Improving Clinical Generalization of Pressure Ulcer Stage Classification Through Saliency-Guided Data Augmentation
Abstract
1. Introduction
Related Works
2. Materials and Methods
2.1. Preparing Decubitus Stage Detection Model
2.2. Phase 1: Standard Augmentation Baseline
| Algorithm 1 Pseudocode of Pressure Ulcer Stage Classification Using YOLOv7 | |
| 1: | function CLASSIFICATION |
| 2: | dataset ← LOADDATASET (Pressure_Ulcer_Image) |
| 3: | classes ← [Stage1, Stage2, Stage3, Stage4] |
| 4: | train_set, val_set ← SPLITDATASET (dataset, retio = 0.8) |
| 5: | models ← [TOLOv7] |
| 6: | for model do |
| 7: | TRAIN (model, train_set) |
| 8: | predictions ← PREDICT (model, val_set) |
| 9: | precision ← COMPUTEPRECISION (prediction, val_set.labels, classes) |
| 10: | recall ← COMPUTERECALL (prediction, val_set.labels, classes) |
| 11: | mAP@0.5 ← COMPUTEMAP (predictions, cal_set.labels, 0.5) |
| 12: | mAP@0.5:0.95 ← COMPUTEMAP (predictions, cal_set.labels, [0.5, 0.95]) |
| 13: | STORERESULTS (precision, recall, mAP@0.5, mAP@0.5:0.95) |
| 14: | end for |
| 15: | COMPARERESULTS |
| 16: | end function |
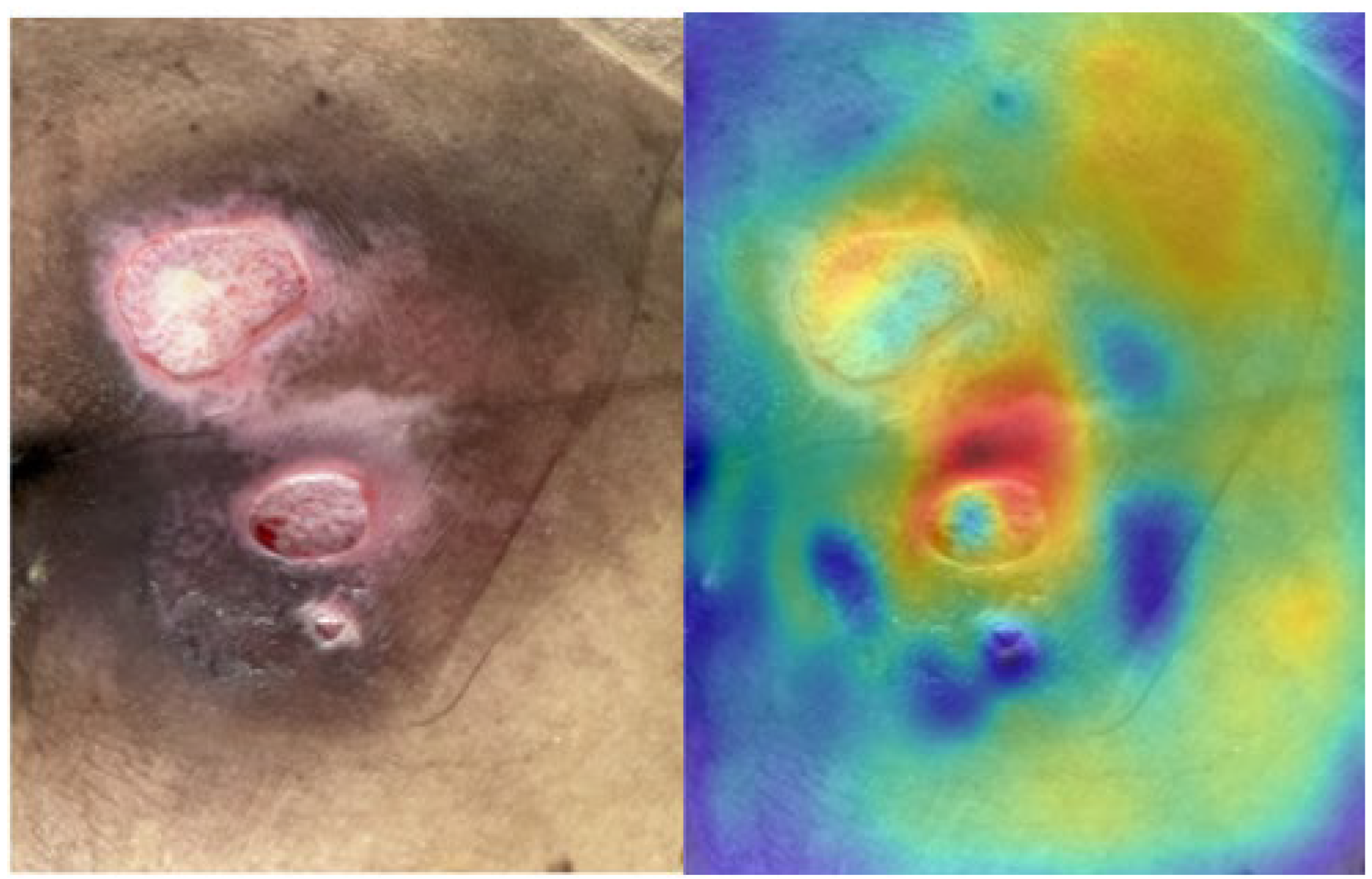
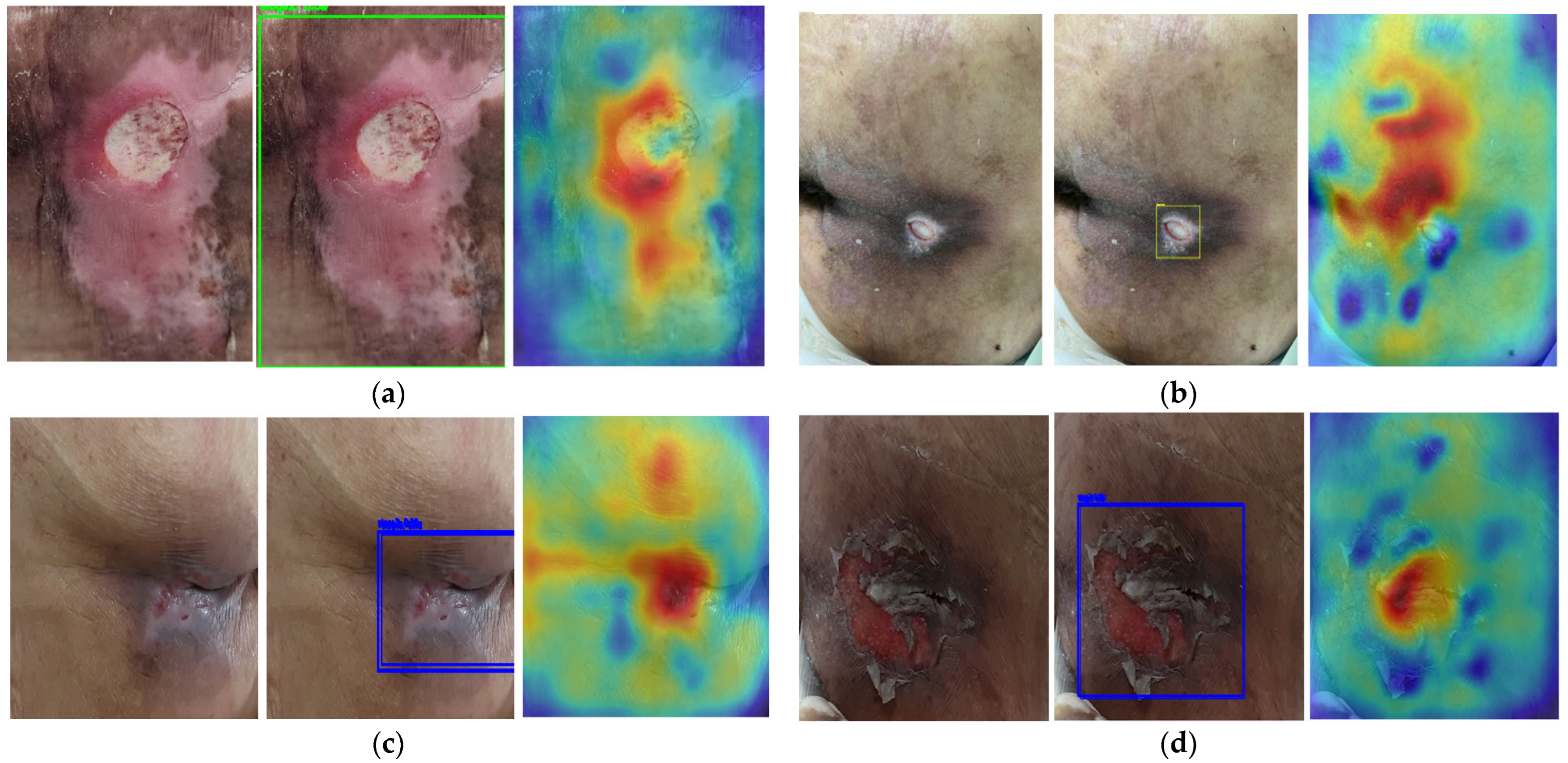
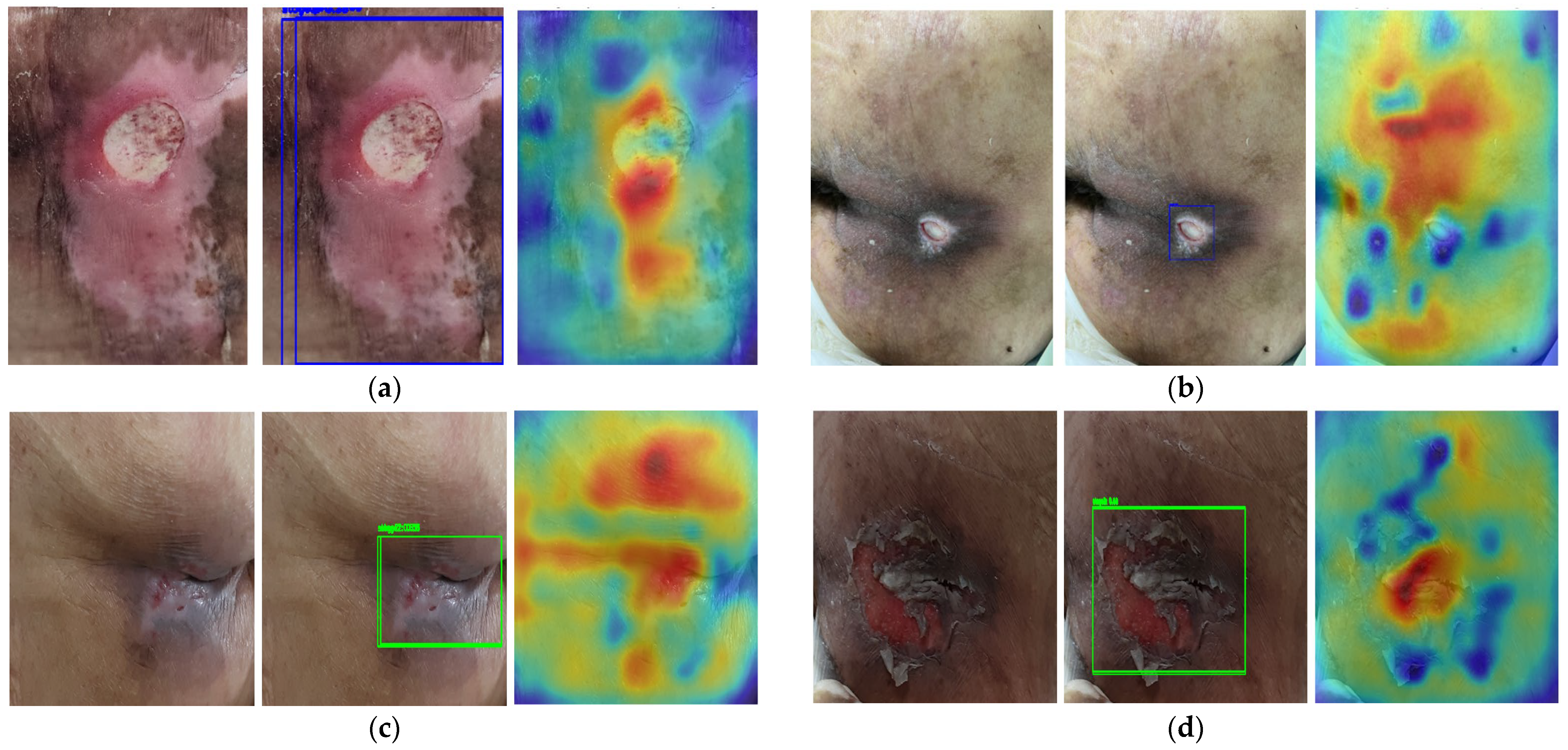
2.3. Saliency Map for Pressure Ulcer Stage Prediction
| Algorithm 2 Pseudocode of Activation-based Saliency | |
| 1: | function SINGLE_LAYER_SALIENCY (image, model, selected_layer, size) |
| 2: | cache ← ATTACH_HOOKS_AND_RUN (model, image, selected_layer) |
