ConvNeXt-Driven Detection of Alzheimer’s Disease: A Benchmark Study on Expert-Annotated AlzaSet MRI Dataset Across Anatomical Planes
Abstract
1. Introduction
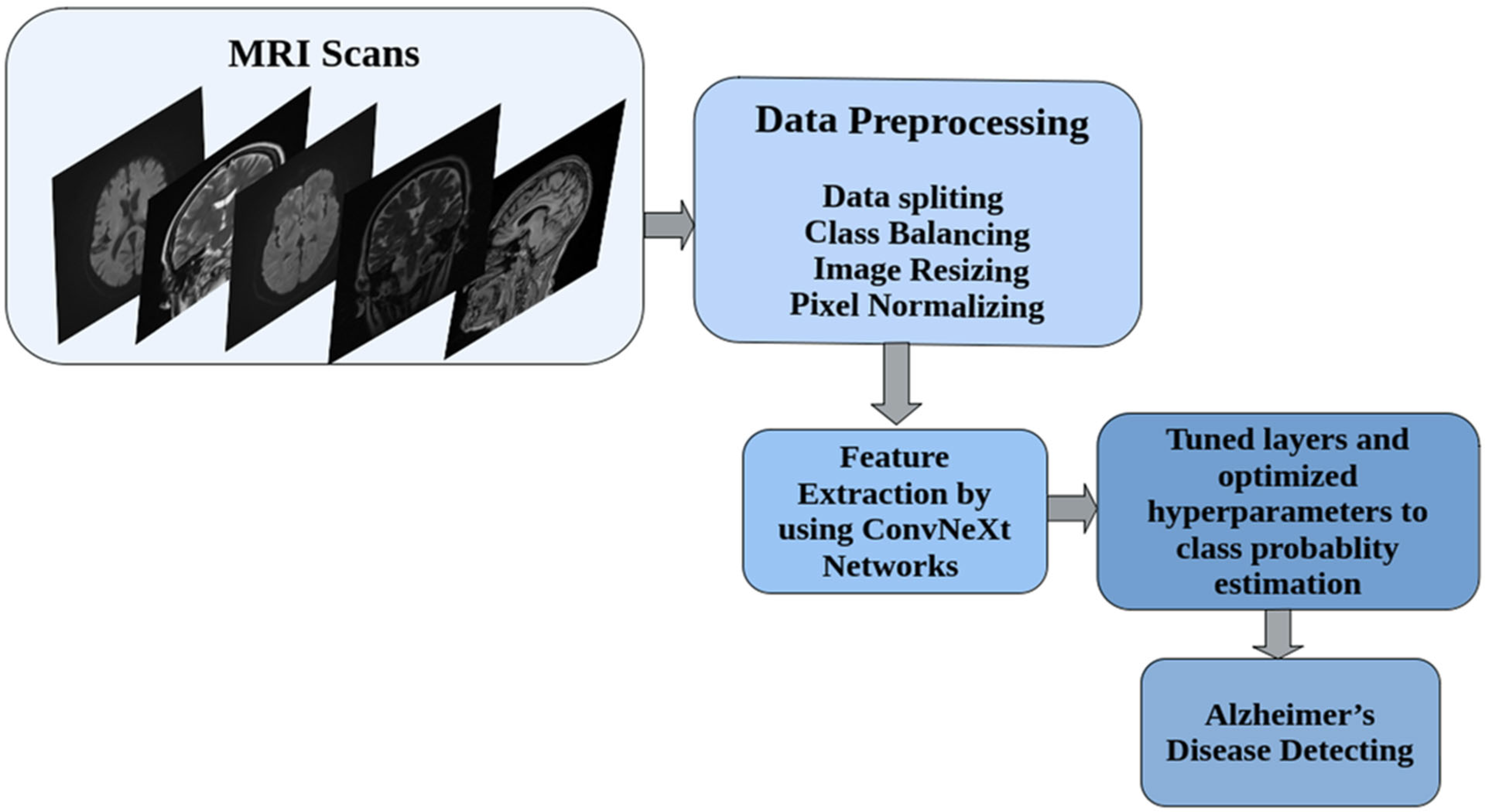
2. Materials and Methods
2.1. Dataset Description: The AlzaSet Cohort
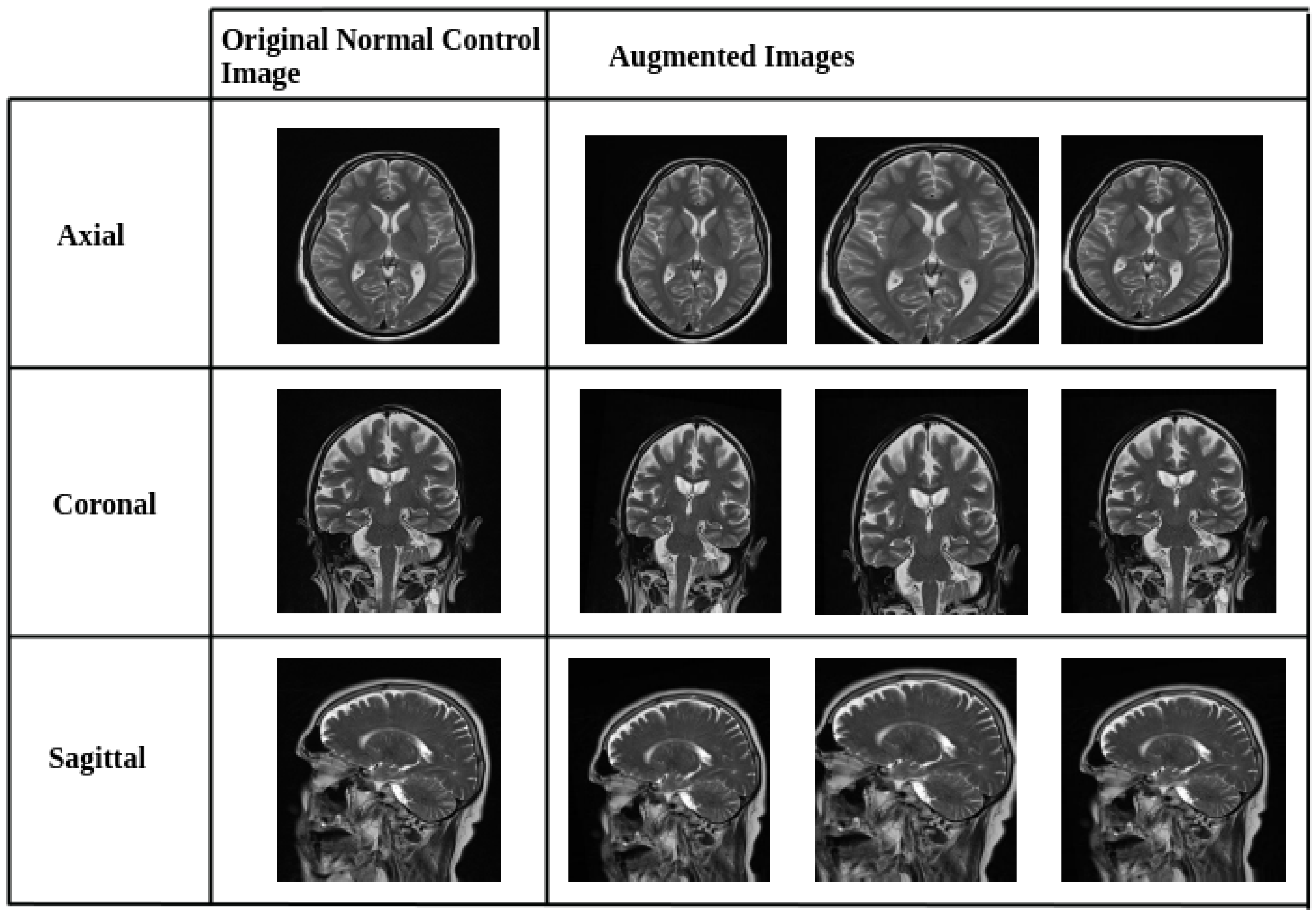
2.2. Data Preprocessing and Augmentation Strategy
2.3. Model Architecture and Implementation of Transfer Learning and Feature Extraction Strategy Using ConvNeXt Architecture
