Urine and Serum miRNA Signatures for the Non-Invasive Diagnosis of Adenomyosis: A Machine Learning-Based Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Sampling
- Serum
- Urine
- Sample preparation
2.3. Preprocessing
2.4. Feature Selection
2.5. Model Selection and Evaluation
- Logistic Regression;
- Decision Tree;
- Random Forest;
- Support Vector Machine (SVM).
3. Results
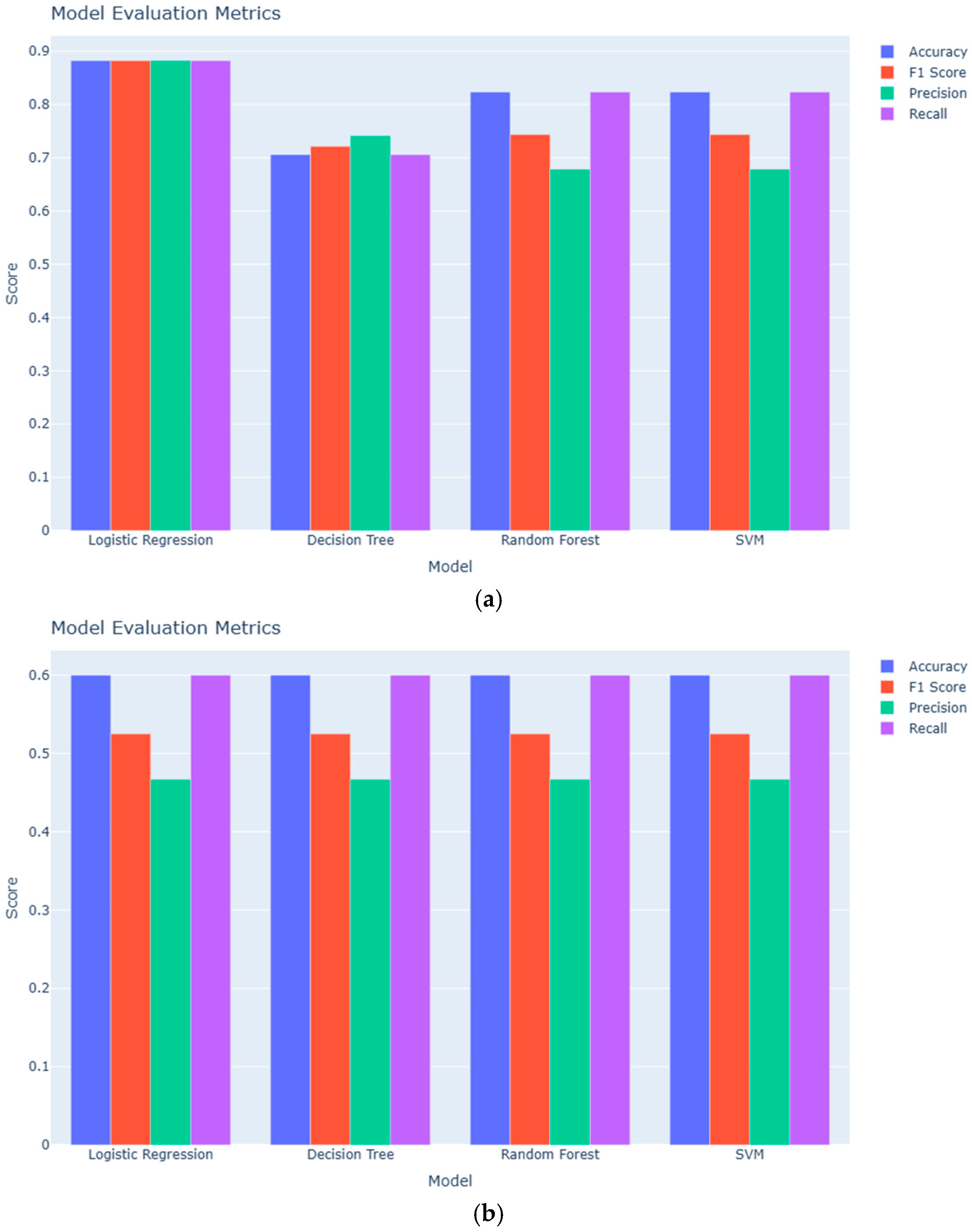
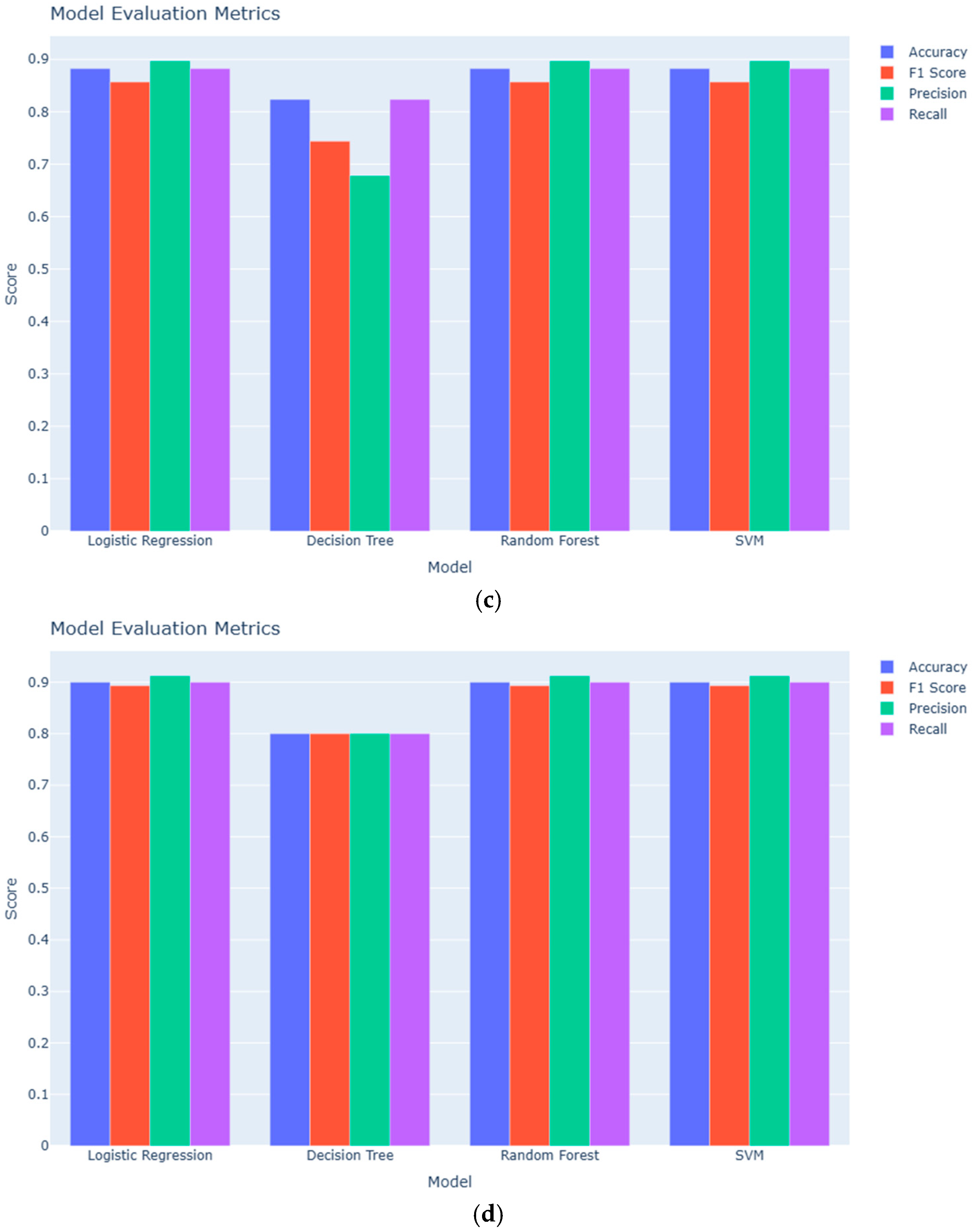
3.1. Classification Performance
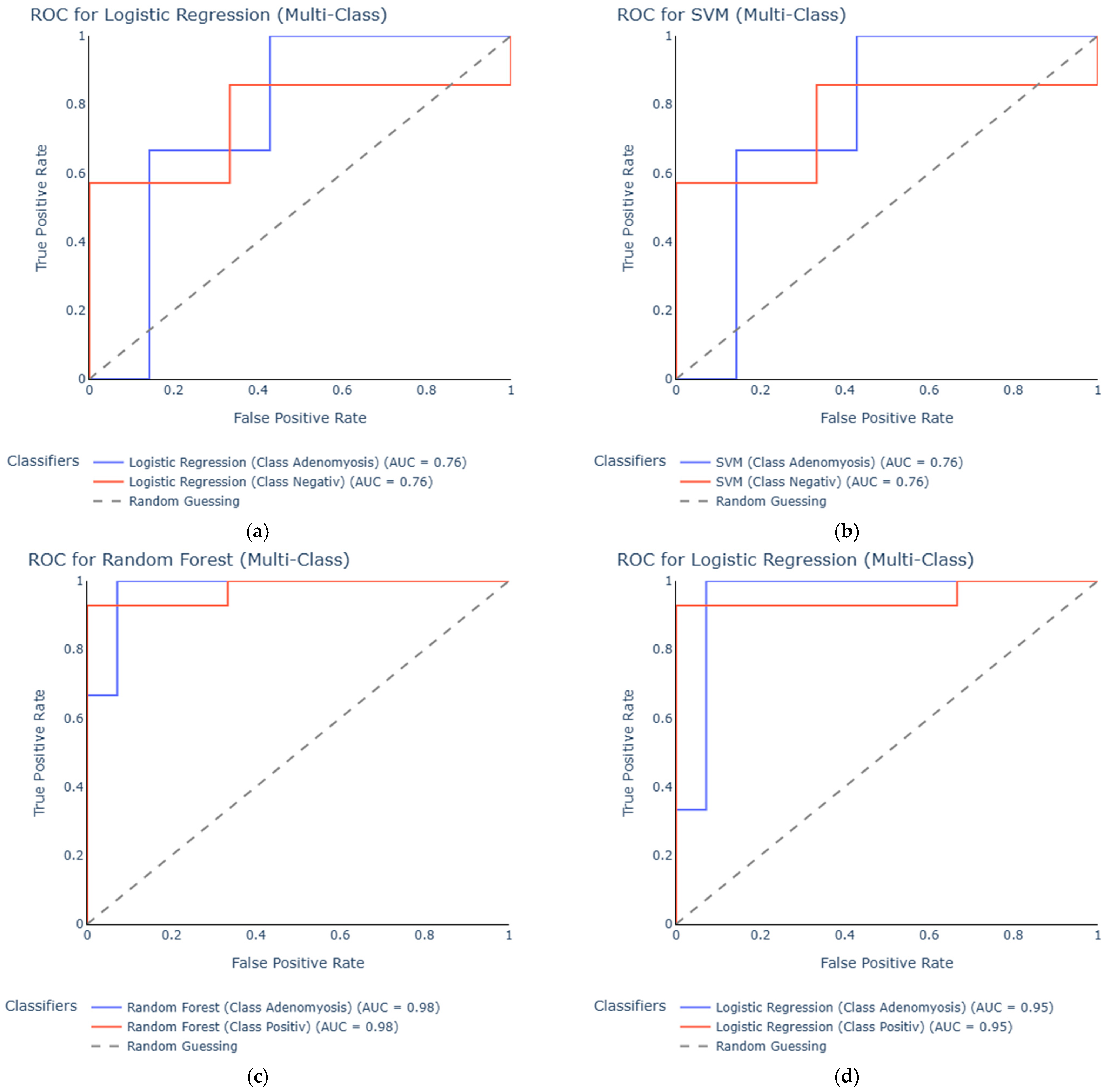
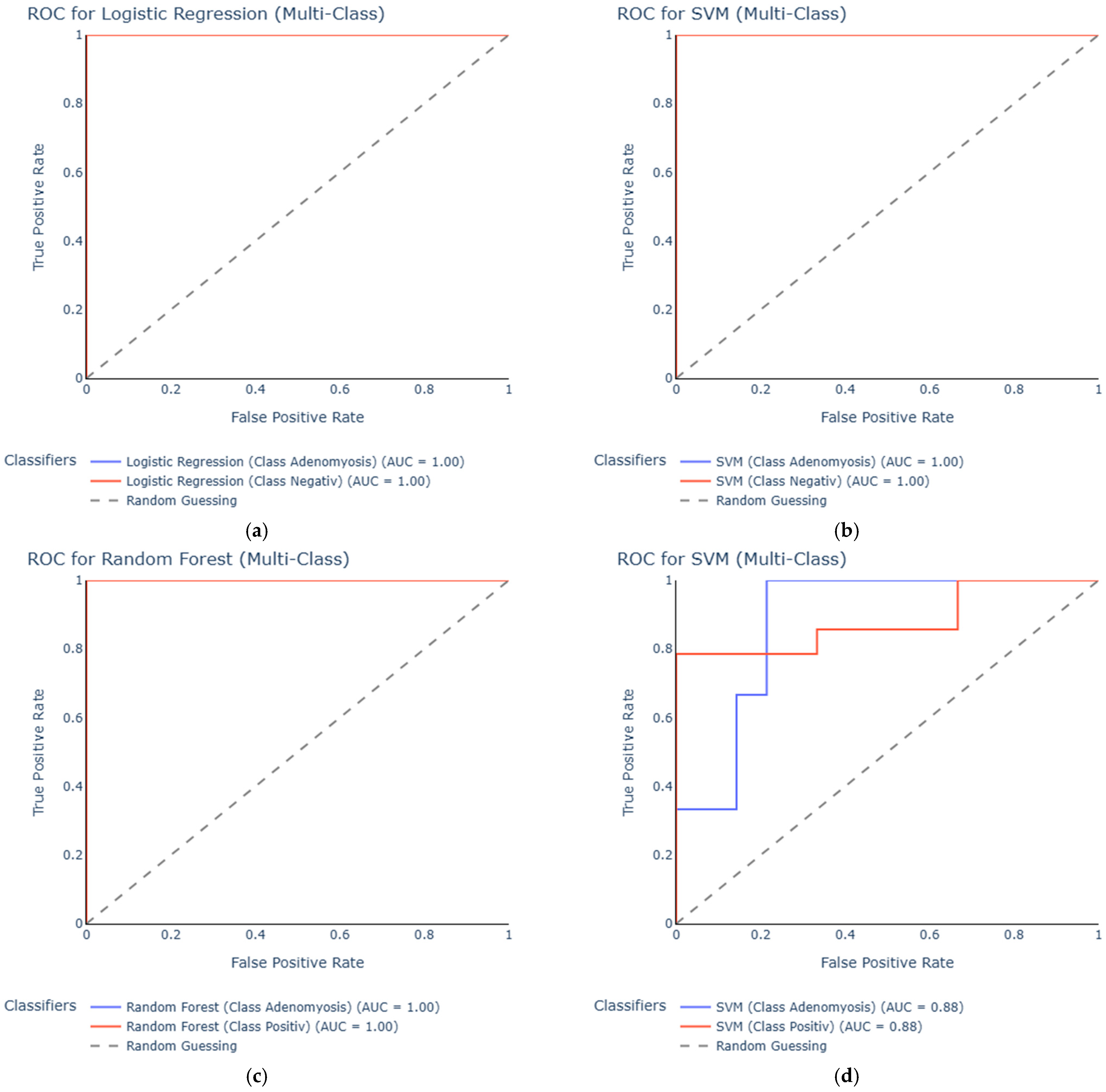
3.2. ROC
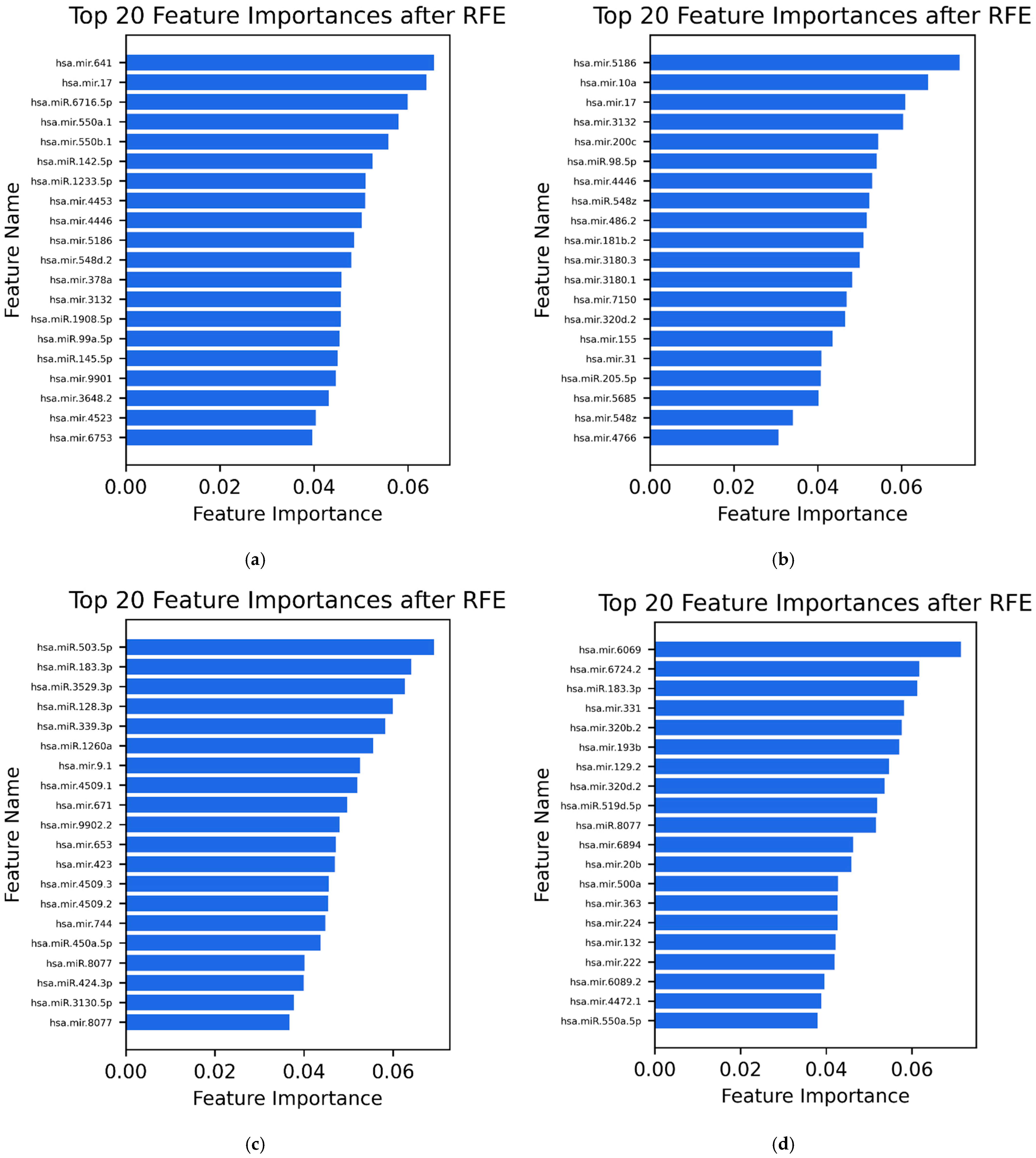
3.3. Selected Features (20 MiRNAs)
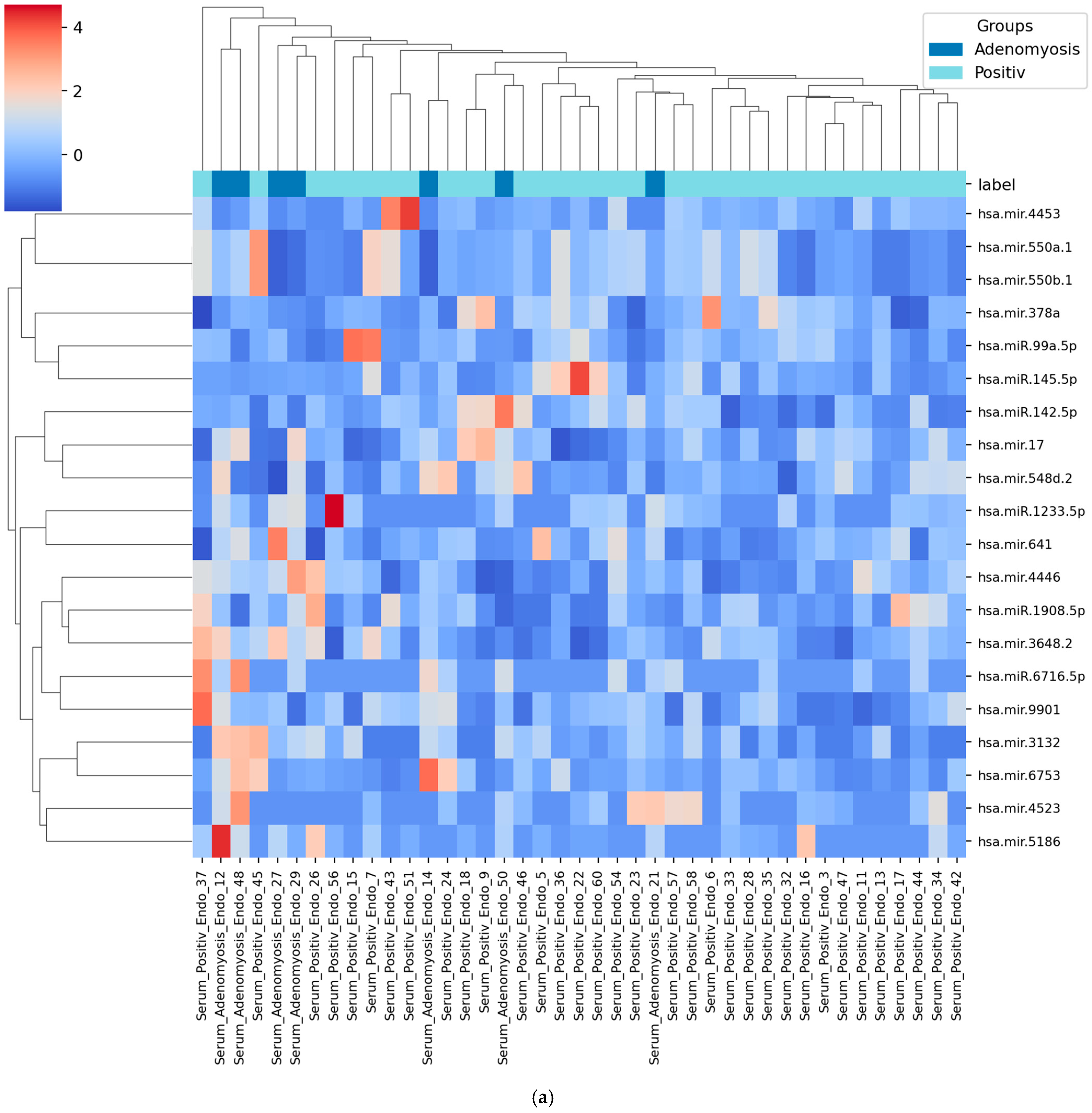
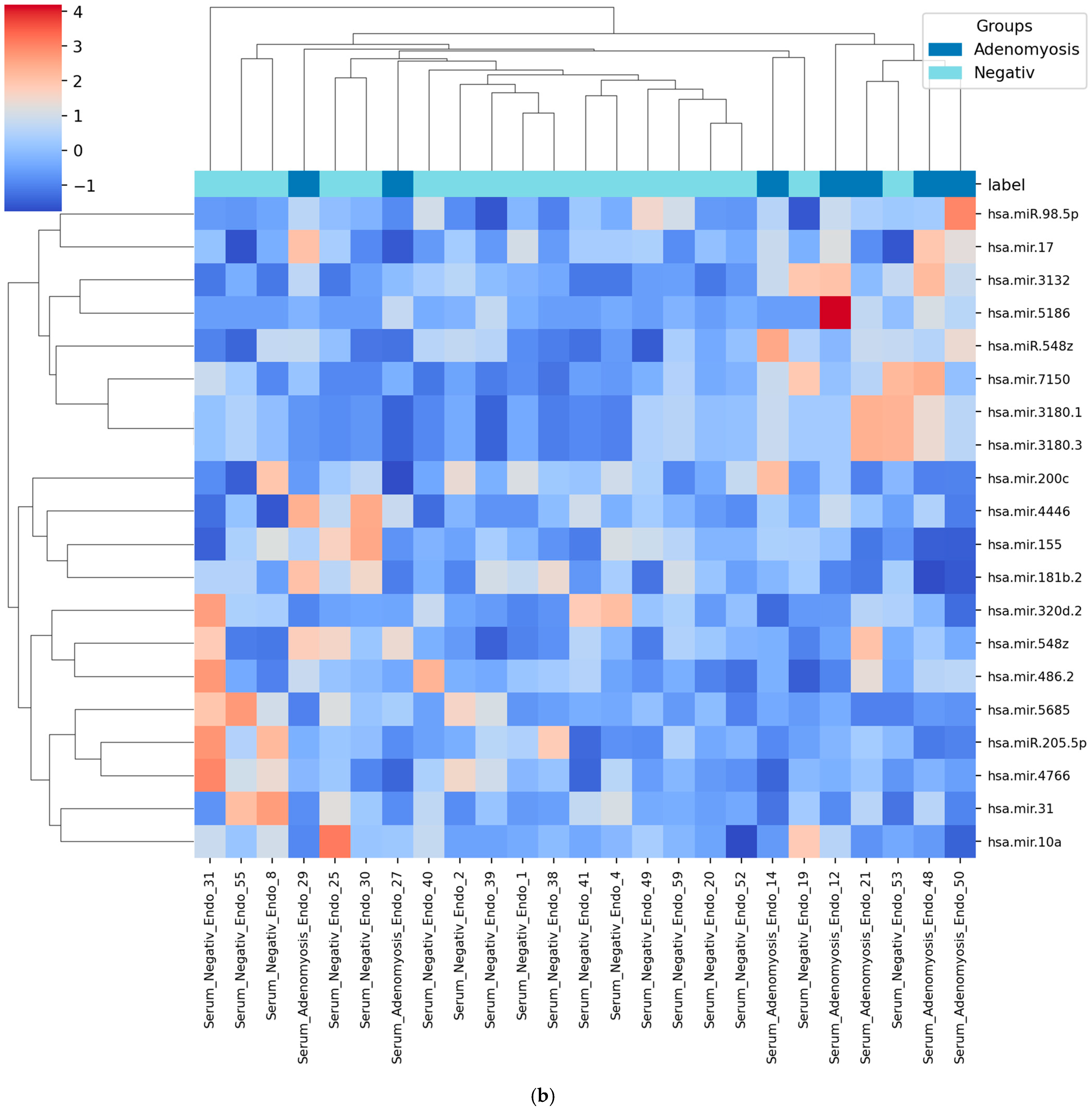
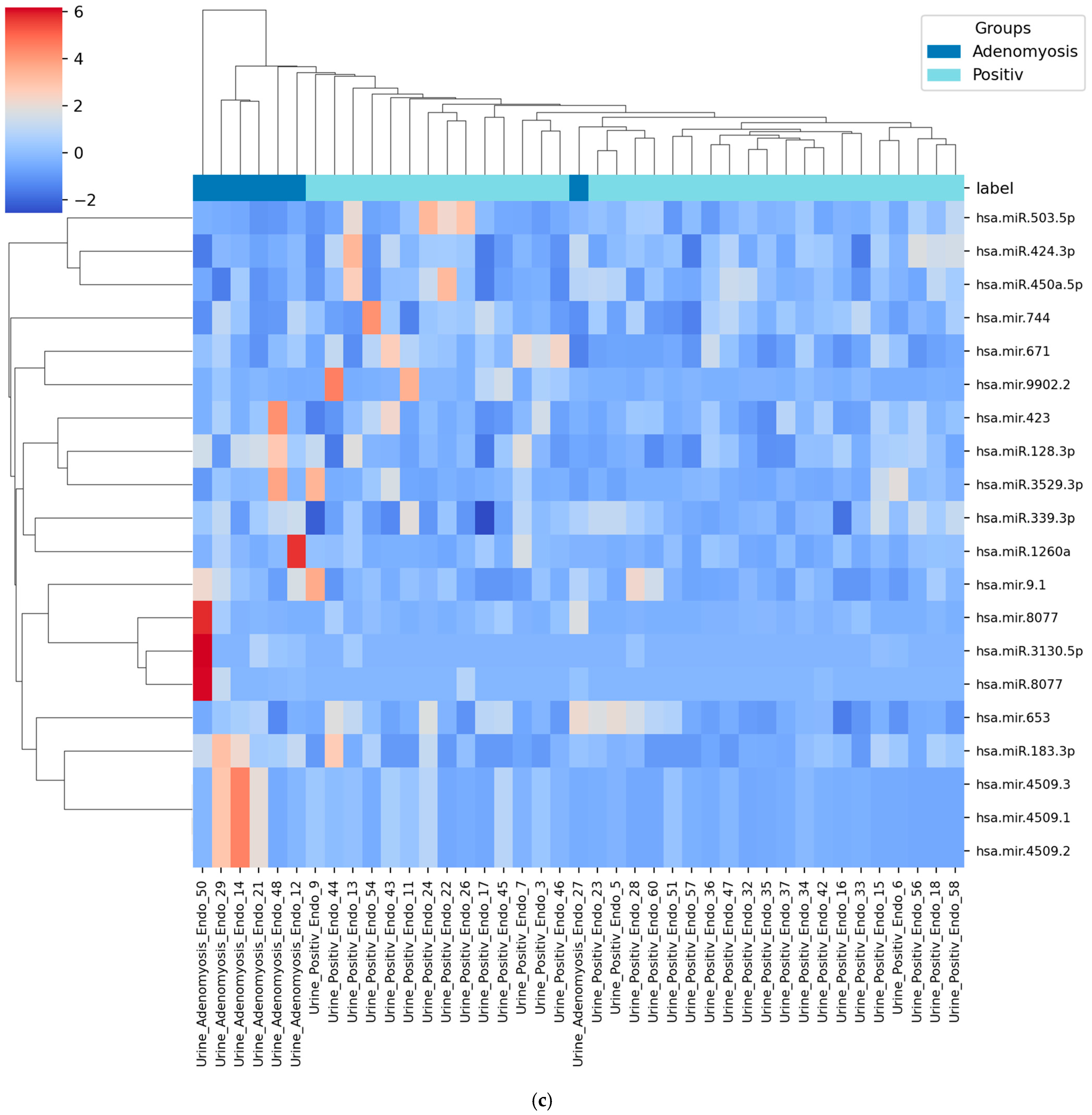
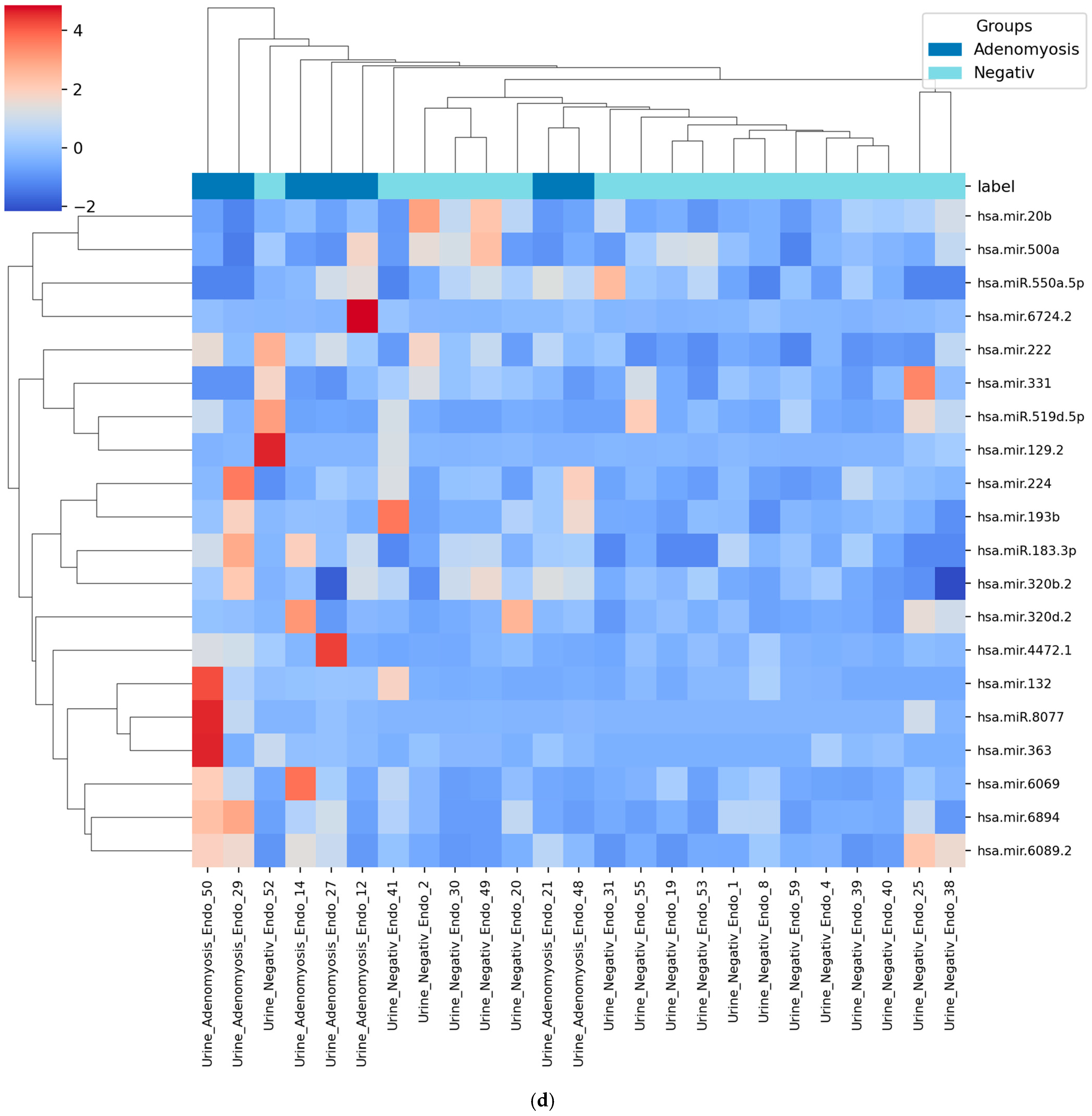
3.4. Heatmap of Selected miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef]
- Upson, K.; Missmer, S.A. Epidemiology of Adenomyosis. Semin. Reprod. Med. 2020, 38, 89–107. [Google Scholar] [CrossRef]
- Chapron, C.; Vannuccini, S.; Santulli, P.; Abrão, M.S.; Carmona, F.; Fraser, I.S.; Gordts, S.; Guo, S.W.; Just, P.A.; Noël, J.C.; et al. Diagnosing adenomyosis: An integrated clinical and imaging approach. Hum. Reprod. Update 2020, 26, 392–411. [Google Scholar] [CrossRef]
- Naftalin, J.; Hoo, W.; Pateman, K.; Mavrelos, D.; Foo, X.; Jurkovic, D. Is adenomyosis associated with menorrhagia? Hum. Reprod. 2014, 29, 473–479. [Google Scholar] [CrossRef]