| 3: | detections ← GET_DETECTIONS (cache) |
| 4: | for bbox/laber overlay only |
| 5: | feature_map ← GET_FEATURE_MAP (cache, selected_layer) |
| 6: | a_single ← AVERAGE_CHANNELS (feature_map) |
| 7: | channel mean |
| 8: | a_single ← NORMALIZE_AND_RESIZE (a_single, size) |
| 9: | VISUALIZE (image, a_single, detections) |
| 10: | return a_single |
| 11: | end function |
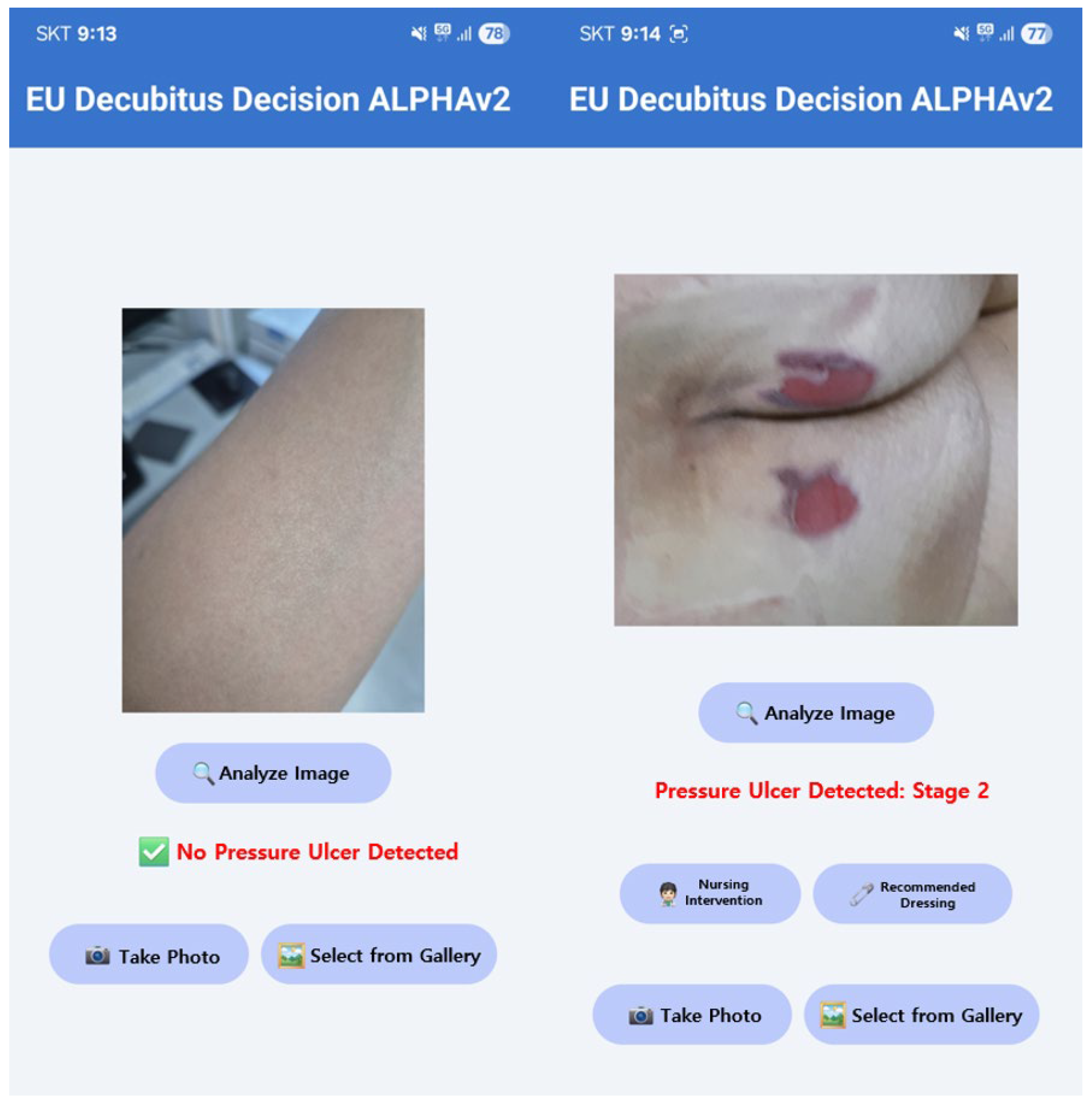
2.4. Phase 2: Saliency-Guided Clinical Overlays
3. Results
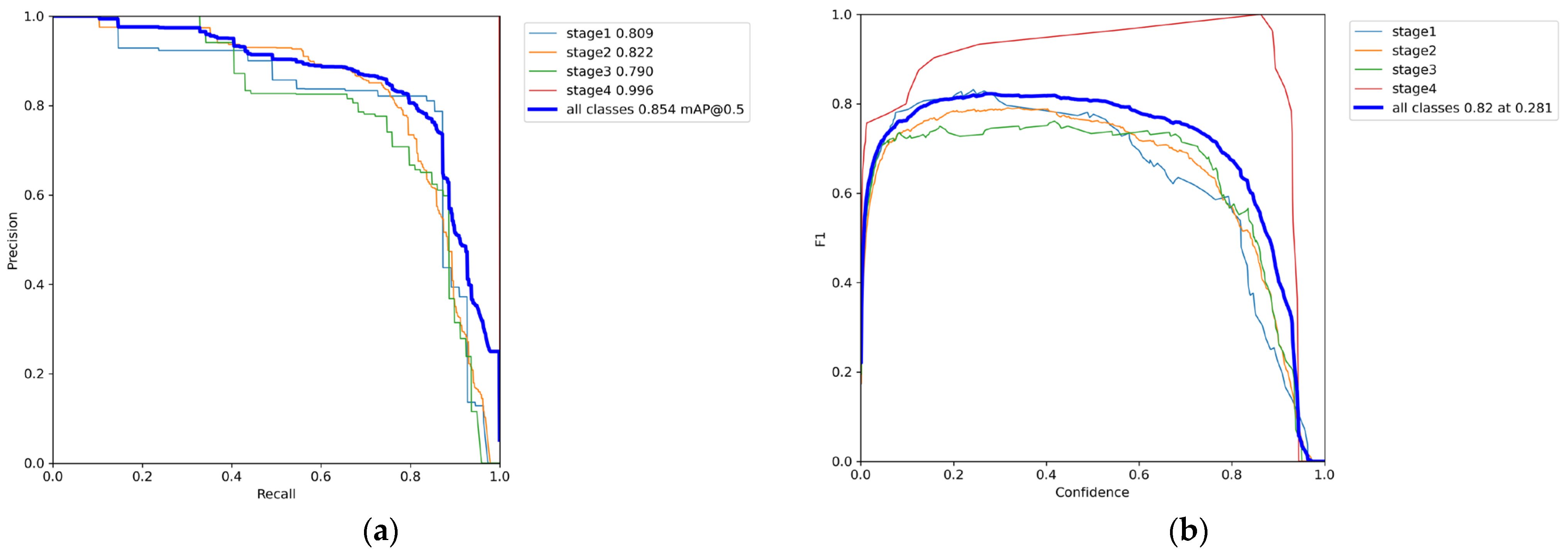
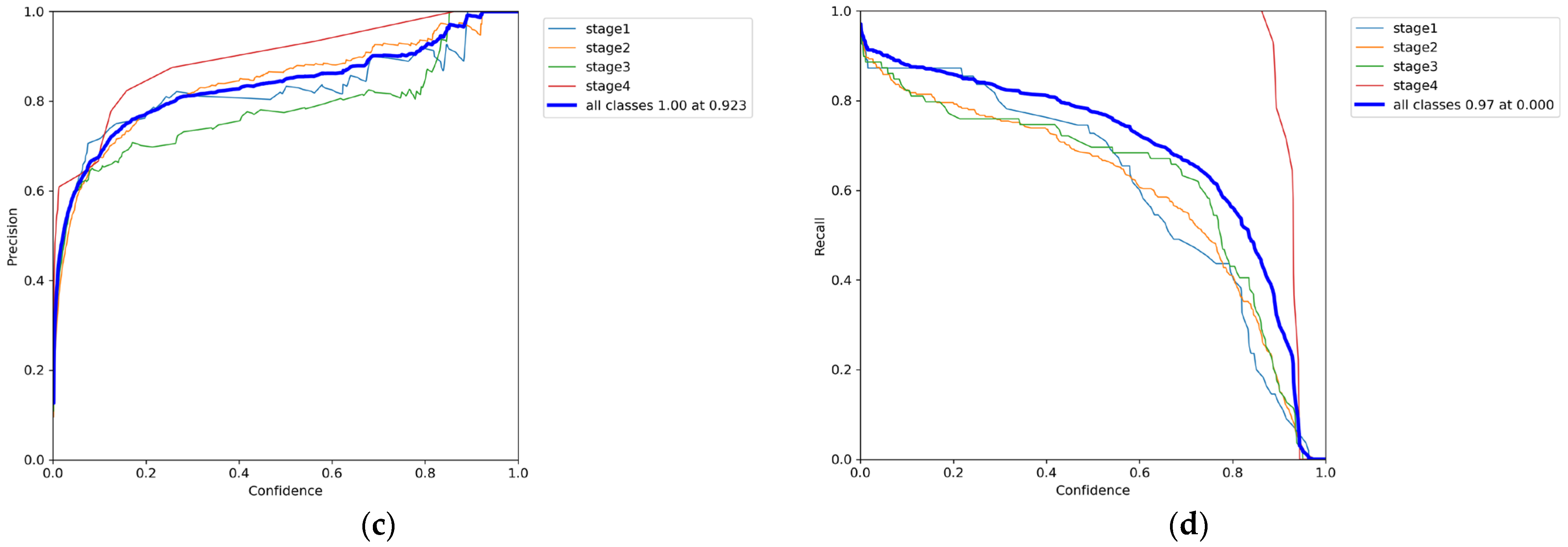
3.1. Phase 1 Training Results
3.2. Phase 2 Training Results
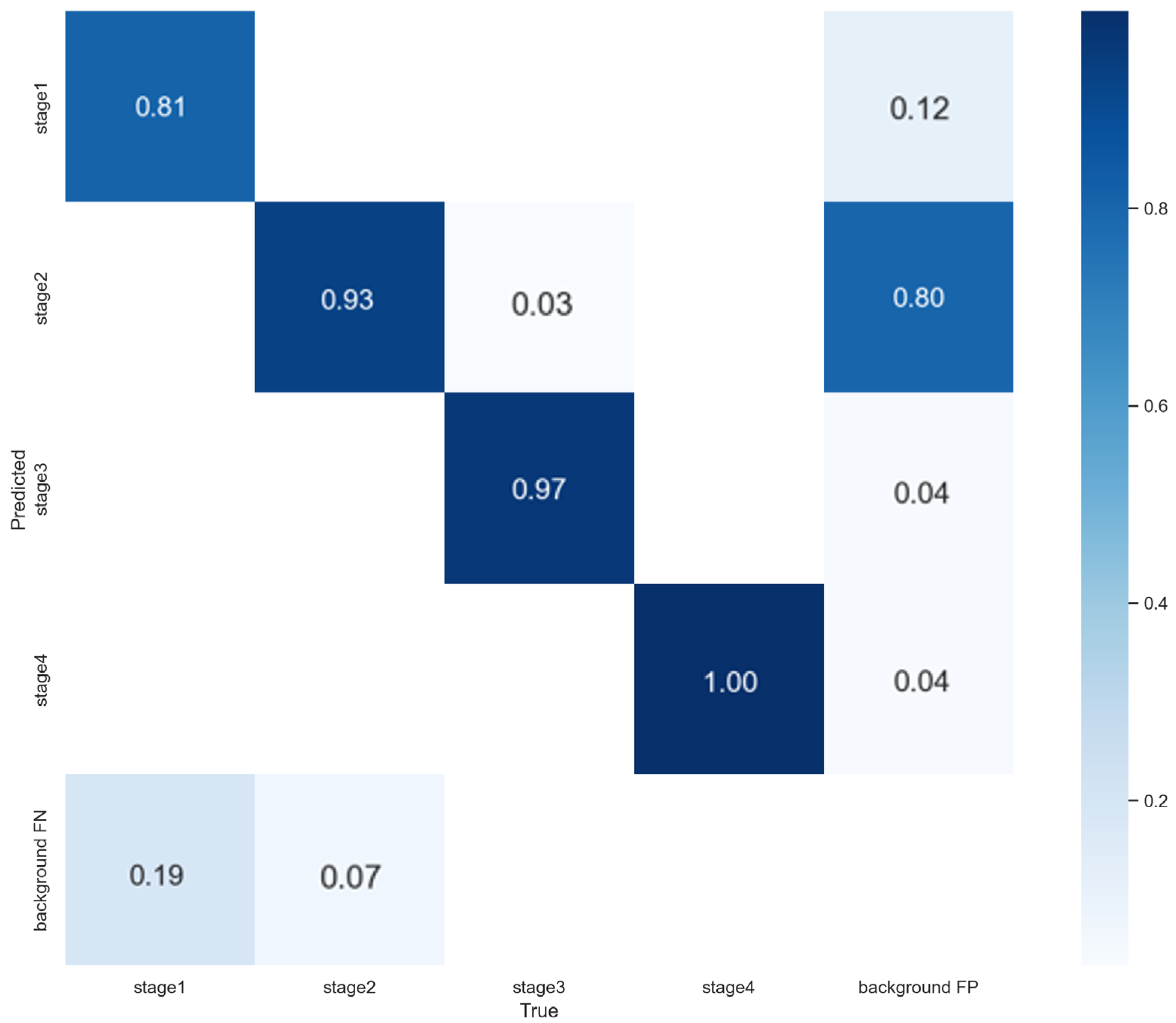
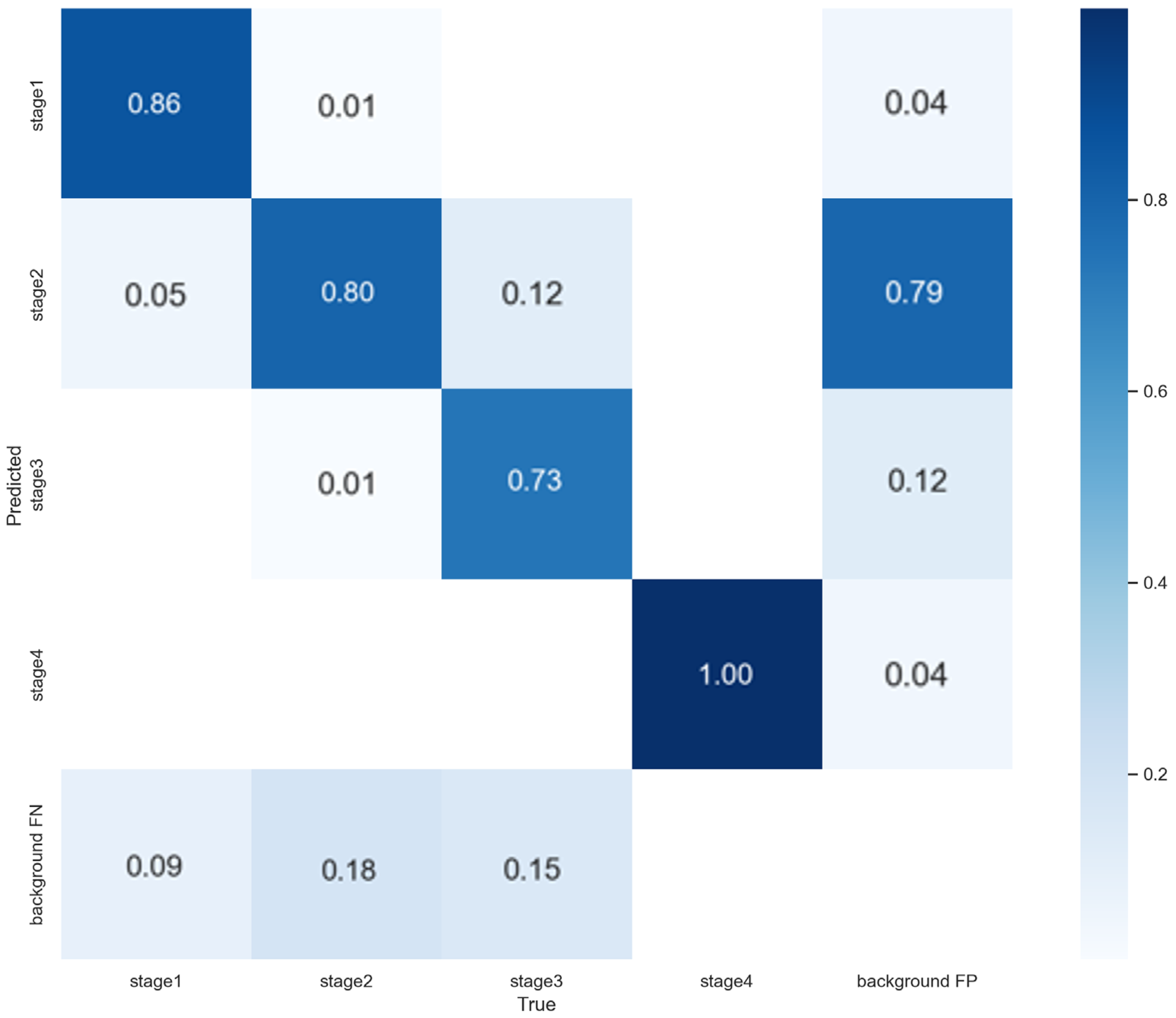
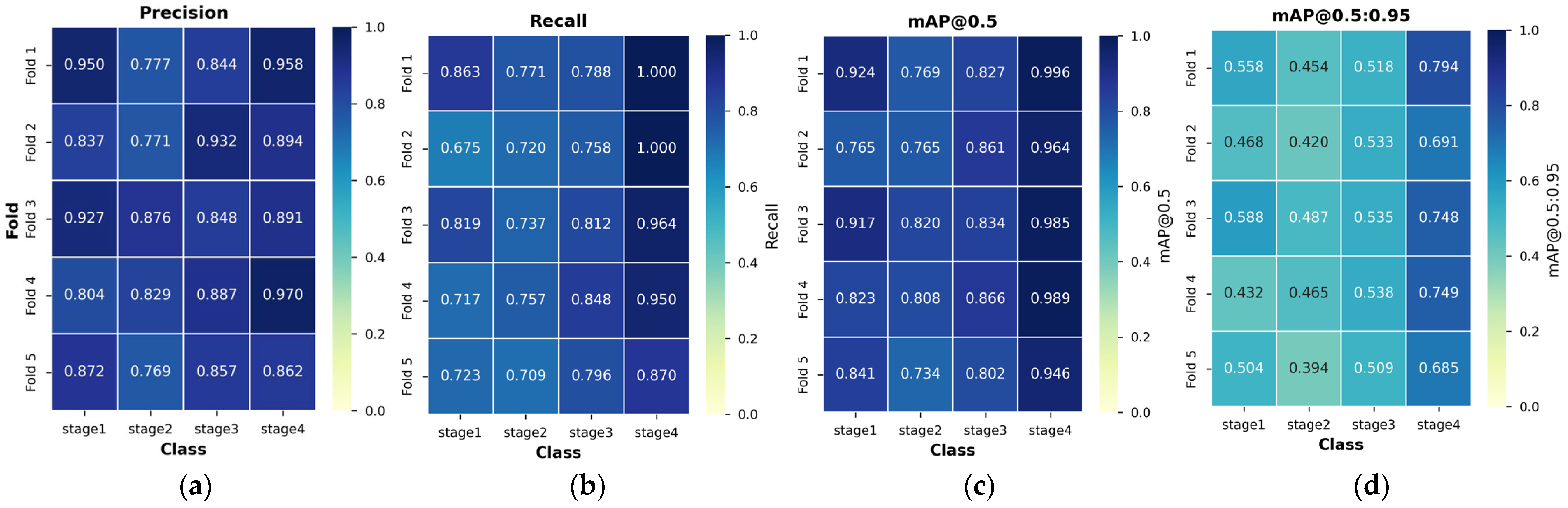
3.3. 5-Fold Cross-Validation Results
3.4. Saliency Map Comparison
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Foi, A.; Trimeche, M.; Katkovnik, V.; Egiazarian, K. Practical Poissonian-Gaussian noise modeling and fitting for single-image raw-data. IEEE Trans. Image Process. 2008, 17, 1737–1754. [Google Scholar] [CrossRef]
- Yang, S.; Xiao, W.; Zhang, M.; Guo, S.; Zhao, J.; Shen, F. Image data augmentation for deep learning: A survey. arXiv 2022, arXiv:2204.08610. [Google Scholar] [CrossRef]
- Garcea, F.; Serra, A.; Lamberti, F.; Morra, L. Data augmentation for medical imaging: A systematic literature review. Comput. Biol. Med. 2023, 152, 106391. [Google Scholar] [CrossRef]