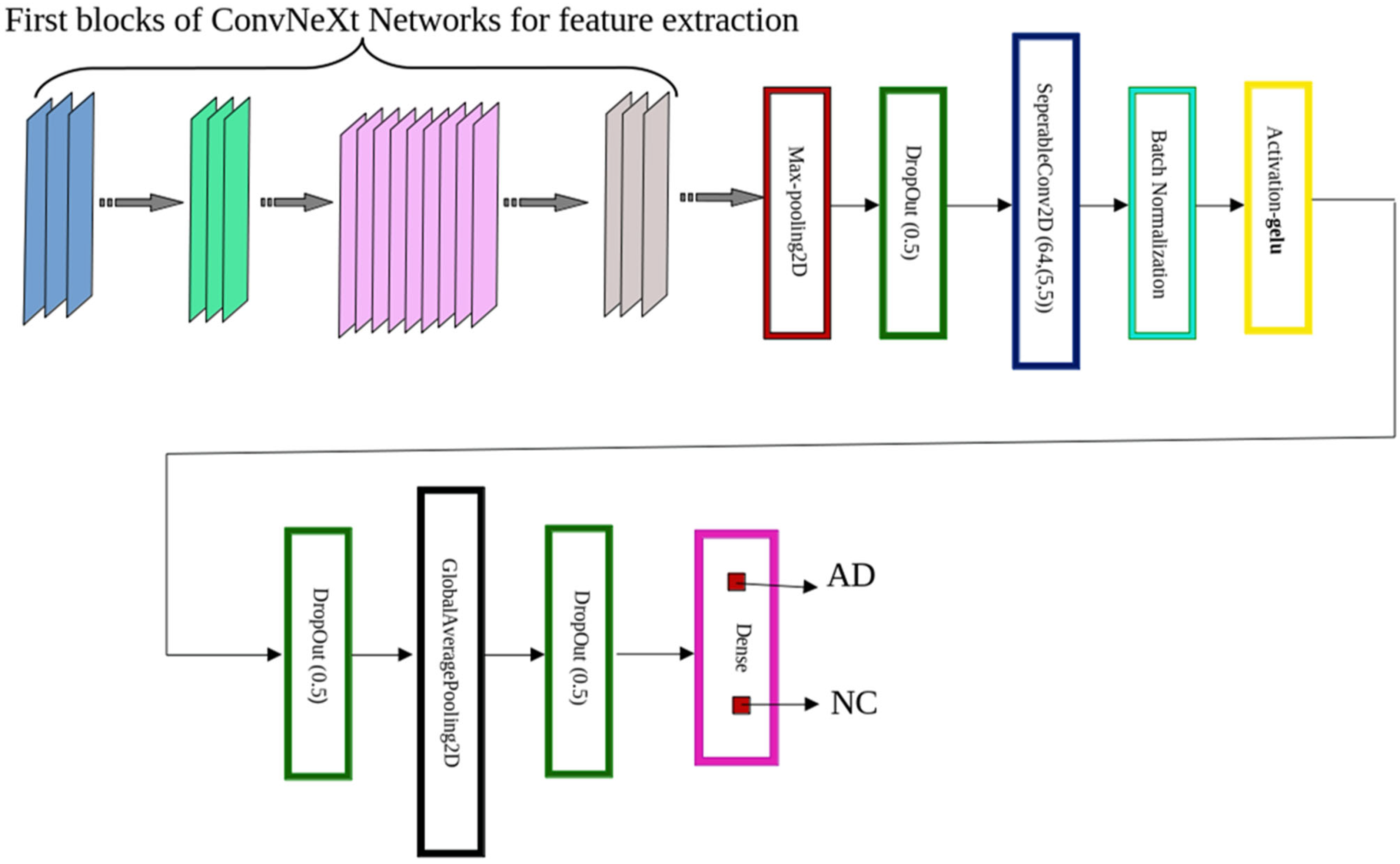
2.4. ConvNeXt-Based Model Design Architecture
- ConvNeXt-Tiny consists of four stages with [3, 3, 9, 3] blocks, respectively. We preserved the first 125 layers and truncated the final 25 layers to prevent overfitting and reduce model complexity. Each block implements the sequence: Input → LayerNorm → PWConv (1 × 1) → GELU → DWConv (7 × 7) → PWConv (1 × 1) → Residual Add, where DWConv denotes depthwise convolution and PWConv denotes pointwise convolution.
