Integrative Long Non-Coding RNA Analysis and Recurrence Prediction in Cervical Cancer Using a Recurrent Neural Network
Abstract
1. Introduction
2. Related Works
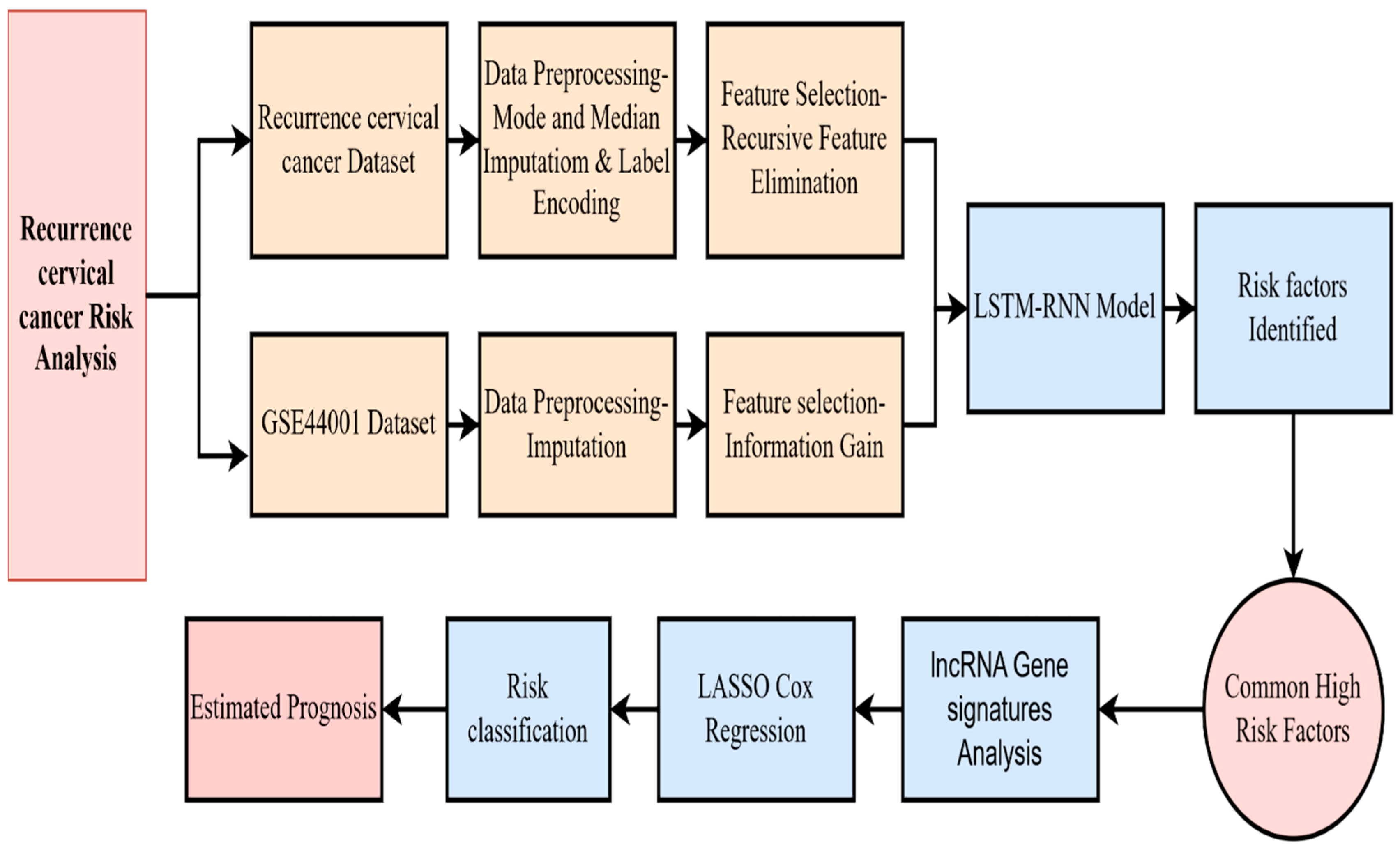
3. Materials and Methods
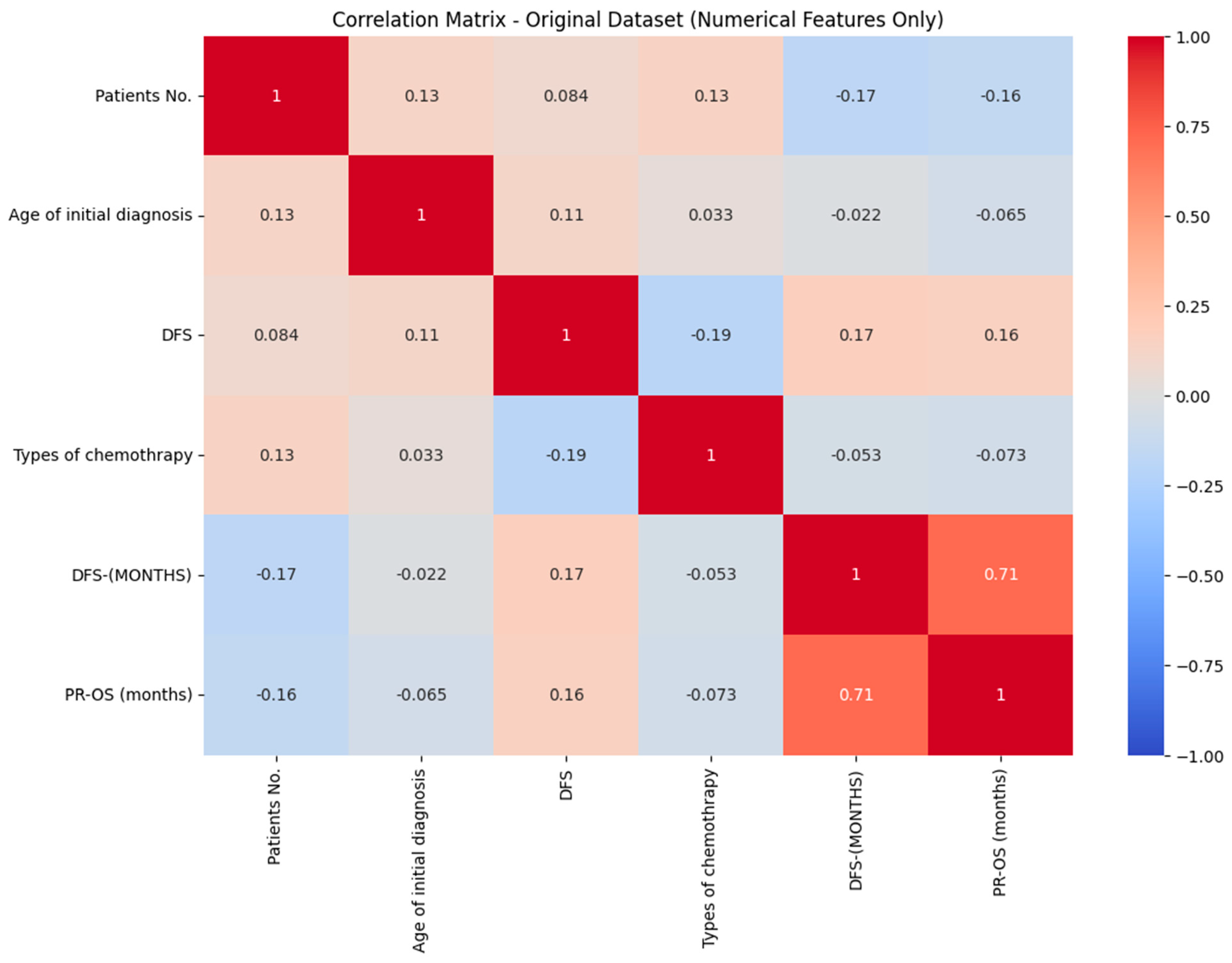
3.1. Data Preprocessing
3.2. Feature Selection
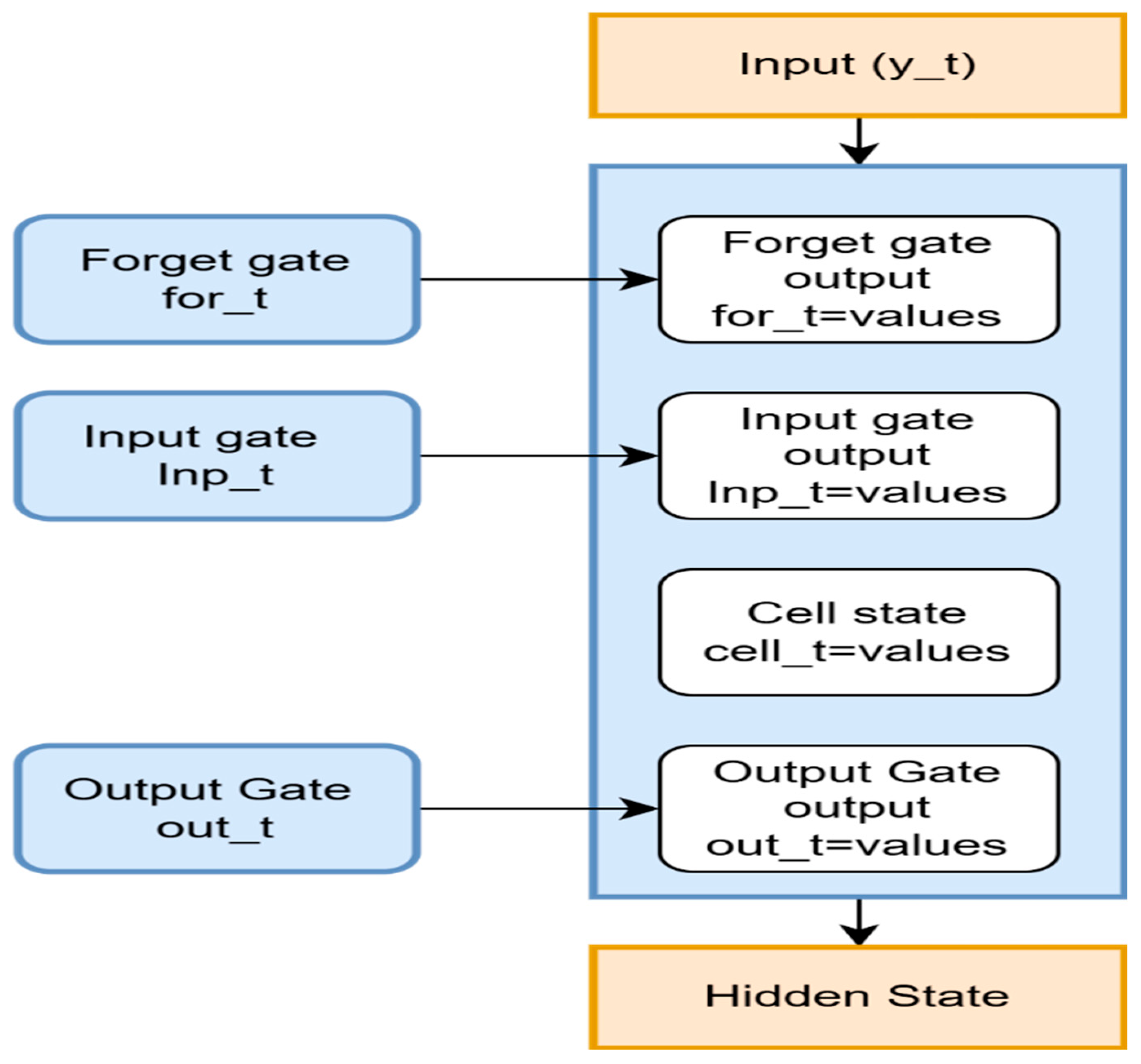
3.3. RNN LSTM
| Algorithm 1. Identification of Relevant Features |
| First stage: Preparing the Dataset and Setting Up Hyperparameters Step 1: Use the matrix method for displaying the expression of genes in the dataset. j = The quantity of chosen lncRNA genes used as input characteristics. . Step 2: Initialization of Hyperparameters Configure LSTM network-specific hyperparameters. One layer of LSTM at first, and up to three layers at most. Use tanh activation in the LSTM’s internal processing. For the binary category of recurrence condition, recurrence versus non-recur rence employs sigmoid activation. Step 4: Splitting of Data. Step 5: The LSTM unit performs subsequent calculations at every step based on the gene expression pattern of each sample. Step 6: Forget gate: The forget gate regulates which data derived from the prior cell state should be kept. &. . . Step 7: Input Gate: Choose which data should be incorporated into the: . . ,, Step 8: Cell state update-Improves the cell state through the combination of data