- Islam, T.; Hafiz, M.S.; Jim, J.R.; Kabir, M.M.; Mridha, M.F. A systematic review of deep learning data augmentation in medical imaging: Recent advances and future research directions. Healthc. Anal. 2024, 5, 100340. [Google Scholar] [CrossRef]
- Etafa, W.; Argaw, Z.; Gemechu, E.; Melese, B. Nurses’ attitude and perceived barriers to pressure ulcer prevention. BMC Nurs. 2018, 17, 14. [Google Scholar] [CrossRef]
- DeVries, T.; Taylor, G.W. Improved regularization of convolutional neural networks with cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar] [CrossRef]
- Brima, Y.; Atemkeng, M. Saliency-driven explainable deep learning in medical imaging: Bridging visual explainability and statistical quantitative analysis. BioData Min. 2024, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Bengio, Y.; Louradour, J.; Collobert, R.; Weston, J. Curriculum learning. In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; pp. 41–48. [Google Scholar] [CrossRef]
- Shrivastava, A.; Gupta, A.; Girshick, R. Training region-based object detectors with online hard example mining. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 761–769. [Google Scholar] [CrossRef]
- Singh, P.; Nampalle, K.B.; Narayan, U.V.; Raman, B. See through the fog: Curriculum learning with progressive occlusion in medical imaging. arXiv 2023, arXiv:2306.15574. [Google Scholar] [CrossRef]