- Van den Bosch, T.; Dueholm, M.; Leone, F.P.; Valentin, L.; Rasmussen, C.K.; Votino, A.; Van Schoubroeck, D.; Landolfo, C.; Installé, A.J.; Guerriero, S.; et al. Terms, definitions and measurements to describe sonographic features of myometrium and uterine masses: A consensus opinion from the Morphological Uterus Sonographic Assessment (MUSA) group. Ultrasound Obstet. Gynecol. 2015, 46, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, M.J.; Van den Bosch, T.; de Leeuw, R.A.; Dueholm, M.; Exacoustos, C.; Valentin, L.; Hehenkamp, W.J.K.; Groenman, F.; De Bruyn, C.; Rasmussen, C.; et al. Consensus on revised definitions of Morphological Uterus Sonographic Assessment (MUSA) features of adenomyosis: Results of modified Delphi procedure. Ultrasound Obstet. Gynecol. 2022, 60, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Andres, M.P.; Borrelli, G.M.; Ribeiro, J.; Baracat, E.C.; Abrão, M.S.; Kho, R.M. Transvaginal Ultrasound for the Diagnosis of Adenomyosis: Systematic Review and Meta-Analysis. J. Minim. Invasive Gynecol. 2018, 25, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, J.L.; Vara, J.; Usandizaga, C.; Ajossa, S.; Pascual, M.; Guerriero, S. Transvaginal ultrasound versus magnetic resonance imaging for diagnosing adenomyosis: A systematic review and head-to-head meta-analysis. Int. J. Gynaecol. Obstet. 2023, 161, 397–405. [Google Scholar] [CrossRef]