from the past and present. . . Step 9: Output gate—Uses the modified cell status for identifying the subsequent concealed state . . . . . . . Step 11: To track convergence and prevent overfitting, the LSTM model is trained with ETrain and compute accuracy, F1 score, and loss. Step 12: Hyperparameter adjustment is made until the kmax value reaches 30. Step 13: After determining the ideal arrangement, test the system on , and estimate final indicators, such as F1 score, accuracy, and ROC-AUC. 3rd Stage: Result analysis and optimal configuration. |
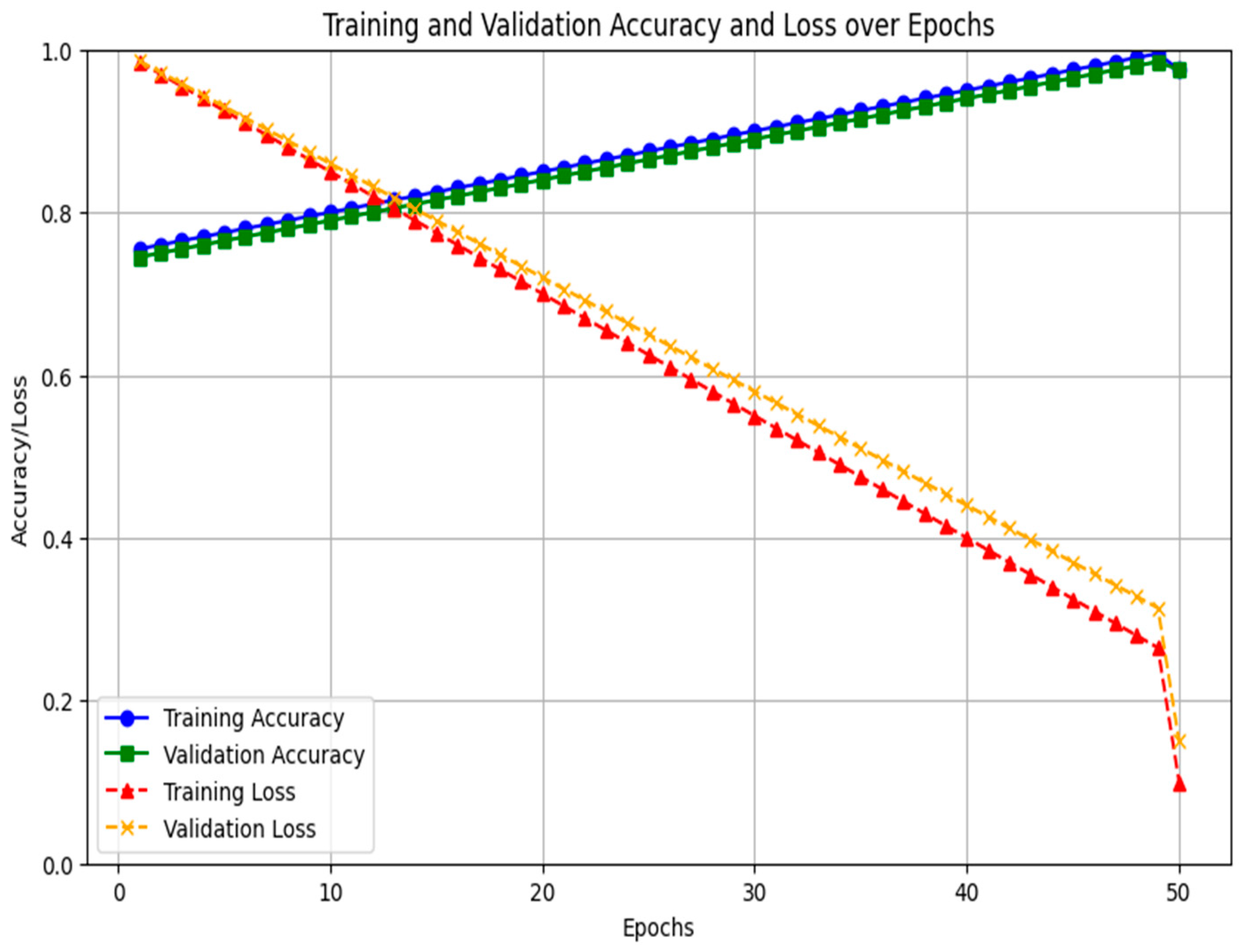
4. Results
Comparison of Common Features
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al Mudawi, N.; Alazeb, A. A Model for Predicting Cervical Cancer Using Machine Learning Algorithms. Sensors 2022, 22, 4132. [Google Scholar] [CrossRef]
- Ghoneim, A.; Muhammad, G.; Hossain, M.S. Cervical cancer classification using convolutional neural networks and extreme learning machines. Future Gener. Comput. Syst. 2020, 102, 643–649. [Google Scholar] [CrossRef]
- Antunes, D.; Cunha, T.M. Recurrent Cervical Cancer: How Can Radiology be Helpful. OMICS J. Radiol. 2013, 2, 138. [Google Scholar] [CrossRef]
- Senthilkumar, G.; Ramakrishnan, J.; Frnda, J.; Ramachandran, M.; Gupta, D.; Tiwari, P.; Shorfuzzaman, M.; Mohammed, M.A. Incorporating Artificial Fish Swarm in Ensemble Classification Framework for Recurrence Prediction of Cervical Cancer. IEEE Access 2021, 9, 83876–83886. [Google Scholar] [CrossRef]