- Park, B.; Cho, Y.; Lee, G.; Lee, S.M.; Cho, Y.H.; Lee, E.S.; Lee, K.H.; Seo, J.B.; Kim, N. A curriculum learning strategy to enhance the accuracy of classification of various lesions in chest-PA X-ray screening for pulmonary abnormalities. Sci. Rep. 2019, 9, 15352. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Suriawinata, A.; Ren, B.; Liu, X.; Lisovsky, M.; Vaickus, L.; Brown, C.; Baker, M.; Nasir-Moin, M.; Tomita, N.; et al. Learn like a pathologist: Curriculum learning by annotator agreement for histopathology image classification. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Virtual, 5–9 January 2021; pp. 2473–2483. [Google Scholar] [CrossRef]
- Wan, J.J.; Zhu, P.C.; Chen, B.L.; Yu, Y.T. A semantic feature enhanced YOLOv5-based network for polyp detection from colonoscopy images. Sci. Rep. 2024, 14, 15478. [Google Scholar] [CrossRef]
- Nie, C.; Xu, C.; Li, Z.; Chu, L.; Hu, Y. Specular reflections detection and removal for endoscopic images based on brightness classification. Sensors 2023, 23, 974. [Google Scholar] [CrossRef]
- Krishnapriya, S.; Karuna, Y. A deep learning model for the localization and extraction of brain tumors from MR images using YOLOv7 and grab cut algorithm. Front. Oncol. 2024, 14, 1347363. [Google Scholar] [CrossRef]
- Lau, C.H.; Yu, K.H.O.; Yip, T.F.; Luk, L.Y.F.; Wai, A.K.C.; Sit, T.Y.; Wong, J.Y.-H.; Ho, J.W.K. An artificial intelligence-enabled smartphone app for real-time pressure injury assessment. Front. Med. Technol. 2022, 4, 905074. [Google Scholar] [CrossRef]
- Aldughayfiq, B.; Ashfaq, F.; Jhanjhi, N.Z.; Humayun, M. Yolo-based deep learning model for pressure ulcer detection and classification. Healthcare 2023, 11, 1222. [Google Scholar] [CrossRef]