- Tellum, T.; Matic, G.V.; Dormagen, J.B.; Nygaard, S.; Viktil, E.; Qvigstad, E.; Lieng, M. Diagnosing adenomyosis with MRI: A prospective study revisiting the junctional zone thickness cutoff of 12 mm as a diagnostic marker. Eur. Radiol. 2019, 29, 6971–6981. [Google Scholar] [CrossRef]
- Champaneria, R.; Abedin, P.; Daniels, J.; Balogun, M.; Khan, K.S. Ultrasound scan and magnetic resonance imaging for the diagnosis of adenomyosis: Systematic review comparing test accuracy. Acta Obstet. Gynecol. Scand. 2010, 89, 1374–1384. [Google Scholar] [CrossRef]
- Gordts, S.; Koninckx, P.; Brosens, I. Pathogenesis of deep endometriosis. Fertil. Steril. 2017, 108, 872–885.e871. [Google Scholar] [CrossRef]
- Hashimoto, A.; Iriyama, T.; Sayama, S.; Okamura, A.; Kato, K.; Fujii, T.; Kubota, K.; Ichinose, M.; Sone, K.; Kumasawa, K.; et al. Differences in the incidence of obstetric complications depending on the extent and location of adenomyosis lesions. J. Matern. Fetal Neonatal Med. 2023, 36, 2226789. [Google Scholar] [CrossRef] [PubMed]
- Vercellini, P.; Viganò, P.; Bandini, V.; Buggio, L.; Berlanda, N.; Somigliana, E. Association of endometriosis and adenomyosis with pregnancy and infertility. Fertil. Steril. 2023, 119, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wu, Y.; Yu, W.; Li, H. Identification of a seven-miRNA signature as prognostic biomarker for lung squamous cell carcinoma. Oncotarget 2016, 7, 81670–81679. [Google Scholar] [CrossRef]
