | 0.96 | 96.94 | |||||
| HR | 0.97 | 0.98 | 0.97 | 97.72 | |||||
| SVM | PHR | 0.91 | 0.89 | 0.90 | 96.23 | 0.94 | 0.94 | 0.94 | 96.79 |
| LR | 0.96 | 0.95 | 0.95 | 96.41 | |||||
| HR | 0.95 | 0.95 | 0.95 | 95.91 | |||||
| AB | PHR | 0.85 | 0.79 | 0.82 | 93.48 | 0.90 | 0.89 | 0.90 | 94.36 |
| LR | 0.90 | 0.93 | 0.92 | 93.69 | |||||
| HR | 0.98 | 0.98 | 0.98 | 98.13 | |||||
| RF | PHR | 0.91 | 0.89 | 0.90 | 96.30 | 0.94 | 0.94 | 0.94 | 96.87 |
| LR | 0.94 | 0.96 | 0.95 | 96.19 | |||||
| HR | 0.98 | 0.98 | 0.98 | 98.30 | |||||
| NN | PHR | 0.92 | 0.92 | 0.92 | 97.09 | 0.95 | 0.95 | 0.95 | 97.47 |
| LR | 0.97 | 0.96 | 0.96 | 97.03 | |||||
| Classifier | CpG | CpG, CAAT-Box | CpG, CAAT-Box | All 88 |
|---|---|---|---|---|
| TATA-Box | TATA-Box, 2-Mer | Features | ||
| DT | 85.09 | 89.09 | 93.26 | 95.05 |
| KNN | 89.65 | 95.06 | 97.22 | 97.46 |
| SVM | 72.21 | 89.83 | 96.79 | 96.79 |
| AB | 80.93 | 83.41 | 92.15 | 94.36 |
| RF | 92.87 | 96.32 | 96.76 | 96.87 |
| NN | 75.53 | 92.24 | 96.8 | 97.47 |
| Study | Method | Number of | Number of | Acc (%) |
| Features | DNA Samples | |||
| Ai et al. [59] | LR | 13 | 199 | 84.3 |
| Study | Method | Number of | # of Genome | Acc (%) |
| Features | Sequences | |||
| Hao et al. [63] | CNN | 1584 | 2582 | 95 |
| Study | Method | # of Protein | Number of | Acc (%) |
| Sequences | HPV Types | |||
| Wang et al. [66] | SVM | 7 | 72 | 95.59 |
| Study | Method | Number of | # of Genome | Acc (%) |
| Features | Sequences | |||
| Tiwary and Karamveer [64] | LR | 750 | 25 carcinogenic | 100 |
| 397 non-carcinogenic | ||||
| Study | Method | Number of | # of Genome | Acc (%) |
| Features | Sequences | |||
| 2004 high-risk | 98.30 | |||
| Proposed Method | KNN | 88 | 879 probable high-risk | 97.14 |
| 1763 low-risk | 96.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çeçen Kaynak, S.; Arslan, H. A Novel Method for Predicting Oncogenic Types of Human Papillomavirus. Diagnostics 2025, 15, 3014. https://doi.org/10.3390/diagnostics15233014
Çeçen Kaynak S, Arslan H. A Novel Method for Predicting Oncogenic Types of Human Papillomavirus. Diagnostics. 2025; 15(23):3014. https://doi.org/10.3390/diagnostics15233014
Chicago/Turabian StyleÇeçen Kaynak, Songül, and Hilal Arslan. 2025. "A Novel Method for Predicting Oncogenic Types of Human Papillomavirus" Diagnostics 15, no. 23: 3014. https://doi.org/10.3390/diagnostics15233014
APA StyleÇeçen Kaynak, S., & Arslan, H. (2025). A Novel Method for Predicting Oncogenic Types of Human Papillomavirus. Diagnostics, 15(23), 3014. https://doi.org/10.3390/diagnostics15233014