- Roszik, J.; Ring, K.L.; Wani, K.M.; Lazar, A.J.; Yemelyanova, A.V.; Soliman, P.T.; Frumovitz, M.; Jazaeri, A.A. Gene Expression Analysis Identifies Novel Targets for Cervical Cancer Therapy. Front. Immunol. 2018, 9, 2102. [Google Scholar] [CrossRef]
- Vistad, I.; Bjorge, L.; Solheim, O.; Fiane, B.; Sachse, K.; Tjugum, J.; Skroppa, S.; Bentzen, A.G.; Stokstad, T.; Iversen, G.A.; et al. A national, prospective observational study of first recurrence after primary treatment for gynecological cancer in Norway. Acta Obs. Gynecol. Scand. 2017, 96, 1162–1169. [Google Scholar] [CrossRef]
- Chang, C.; Chen, J.; Chang, W.Y.; Chiang, A.J. Tumor Size Has a Time-Varying Effect on Recurrence in Cervical Cancer. J. Low. Genit. Tract Dis. 2016, 20, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.-J.; Lu, C.-J.; Chang, C.-C.; Chen, G.-D. Application of Machine Learning to Predict the Recurrence-Proneness for Cervical Cancer. Neural Comput. Appl. 2014, 24, 1311–1316. [Google Scholar] [CrossRef]
- Chang, C.C.; Cheng, S.L.; Lu, C.J.; Liao, K.H. Prediction of Recurrence in Patients with Cervical Cancer Using MARS and Classification. Int. J. Mach. Learn. Comput. 2013, 3, 75–78. [Google Scholar] [CrossRef]