- Yang, Y.; Mei, Q. miRNA signature identification of retinoblastoma and the correlations between differentially expressed miRNAs during retinoblastoma progression. Mol. Vis. 2015, 21, 1307–1317. [Google Scholar]
- Zhang, Y.; Sun, X.; Li, Z.; Han, X.; Wang, W.; Xu, P.; Liu, Y.; Xue, Y.; Wang, Z.; Xu, S.; et al. Interactions between miRNAs and the Wnt/β-catenin signaling pathway in endometriosis. Biomed. Pharmacother. 2024, 171, 116182. [Google Scholar] [CrossRef]
- Watrowski, R.; Kostov, S.; Palumbo, M.; Rosati, A.; Sparić, R.; Alkatout, I.; Juhasz-Böss, I.; Vitale, S.G.; Mereu, L. Non-Coding RNAs (microRNAs, lncRNAs, circRNAs) in Adenomyosis: A Systematic Review of Mechanistic and Translational Evidence. Int. J. Mol. Sci. 2025, 26, 10713. [Google Scholar] [CrossRef]
- Fortunato, O.; Verri, C.; Pastorino, U.; Sozzi, G.; Boeri, M. MicroRNA Profile of Lung Tumor Tissues Is Associated with a High Risk Plasma miRNA Signature. Microarrays 2016, 5, 18. [Google Scholar] [CrossRef]
- Ritter, A.; Hirschfeld, M.; Berner, K.; Jaeger, M.; Grundner-Culemann, F.; Schlosser, P.; Asberger, J.; Weiss, D.; Noethling, C.; Mayer, S.; et al. Discovery of potential serum and urine-based microRNA as minimally-invasive biomarkers for breast and gynecological cancer. Cancer Biomark. 2020, 27, 225–242. [Google Scholar] [CrossRef]