- ConvNeXt-Small shares the same block structure as Tiny but incorporates deeper feature extraction in Stage 3, with [3, 3, 27, 3] blocks. We retained the first 240 layers and removed the final 50 layers for this configuration. Furthermore, the deeper feature extraction results from the iterative application of blocks rather than being an inherent component of the architecture itself.
- ConvNeXt-Base expands upon the Small variant by increasing channel widths for greater representational capacity. We extracted features from the first 245 layers, truncating the final 45 layers.
- MaxPooling2D—A spatial pooling operation (e.g., 2 × 2) that downsamples the feature map by selecting the maximum activation within each region. This reduces dimensionality, accelerates computation, and mitigates overfitting by emphasizing salient features [28].
- Dropout (rate = 0.5)—A regularization technique that randomly deactivates 50% of neurons during training, improving generalization by preventing co-adaptation of feature detectors [29].
- SeparableConv2D—Implements a two-step process: (i) depth-wise convolution, which applies a spatial filter independently to each input channel, followed by (ii) pointwise convolution (1 × 1) to mix inter-channel information. This architecture offers significant computational savings while retaining competitive performance [30].
- Batch Normalization—Normalizes each feature map by applying consistent mean and variance across spatial dimensions. This stabilizes and accelerates training, especially in deep networks [31].
- GELU Activation—The Gaussian Error Linear Unit (GELU) activation, defined as:where represents the standard Gaussian cumulative distribution function and erf denotes the error function. The GELU outperforms other activation functions such as ReLU and ELU across multiple tasks, as demonstrated in the experiments [32].
- GlobalAveragePooling2D—Reduces each feature map to a single scalar by averaging across all spatial dimensions (height and width), effectively summarizing global context while minimizing parameter count [33].
- Dense Layer with Softmax Activation—A fully connected output layer containing two neurons, representing the binary classes (AD and NC). The softmax activation function generates normalized probability scores for final classification.
2.5. Training Configuration and Hyperparameters
- True Positives (tp): Correctly identified instances of the class.
- True Negatives (tn): Correctly rejected instances that do not belong to the class.
- False Positives (fp): Instances wrongly classified as part of the class.
3. Results
3.1. Model Performance Across Anatomical Planes
3.2. Comparative Benchmarking Against Established Models
4. Discussion
4.1. Interpretation of Findings and Architectural Insights
4.2. Integration with Prior Literature
4.3. Comparative Positioning Relative to Existing ConvNeXt-Based AD Classifiers
4.4. Clinical Implications and Future Directions
4.5. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Dementia. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 17 November 2025).
- Cataldi, R.; Chowdhary, N.; Seeher, K.; Moorthy, V.; Dua, T. A blueprint for the worldwide research response to dementia. Lancet Neurol. 2022, 21, 690–691. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.E.; Canning, D.; Lubet, A. Global Population Aging: Facts, Challenges, Solutions & Perspectives. Dædalus J. Am. Acad. Arts Sci. 2015, 144, 80–92. [Google Scholar] [CrossRef]
- Awad, M.H.; Sanchez, M.; Abikenari, M.A. The values work of restorative ventures: The role of founders’ embodied embeddedness with at-risk social groups. J. Bus. Ventur. Insights 2022, 18, e00337. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Wiste, H.J.; Vemuri, P.; Weigand, S.D.; Senjem, M.L.; Zeng, G.; Bernstein, M.A.; Gunter, J.L.; Pankratz, V.S.; Aisen, P.S.; et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain 2010, 133, 3336–3348. [Google Scholar] [CrossRef]
- Oughli, H.A.; Siddarth, P.; Lum, M.; Tang, L.; Ito, B.; Abikenari, M.; Cappelleti, M.; Khalsa, D.S.; Nguyen, S.; Lavretsky, H. Peripheral Alzheimer’s disease biomarkers are related to change in subjective memory in older women with cardiovascular risk factors in a trial of yoga vs memory training. Can. J. Psychiatry 2025, 07067437251343291. [Google Scholar] [CrossRef]
- Abikenari, M.; Jain, B.; Xu, R.; Jackson, C.; Huang, J.; Bettegowda, C.; Lim, M. Bridging imaging and molecular biomarkers in trigeminal neuralgia: Toward precision diagnosis and prognostication in neuropathic pain. Med. Res. Arch. 2025, 13. [Google Scholar] [CrossRef]
- Siddarth, P.; Abikenari, M.; Grzenda, A.; Cappelletti, M.; Oughli, H.; Liu, C.; Millillo, M.M.; Lavretsky, H. Inflammatory markers of geriatric depression response to Tai Chi or health education adjunct interventions. Am. J. Geriatr. Psychiatry 2023, 31, 22–32. [Google Scholar] [CrossRef]