- Tusar, M.H.; Fayyazbakhsh, F.; Zendehdel, N.; Mochalin, E.; Melnychuk, I.; Gould, L.; Leu, M.C. AI-Powered Image-Based Assessment of Pressure Injuries Using You Only Look Once Version 8 (YOLOv8) Models. Adv. Wound Care 2025. Ahead of Print. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Kim, J.H.; Shin, H.W.; Ha, C.; Lee, S.Y.; Go, T. Diagnosis of pressure ulcer stage using on-device AI. Appl. Sci. 2024, 14, 7124. [Google Scholar] [CrossRef]
- Bae, H.J.; Kim, C.W.; Kim, N.; Park, B.; Kim, N.; Seo, J.B.; Lee, S.M. A Perlin noise-based augmentation strategy for deep learning with small data samples of HRCT images. Sci. Rep. 2018, 8, 17687. [Google Scholar] [CrossRef]
- Momeny, M.; Neshat, A.A.; Hussain, M.A.; Kia, S.; Marhamati, M.; Jahanbakhshi, A.; Hamarneh, G. Learning-to-augment strategy using noisy and denoised data: Improving generalizability of deep CNN for the detection of COVID-19 in X-ray images. Comput. Biol. Med. 2021, 136, 104704. [Google Scholar] [CrossRef]
- Mustaqim, T.; Fatichah, C.; Suciati, N.; Obi, T.; Lee, J.S. FocusAugMix: A data augmentation method for enhancing Acute Lymphoblastic Leukemia classification. Intell. Syst. Appl. 2025, 26, 200512. [Google Scholar] [CrossRef]
- Hammoudi, K.; Cabani, A.; Slika, B.; Benhabiles, H.; Dornaika, F.; Melkemi, M. Superpixelgridmasks data augmentation: Application to precision health and other real-world data. J. Healthc. Inform. Res. 2022, 6, 442–460. [Google Scholar] [CrossRef] [PubMed]