- Bendifallah, S.; Suisse, S.; Puchar, A.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Jornea, L.; Bouteiller, D.; Touboul, C.; et al. Salivary MicroRNA Signature for Diagnosis of Endometriosis. J. Clin. Med. 2022, 11, 612. [Google Scholar] [CrossRef]
- Kupec, T.; Bleilevens, A.; Iborra, S.; Najjari, L.; Wittenborn, J.; Maurer, J.; Stickeler, E. Stability of circulating microRNAs in serum. PLoS ONE 2022, 17, e0268958. [Google Scholar] [CrossRef] [PubMed]
- Kupec, T.; Bleilevens, A.; Klein, B.; Hansen, T.; Najjari, L.; Wittenborn, J.; Stickeler, E.; Maurer, J. Comparison of Serum and Urine as Sources of miRNA Markers for the Detection of Ovarian Cancer. Biomedicines 2023, 11, 2508. [Google Scholar] [CrossRef] [PubMed]
- Bendifallah, S.; Puchar, A.; Suisse, S.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Touboul, C.; Dabi, Y.; Daraï, E. Machine learning algorithms as new screening approach for patients with endometriosis. Sci. Rep. 2022, 12, 639. [Google Scholar] [CrossRef] [PubMed]
- Bendifallah, S.; Roman, H.; Suisse, S.; Spiers, A.; Petit, E.; Delbos, L.; Dabi, Y.; Touboul, C.; Dennis, T.; Merlot, B.; et al. Validation of a Saliva Micro–RNA Signature for Endometriosis. NEJM Evid. 2025, 4, EVIDoa2400195. [Google Scholar] [CrossRef]
- Becker, C.M.; Bokor, A.; Heikinheimo, O.; Horne, A.; Jansen, F.; Kiesel, L.; King, K.; Kvaskoff, M.; Nap, A.; Petersen, K.; et al. ESHRE guideline: Endometriosis. Hum. Reprod. Open 2022, 2022, hoac009. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bourdon, M.; Santulli, P.; Jeljeli, M.; Vannuccini, S.; Marcellin, L.; Doridot, L.; Petraglia, F.; Batteux, F.; Chapron, C. Immunological changes associated with adenomyosis: A systematic review. Hum. Reprod. Update 2021, 27, 108–129. [Google Scholar] [CrossRef]