- Hasan Saif, F.; Al-Andoli, M.N.; Wan Bejuri, W.M.Y. Explainable AI for Alzheimer Detection: A Review of Current Methods and Applications. Appl. Sci. 2024, 14, 10121. [Google Scholar] [CrossRef]
- Al-Bakri, F.H.; Wan Bejuri, W.M.Y.; Al-Andoli, M.N.; Ikram, R.R.R.; Khor, H.M.; Tahir, Z. A Meta-Learning-Based Ensemble Model for Explainable Alzheimer’s Disease Diagnosis. Diagnostics 2025, 15, 1642. [Google Scholar] [CrossRef]
- Al-Bakri, F.H.; Wan Bejuri, W.M.Y.; Al-Andoli, M.N.; Ikram, R.R.R.; Khor, H.M.; Tahir, Z. A Feature-Augmented Explainable Artificial Intelligence Model for Diagnosing Alzheimer’s Disease from Multimodal Clinical and Neuroimaging Data. Diagnostics 2025, 15, 2060. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Thibeau-Sutre, E.; Diaz-Melo, M.; Samper-González, J.; Routier, A.; Bottani, S.; Dormont, D.; Durrleman, S.; Burgos, N.; Colliot, O. Convolutional neural networks for classification of Alzheimer’s disease: Overview and reproducible evaluation. Med. Image Anal. 2020, 63, 101694. [Google Scholar] [CrossRef] [PubMed]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; Volume 2. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014. [Google Scholar] [CrossRef]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Eroglu, Y.; Yildirim, M.; Cinar, A. mRMR-based hybrid convolutional neural network model for classification of Alzheimer’s disease on brain magnetic resonance images. Int. J. Imaging Syst. Technol. 2022, 32, 517–527. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022. [Google Scholar] [CrossRef]
- Nanni, L.; Interlenghi, M.; Brahnam, S.; Salvatore, C.; Papa, S.; Nemni, R.; Castiglioni, I.; Alzheimer’s Disease Neuroimaging Initiative. Comparison of Transfer Learning and Conventional Machine Learning Applied to Structural Brain MRI for the Early Diagnosis and Prognosis of Alzheimer’s Disease. Front. Neurol. 2020, 11, 576194. [Google Scholar] [CrossRef]
- Mehmood, A.; Yang, S.; Feng, Z.; Wang, M.; Ahmad, A.S.; Khan, R.; Maqsood, M.; Yaqub, M. A Transfer Learning Approach for Early Diagnosis of Alzheimer’s Disease on MRI Images. Neuroscience 2021, 460, 43–52. [Google Scholar] [CrossRef]
- Khan, R.; Akbar, S.; Mehmood, A.; Shahid, F.; Munir, K.; Ilyas, N.; Asif, M.; Zheng, Z. A transfer learning approach for multiclass classification of Alzheimer’s disease using MRI images. Front. Neurosci. 2022, 16, 1050777. [Google Scholar] [CrossRef]
- Mahmud, T.; Barua, K.; Barua, A.; Das, S.; Basnin, N.; Hossain, M.S.; Andersson, K.; Kaiser, M.S.; Sharmen, N. Exploring Deep Transfer Learning Ensemble for Improved Diagnosis and Classification of Alzheimer’s Disease. In Lecture Notes in Computer Science, Proceedings of the 16th International Conference, BI 2023, Hoboken, NJ, USA, 1–3 August 2023; Springer: Berlin/Heidelberg, Germany, 2023. [Google Scholar] [CrossRef]
- Reddy C, K.K.; Rangarajan, A.; Rangarajan, D.; Shuaib, M.; Jeribi, F.; Alam, S. A Transfer Learning Approach: Early Prediction of Alzheimer’s Disease on US Healthy Aging Dataset. Mathematics 2024, 12, 2204. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, Y.; Xiao, L. Alzheimer’s disease diagnosis by 3D-SEConvNeXt. J. Big Data 2025, 12, 15. [Google Scholar] [CrossRef]
- Jin, W.; Yin, Y.; Bai, J.; Zhen, H. CA-ConvNeXt: Coordinate Attention on ConvNeXt for Early Alzheimer’s disease classification. In Proceedings of the Intelligence Science IV, ICIS 2022, IFIP Advances in Information and Communication Technology, Xi’an, China, 28–31 October 2022; Springer: Cham, Switzerland; Volume 659. [Google Scholar] [CrossRef]
- Angriawan, M. Transfer Learning Strategies for Fine-Tuning Pretrained Convolutional Neural Networks in Medical Imaging. Res. J. Comput. Syst. Eng. 2023, 4, 73–88. [Google Scholar] [CrossRef]
- Sharma, S.; Mehra, R. Implications of Pooling Strategies in Convolutional Neural Networks: A Deep Insight. Found. Comput. Decis. Sci. 2019, 44, 303–330. [Google Scholar] [CrossRef]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the ICML’15: 32nd International Conference on International Conference on Machine Learning, Lille, France, 6–11 July 2015; Volume 37, pp. 448–456. [Google Scholar] [CrossRef]
- Hendrycks, D.; Gimpel, K. Gaussian Error Linear Units (GELUs). arXiv 2016. [Google Scholar] [CrossRef]
- Lin, M.; Chen, Q.; Yan, S. Network in Network. arXiv 2013. [Google Scholar] [CrossRef]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Kohavi, R.; Provost, F. Glossary of Terms; Stanford University Technical Report; Stanford University: Stanford, CA, USA, 1998; Available online: https://ai.stanford.edu/~ronnyk/glossary.html (accessed on 17 November 2025).