- Guo, C.; Wang, J.; Wang, Y.; Qu, X.; Shi, Z.; Meng, Y.; Qiu, J.; Hua, K. Novel artificial intelligence machine learning approaches to precisely predict survival and site-specific recurrence in cervical cancer: A multiinstitutional study. Transl. Oncol. 2021, 14, 101032. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, G.; Luo, J.; Yan, S.; Ye, P.; Wang, J.; Luo, M. Cervical cancer prognosis and related risk factors for patients with cervical cancer: A long-term retrospective cohort study. Sci. Rep. 2022, 12, 13994. [Google Scholar] [CrossRef]
- Taarnhoj, G.A.; Christensen, I.J.; Lajer, H.; Fuglsang, K.; Jeppesen, M.M.; Kahr, H.S.; Hogdall, C. Risk of recurrence, prognosis, and follow-up for Danish women with cervical cancer in 2005–2013: A national cohort study. Cancer 2018, 24, 943–951. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Zhu, H.; Liu, Y.; Cao, J.; Li, D.; Ding, B.; Yan, W.; Jin, H.; Wang, S. Identification of Potential Prognostic Long Non-coding RNA Biomarkers for Predicting Recurrence in Patients with Cervical Cancer. Cancer Manag. Res. 2020, 12, 719–730. [Google Scholar] [CrossRef]
- Chao, X.; Fan, J.; Song, X.; You, Y.; Wu, H.; Wu, M.; Li, L. Diagnostic Strategies for Recurrent Cervical Cancer: A Cohort Study. Front. Oncol. 2020, 10, 591253. [Google Scholar] [CrossRef]