- Jin, T.; Li, M.; Li, T.; Yan, S.; Ran, Q.; Chen, W. The Inactivation of Hippo Signaling Pathway Promotes the Development of Adenomyosis by Regulating EMT, Proliferation, and Apoptosis of Cells. Reprod. Sci. 2023, 30, 2715–2727. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, J.; Jia, C.; Liao, Y. SKP2 regulates ZEB1 expression and stimulates eutopic endometrial stromal cell invasion and proliferation of adenomyosis. Reprod. Biol. 2022, 22, 100578. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, L. miR-183 inhibits the viability, migration and invasion of epithelium on adenomyosis via targeting MMP-9. J. Gynecol. Obstet. Hum. Reprod. 2022, 51, 102349. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Zhang, W.; Wang, H.; Shi, W.; Wang, L.; Ma, L. Levonorgestrel Ameliorates Adenomyosis via lncRNA H19/miR-17/TLR4 Pathway. Drug Des. Devel. Ther. 2020, 14, 3449–3460. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Fuse, M.; Goto, Y.; Kaga, K.; Kurozumi, A.; Yamada, Y.; Sugawara, S.; Okato, A.; Ichikawa, T.; Yamanishi, T.; et al. Molecular pathogenesis of interstitial cystitis based on microRNA expression signature: miR-320 family-regulated molecular pathways and targets. J. Hum. Genet. 