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Techa, C.; Ridouani, M.; Hassouni, L.; Anoun, H. Automated Alzheimer’s disease classification from brain MRI scans using ConvNeXt and ensemble of machine learning classifiers. In Proceedings of the International Conference on Soft Computing and Pattern Recognition, Online, 14–16 December 2022; Springer Nature: Cham, Switzerland, 2022. [Google Scholar]
- Li, H.; Habes, M.; Wolk, D.A.; Fan, Y.; Alzheimer’s Disease Neuroimaging Initiative and the Australian Imaging Biomarkers and Lifestyle Study of Aging. A deep learning model for early prediction of Alzheimer’s disease dementia based on hippocampal magnetic resonance imaging data. Alzheimers Dement. 2019, 15, 1059–1070. [Google Scholar] [CrossRef]
- Moscoso, A.; Heeman, F.; Raghavan, S.; Costoya-Sánchez, A.; van Essen, M.; Mainta, I.; Camacho, V.; Rodríguez-Fonseca, O.; Silva-Rodríguez, J.; Perissinotti, A.; et al. Frequency and Clinical Outcomes Associated with Tau Positron Emission Tomography Positivity. JAMA 2025, 334, 229–242. [Google Scholar] [CrossRef]
- Abikenari, M.; Awad, M.H.; Korouri, S.; Mohseni, K.; Abikenari, D.; Freichel, R.; Mukadam, Y.; Tanzim, U.; Habib, A.; Kerwan, A. Reframing Clinical AI Evaluation in the Era of Generative Models: Toward Multidimensional, Stakeholder-Informed, and Safety-Centric Frameworks for Real-World Health Care Deployment. Prem. J. Sci. 2025, 11, 100089. [Google Scholar] [CrossRef]
- Luo, X.; Wang, X.; Eweje, F.; Zhang, X.; Yang, S.; Quinton, R.; Xiang, J.; Li, Y.; Ji, Y.; Li, Z.; et al. Ensemble learning of foundation models for precision oncology. arXiv 2025. [Google Scholar] [CrossRef]
- Basereh, M.; Abikenari, M.; Sadeghzadeh, S.; Dunn, T.; Freichel, R.; Siddarth, P.; Ghahremani, D.; Lavretsky, H.; Buch, V.P. ConvNeXt-Driven Detection of Alzheimer’s Disease: A Benchmark Study on Expert-Annotated AlzaSet MRI Dataset Across Anatomical Planes. bioRxiv 2025. [Google Scholar] [CrossRef]
| Participant Status | Age | Gender (F/M)% |
|---|---|---|
| AD | 37/63 | |
| NC | 54/46 |
| Participants Status | Axial | Coronal | Sagittal | Total |
|---|---|---|---|---|
| AD | 4876 | 1801 | 4053 | 10,730 |
| NC | 1283 | 283 | 651 | 2217 |