- Peiretti, M.; Zapardiel, I.; Zanagnolo, V.; Landoni, F.; Morrow, C.P.; Maggioni, A. Management of recurrent cervical cancer: A review of the literature. Surg. Oncol. 2012, 21, e59–e66. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, Y.; Lu, X.; Wang, Y.; Qiao, H.; Liu, M. Upregulation of long noncoding RNA SPRY4-IT1 correlates with tumor progression and poor prognosis in cervical cancer. FEBS Open Bio 2016, 6, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Babichev, S.; Liakh, I.; Kalinina, I. Applying a Recurrent Neural Network-Based Deep Learning Model for Gene Expression Data Classification. Appl. Sci. 2023, 13, 11823. [Google Scholar] [CrossRef]
- Deng, S.P.; Zhu, L.; Huang, D.S. Predicting Hub Genes Associated with Cervical Cancer through Gene Co-Expression Networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016, 13, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhao, K.; Cao, J.; Ma, H. Prediction research of cervical cancer clinical events based on recurrent neural network. Procedia Comput. Sci. 2021, 183, 221–229. [Google Scholar] [CrossRef]
- Annapurna, S.D.; Pasumarthi, D.; Pasha, A.; Doneti, R.; Sheela, B.; Botlagunta, M.; Vijaya Lakshmi, B.; Pawar, S.C. Identification of Differentially Expressed Genes in Cervical Cancer Patients by Comparative Transcriptome Analysis. Biomed. Res. Int. 2021, 2021, 8810074. [Google Scholar]
- Mao, Y.; Dong, L.; Zheng, Y.; Dong, J.; Li, X. Prediction of Recurrence in Cervical Cancer Using a Nine-lncRNA Signature. Front. Genet. 2019, 10, 284. [Google Scholar] [CrossRef]