- VanGilder, C.A.; Cox, J.; Edsberg, L.E.; Koloms, K. Pressure injury prevalence in acute care hospitals with unit-specific analysis: Results from the International Pressure Ulcer Prevalence (IPUP) survey database. J. Wound Ostomy Cont. Nurs. 2021, 48, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Scafide, K.N.; Narayan, M.C.; Arundel, L. Bedside technologies to enhance the early detection of pressure injuries: A systematic review. J. Wound Ostomy Cont. Nurs. 2020, 47, 128–136. [Google Scholar] [CrossRef]
- Onuh, O.C.; Brydges, H.T.; Nasr, H.; Savage, E.; Gorenstein, S.; Chiu, E. Capturing essentials in wound photography past, present, and future: A proposed algorithm for standardization. Adv. Ski. Wound Care 2022, 35, 483–492. [Google Scholar] [CrossRef]
- Brüngel, R.; Friedrich, C.M. Comparing DETR and YOLO for Diabetic Foot Ulcer Object Detection. In Proceedings of the 34th IEEE International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 230–238. [Google Scholar] [CrossRef]
- Terven, J.; Córdova-Esparza, D.M.; Romero-González, J.A. A comprehensive review of YOLO architectures in computer vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H.-Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar] [CrossRef]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. Score-CAM: Score-weighted visual explanations for convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 24–25. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar] [CrossRef]