| Total | 6159 | 2084 | 4704 | 12,947 |
| Axial | Train | Validation | Test | Coronal | Train | Validation | Test | Sagittal | Train | Validation | Test |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | 3459 | 954 | 463 | AD | 1247 | 449 | 184 | AD | 2823 | 826 | 404 |
| NC | 913 | 240 | 130 | NC | 184 | 66 | 32 | NC | 408 | 160 | 83 |
| Axial | Train | Validation | Test | Coronal | Train | Validation | Test | Sagittal | Train | Validation | Test |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | 3459 | 954 | 463 | AD | 1247 | 449 | 184 | AD | 2823 | 826 | 404 |
| NC | 3648 | 931 | 130 | NC | 1267 | 443 | 32 | NC | 2253 | 776 | 83 |
| Hyperparameter | Value | Description |
|---|---|---|
| Batch Size | 32 | Chosen due to GPU memory constraints. |
| Loss Function | Categorical Cross-Entropy | Selected due to its ability to measure the dissimilarity between predicted probabilities and true class labels, thereby simplifying optimization and enhancing model performance. |
| Epochs | 35 | Selected through validation (tested epochs: 20, 35, 50, 100) with 35 demonstrating model convergence. |
| Optimizer | Adam | Based on its demonstrated advantages in both optimization efficiency and generalization performance. |
| Learning Rate | 1 × 10−4 | The tested range (1 × 10−3 to 1 × 10−5) and final selection 1 × 10−4. |
| Predicted Positives | Predicted Negatives | |
|---|---|---|
| Actual Positives | tp | fn |
| Actual Negatives | fp | tn |
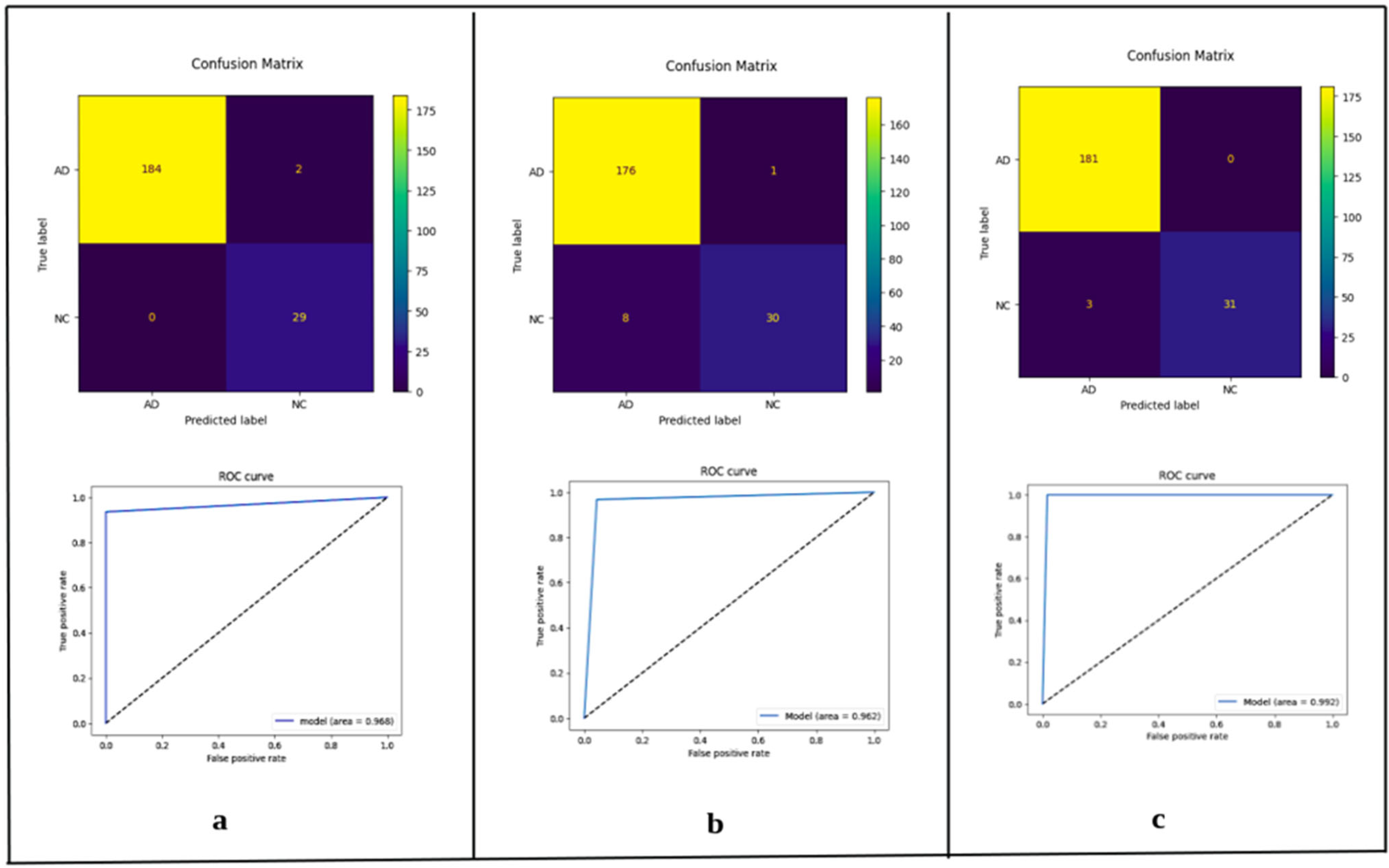
| Axial | Accuracy | Loss | Precision Macro Avg | Recall Macro Avg | F1-Score Macro Avg | AUC |
|---|---|---|---|---|---|---|
| ConvNeXt-Tiny | 0.9303 | 0.2590 | 0.9400 | 0.8500 | 0.8800 | 0.8470 |
| ConvNeXt-Small | 0.8877 | 0.3388 | 0.9200 | 0.7500 | 0.8000 | 0.7480 |
| ConvNeXt-Base | 0.9208 | 0.2176 | 0.9500 | 0.8300 | 0.8700 | 0.8290 |
| Coronal | Accuracy | Loss | Precision Macro Avg | Recall Macro Avg | F1-Score Macro Avg | AUC |
|---|---|---|---|---|---|---|
| ConvNeXt-Tiny | 0.9896 | 0.1015 | 0.9900 | 0.9700 | 0.9800 | 0.9680 |
| ConvNeXt-Small | 0.9579 | 0.1339 | 0.8900 | 0.9600 | 0.9200 | 0.9620 |
| ConvNeXt-Base | 0.9837 | 0.0924 | 0.9600 | 0.9900 | 0.9700 | 0.9920 |