2018, 63, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Duan, H.; Wang, S.; Wang, S.; Lin, Q.; Li, Y. RNA-seq reveals co-dysregulated circular RNAs in the adenomyosis eutopic endometrium and endometrial-myometrial interface. BMC Womens Health 2022, 22, 293. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Shah, P.; Kendall, F.; Khozin, S.; Goosen, R.; Hu, J.; Laramie, J.; Ringel, M.; Schork, N. Artificial intelligence and machine learning in clinical development: A translational perspective. NPJ Digit. Med. 2019, 2, 69. [Google Scholar] [CrossRef]
- Kupec, T.; Wittenborn, J.; Kuo, C.C.; Najjari, L.; Senger, R.; Meyer-Wilmes, P.; Stickeler, E.; Maurer, J. Diagnostic Potential of Serum Circulating miRNAs for Endometriosis in Patients with Chronic Pelvic Pain. J. Clin. Med. 2025, 14, 5154. [Google Scholar] [CrossRef]
| Median (Range) | |
|---|---|
| Age | 27 (18–34) |
| Location of Endometriosis (#ENZIAN) | Number of Patients n |
| #ENZIAN P (Peritoneal) | 29 |
| #ENZIAN O (Ovary) | 6 |
| #ENZIAN T (Tube) | 2 |
| Deep infiltrating endometriosis | 17 |
| #ENZIAN A | 5 |
| #ENZIAN B | 12 |
| #ENZIAN C | 0 |
| #ENZIAN FB | 2 |
| #ENZIAN FI | 1 |
| Model | Accuracy (Serum: Adenomyosis vs. Positive Controls) | Accuracy (Serum: Adenomyosis vs. Negative Controls) | Accuracy (Urine: Adenomyosis vs. Positive Controls) | Accuracy (Urine: Adenomyosis vs. Negative Controls) |
|---|---|---|---|---|
| Logistic Regression | 0.88 | 0.60 | 0.88 | 0.90 |