- Attia, M.; Hossny, M.; Zhou, H.; Nahavandi, S.; Asadi, H.; Yazdabadi, A. Realistic hair simulator for skin lesion images: A novel benchemarking tool. Artif. Intell. Med. 2020, 108, 101933. [Google Scholar] [CrossRef]
- Mathew, S.; Nadeem, S.; Kaufman, A. CLTS-GAN: Color-lighting-texture-specular reflection augmentation for colonoscopy. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer Nature: Cham, Switzerland, 2022; pp. 519–529. [Google Scholar] [CrossRef]
- Nunez, J.A.; Vazquez, F.; Adame, D.; Fu, X.; Gu, P.; Fu, B. White Light Specular Reflection Data Augmentation for Deep Learning Polyp Detection. In Proceedings of the 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI), Houston, TX, USA, 14–17 April 2025; IEEE: New York, NY, USA, 2025; pp. 1–5. [Google Scholar] [CrossRef]
- Jurgas, A.; Wodzinski, M.; Celniak, W.; Atzori, M.; Müller, H. Artifact Augmentation for Learning-based Quality Control of Whole Slide Images. In Proceedings of the 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, Australia, 24–27 July 2023; IEEE: New York, NY, USA, 2023; pp. 1–4. [Google Scholar] [CrossRef]
- Wu, C.C.; Hsu, C.H.; Wang, P.C.; Tu, T.W.; Hsu, Y.Y. Influence of Rician Noise on Cardiac MR Image Segmentation Using Deep Learning. In Proceedings of the International Conference on Intelligent Systems Design and Applications, Olten, Switzerland, 11–13 December 2023; Springer Nature: Cham, Switzerland, 2023; pp. 223–232. [Google Scholar] [CrossRef]