| Sagittal | Accuracy | Loss | Precision Macro Avg | Recall Macro Avg | F1-Score Macro Avg | AUC |
|---|---|---|---|---|---|---|
| ConvNeXt-Tiny | 0.9077 | 0.3011 | 0.9200 | 0.7400 | 0.8000 | 0.7430 |
| ConvNeXt-Small | 0.8754 | 0.3208 | 0.8000 | 0.6800 | 0.7200 | 0.6830 |
| ConvNeXt-Base | 0.9209 | 0.1791 | 0.9300 | 0.7800 | 0.8300 | 0.7790 |
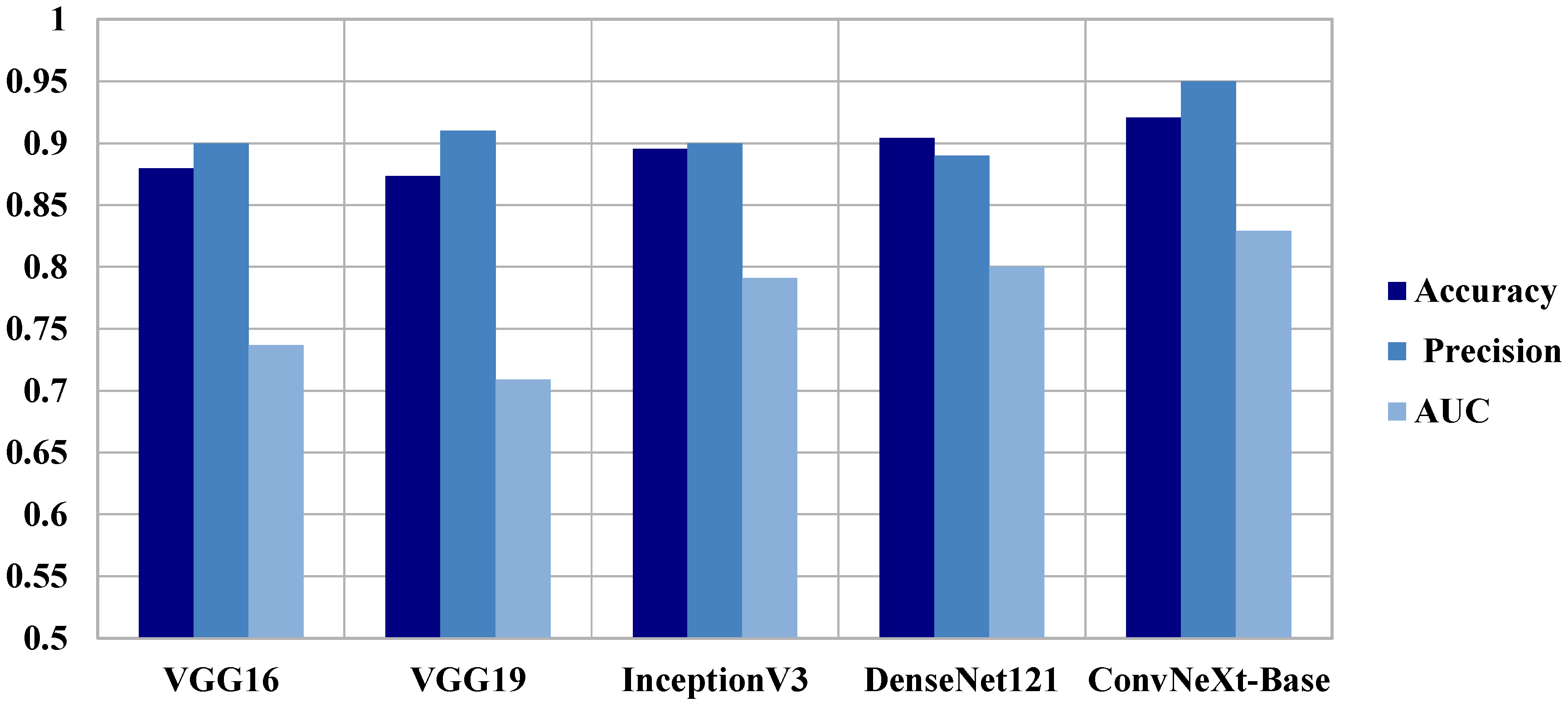
| Axial | Accuracy | Loss | Precision Macro Avg | Recall Macro Avg | F1-Score Macro Avg | AUC |
|---|---|---|---|---|---|---|
| VGG16 | 0.8799 | 0.2771 | 0.9000 | 0.7400 | 0.7800 | 0.7370 |
| VGG19 | 0.8734 | 0.3564 | 0.9100 | 0.7100 | 0.7600 | 0.7090 |
| InceptionV3 | 0.8954 | 0.2006 | 0.9000 | 0.7900 | 0.8300 | 0.7910 |
| DenseNet121 | 0.9043 | 0.2606 | 0.8900 | 0.8000 | 0.8300 | 0.8002 |
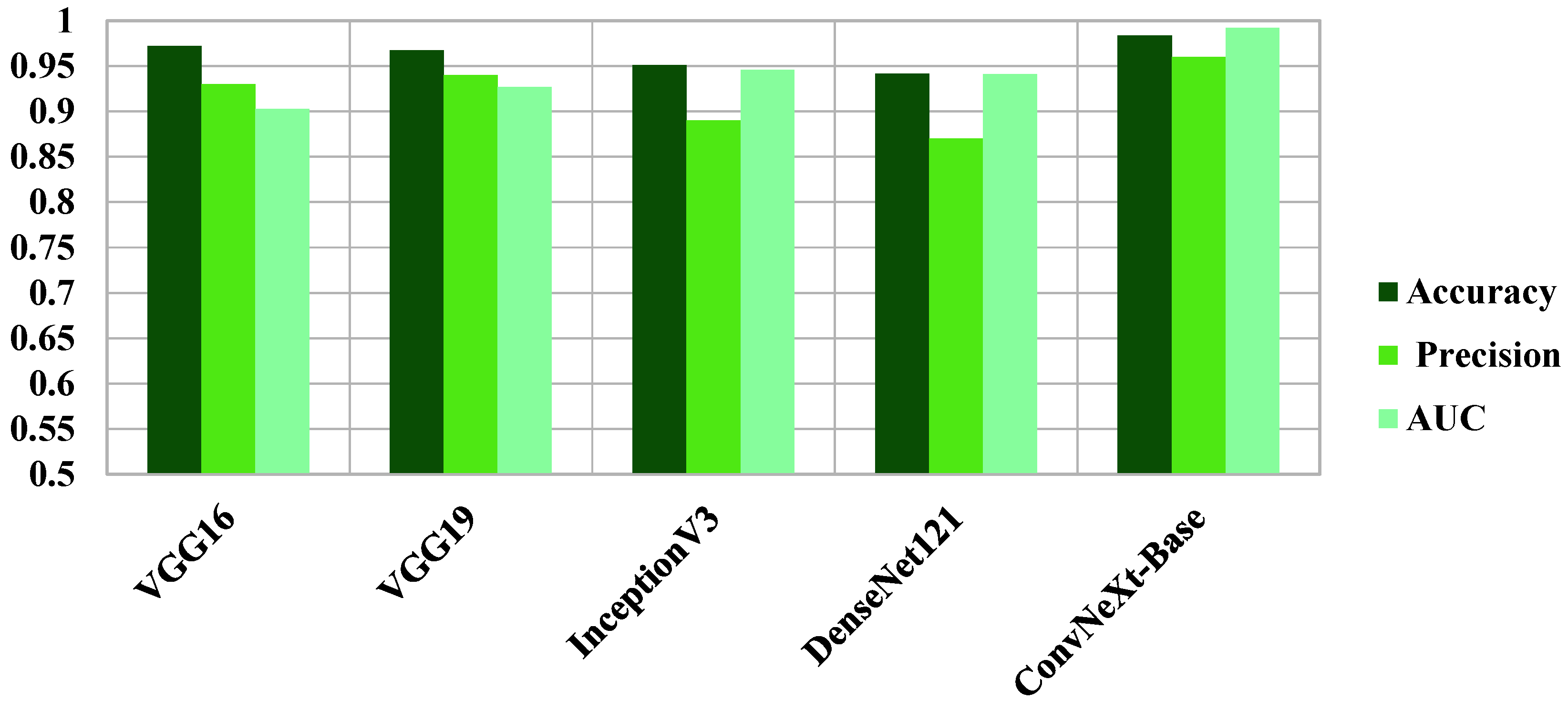
| Coronal | Accuracy | Loss | Precision Macro Avg | Recall Macro Avg | F1-Score Macro Avg | AUC |
|---|---|---|---|---|---|---|
| VGG16 | 0.9722 | 0.1780 | 0.9800 | 0.9400 | 0.9000 | 0.9030 |
| VGG19 | 0.9678 | 0.2133 | 0.9400 | 0.9300 | 0.9300 | 0.9270 |
| InceptionV3 | 0.9513 | 0.2966 | 0.8900 | 0.9500 | 0.9100 | 0.9460 |
| DenseNet121 | 0.9415 | 0.2910 | 0.8700 | 0.9400 | 0.9000 | 0.9410 |
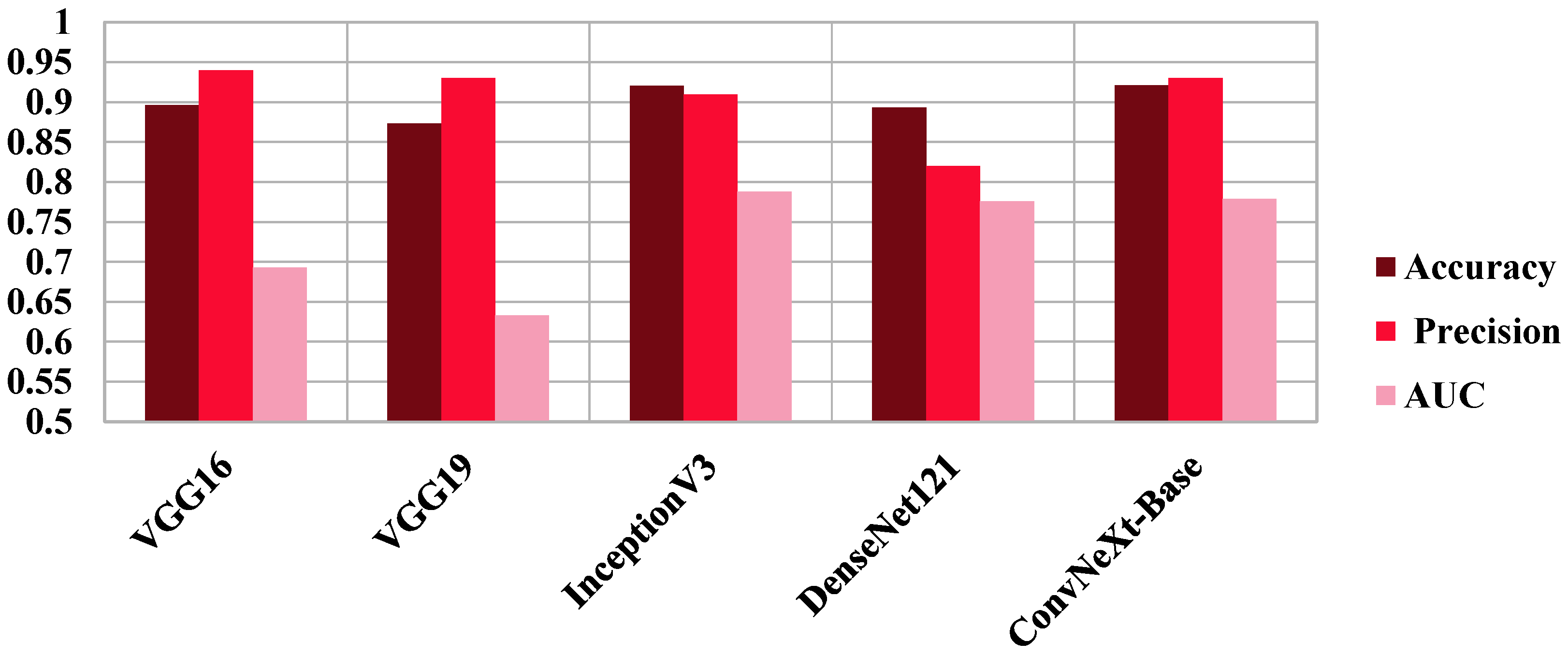
| Sagittal | Accuracy | Loss | Precision Macro Avg | Recall Macro Avg | F1-Score Macro Avg | AUC |
|---|---|---|---|---|---|---|
| VGG16 | 0.8963 | 0.2573 | 0.9400 | 0.6900 | 0.7500 | 0.6930 |
| VGG19 | 0.8731 | 0.2616 | 0.9300 | 0.6300 | 0.6700 | 0.6330 |
| InceptionV3 | 0.9204 | 0.2003 | 0.9100 | 0.7900 | 0.8300 | 0.7880 |
| DenseNet121 | 0.8936 | 0.2874 | 0.8200 | 0.7800 | 0.7900 | 0.7760 |
| Study/Model (Ref) | Dataset Source | Task/Classes | Architecture Style | Reported Accuracy | Notes |
|---|---|---|---|---|---|
| Hu et al., 2025 [25] (“3D-SEConvNeXt”) | Public AD MRI cohort (3D volumes) | AD vs. NC | 3D ConvNeXt + Squeeze-and-Excitation attention | 94.23% | High-capacity 3D model with SE attention; higher compute cost. |
| Jin et al., 2022 [26] (“CA-ConvNeXt”) | ADNI | Multi-stage AD classification/staging | ConvNeXt + coordinate attention; compared with ResNet50, Swin Transformer | 95.3% | Uses attention augmentation and compares to transformer baselines. |