| Decision Tree | 0.71 | 0.60 | 0.82 | 0.80 |
| Random Forest | 0.82 | 0.60 | 0.88 | 0.90 |
| SVM | 0.82 | 0.60 | 0.88 | 0.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kupec, T.; Wittenborn, J.; Kuo, C.-C.; Senger, R.; Meyer-Wilmes, P.; Najjari, L.; Stickeler, E.; Maurer, J. Urine and Serum miRNA Signatures for the Non-Invasive Diagnosis of Adenomyosis: A Machine Learning-Based Pilot Study. Diagnostics 2025, 15, 3012. https://doi.org/10.3390/diagnostics15233012
Kupec T, Wittenborn J, Kuo C-C, Senger R, Meyer-Wilmes P, Najjari L, Stickeler E, Maurer J. Urine and Serum miRNA Signatures for the Non-Invasive Diagnosis of Adenomyosis: A Machine Learning-Based Pilot Study. Diagnostics. 2025; 15(23):3012. https://doi.org/10.3390/diagnostics15233012
Chicago/Turabian StyleKupec, Tomas, Julia Wittenborn, Chao-Chung Kuo, Rebecca Senger, Philipp Meyer-Wilmes, Laila Najjari, Elmar Stickeler, and Jochen Maurer. 2025. "Urine and Serum miRNA Signatures for the Non-Invasive Diagnosis of Adenomyosis: A Machine Learning-Based Pilot Study" Diagnostics 15, no. 23: 3012. https://doi.org/10.3390/diagnostics15233012
APA StyleKupec, T., Wittenborn, J., Kuo, C.-C., Senger, R., Meyer-Wilmes, P., Najjari, L., Stickeler, E., & Maurer, J. (2025). Urine and Serum miRNA Signatures for the Non-Invasive Diagnosis of Adenomyosis: A Machine Learning-Based Pilot Study. Diagnostics, 15(23), 3012. https://doi.org/10.3390/diagnostics15233012