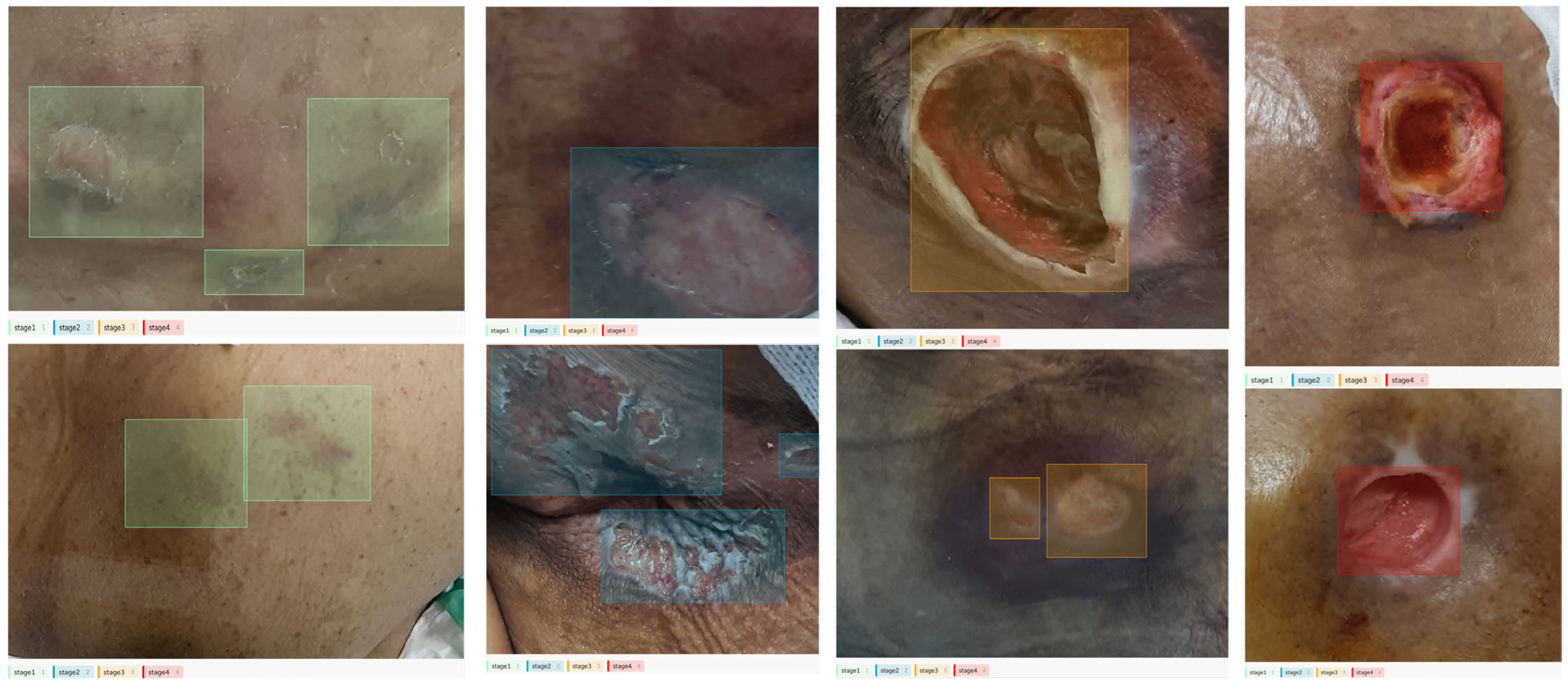

| Stage 1 | Stage 2 | Stage 3 | Stage 4 | |
|---|---|---|---|---|
| Total | 151 | 714 | 319 | 98 |
| Train | 121 | 571 | 255 | 78 |
| Validation | 30 | 143 | 64 | 20 |
| (a) | |||||
| mAP@0.5 | mAP@0.5:0.95 | F1 | Precision | Recall | |
| Phase 1 | 0.96 | 0.68 | 0.93 | 0.94 | 0.92 |
| Phase 2 | 0.85 | 0.56 | 0.81 | 0.84 | 0.86 |
| (b) | |||||
| Accuracy | |||||
| Phase 1 | 0.75 | ||||
| Phase 2 | 0.89 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.-W.; Rhee, W.L.; Han, D.-H.; Kang, M. Improving Clinical Generalization of Pressure Ulcer Stage Classification Through Saliency-Guided Data Augmentation. Diagnostics 2025, 15, 2951. https://doi.org/10.3390/diagnostics15232951
Choi J-W, Rhee WL, Han D-H, Kang M. Improving Clinical Generalization of Pressure Ulcer Stage Classification Through Saliency-Guided Data Augmentation. Diagnostics. 2025; 15(23):2951. https://doi.org/10.3390/diagnostics15232951
Chicago/Turabian StyleChoi, Jun-Woo, Won Lo Rhee, Dong-Hun Han, and Minsoo Kang. 2025. "Improving Clinical Generalization of Pressure Ulcer Stage Classification Through Saliency-Guided Data Augmentation" Diagnostics 15, no. 23: 2951. https://doi.org/10.3390/diagnostics15232951
APA StyleChoi, J.-W., Rhee, W. L., Han, D.-H., & Kang, M. (2025). Improving Clinical Generalization of Pressure Ulcer Stage Classification Through Saliency-Guided Data Augmentation. Diagnostics, 15(23), 2951. https://doi.org/10.3390/diagnostics15232951