| Techa et al., 2022 [38] | Kaggle MRI dataset | Multiclass dementia staging | ConvNeXt feature extractor + stacked ML ensemble (SVM, MLP, etc.) | 92.2% | Ensemble/post hoc classifiers; heterogeneous public data. |
| Li et al., 2019 [39] | Public/open MRI datasets | AD vs. NC | ConvNeXt-based deep model | “High performance” (~92–95% range as reported) | Reported strong ConvNeXt performance on open data. |
| This work (ConvNeXt-Base) | AlzaSet (expert-labeled clinical MRI) | AD vs. NC (binary) | End-to-end 2D ConvNeXt-Base on coronal slices | 98.37% | AUC 0.992; orientation-specific coronal training targeting medial temporal lobe anatomy; computationally lightweight. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basereh, M.; Abikenari, M.A.; Sadeghzadeh, S.; Dunn, T.; Freichel, R.; Siddarth, P.; Ghahremani, D.; Lavretsky, H.; Buch, V.P. ConvNeXt-Driven Detection of Alzheimer’s Disease: A Benchmark Study on Expert-Annotated AlzaSet MRI Dataset Across Anatomical Planes. Diagnostics 2025, 15, 2997. https://doi.org/10.3390/diagnostics15232997
Basereh M, Abikenari MA, Sadeghzadeh S, Dunn T, Freichel R, Siddarth P, Ghahremani D, Lavretsky H, Buch VP. ConvNeXt-Driven Detection of Alzheimer’s Disease: A Benchmark Study on Expert-Annotated AlzaSet MRI Dataset Across Anatomical Planes. Diagnostics. 2025; 15(23):2997. https://doi.org/10.3390/diagnostics15232997
Chicago/Turabian StyleBasereh, Mahdiyeh, Matthew Alexander Abikenari, Sina Sadeghzadeh, Trae Dunn, René Freichel, Prabha Siddarth, Dara Ghahremani, Helen Lavretsky, and Vivek P. Buch. 2025. "ConvNeXt-Driven Detection of Alzheimer’s Disease: A Benchmark Study on Expert-Annotated AlzaSet MRI Dataset Across Anatomical Planes" Diagnostics 15, no. 23: 2997. https://doi.org/10.3390/diagnostics15232997
APA StyleBasereh, M., Abikenari, M. A., Sadeghzadeh, S., Dunn, T., Freichel, R., Siddarth, P., Ghahremani, D., Lavretsky, H., & Buch, V. P. (2025). ConvNeXt-Driven Detection of Alzheimer’s Disease: A Benchmark Study on Expert-Annotated AlzaSet MRI Dataset Across Anatomical Planes. Diagnostics, 15(23), 2997. https://doi.org/10.3390/diagnostics15232997