- Ding, H.; Zhang, L.; Zhang, C.; Song, J.; Jiang, Y. Screening of Significant Biomarkers Related to Prognosis of Cervical Cancer and Functional Study Based on lncRNA-associated ceRNA Regulatory Network. Comb. Chem. High. Throughput Screen. 2021, 24, 472–482. [Google Scholar] [CrossRef]
- Geeitha, S.; Prabha, K.R.; Cho, J.; Easwaramoorthy, S.V. Bidirectional recurrent neural network approach for predicting cervical cancer recurrence and survival. Sci. Rep. 2024, 14, 31641. [Google Scholar] [CrossRef]
- Wu, W.; Sui, J.; Liu, T.; Yang, S.; Xu, S.; Zhang, M.; Huang, S.; Yin, L.; Pu, Y.; Liang, G. Integrated analysis of two-lncRNA signature as a potential prognostic biomarker in cervical cancer: A study based on public database. PeerJ 2019, 7, e6761. [Google Scholar] [CrossRef]
- Geeitha, S.; Renuka, P.; Thilagavathi, C.; Ananth, S.; Ramya, S.; Sinduja, K. LSTM—Recurrent Neural Network Model to Forecast the Risk Factors in Recurrent Cervical Carcinoma. In Proceedings of the 2nd International Conference on Self-Sustainable Artificial Intelligence Systems, Erode, India, 23–25 October 2024; pp. 132–137. [Google Scholar]
- Wang, B.; Wang, W.; Zhou, W.; Zhao, Y.; Liu, W. Cervical cancer-specific long non-coding RNA landscape reveals the favorable prognosis predictive performance of an ion-channel-related signature model. Cancer Med. 2024, 13, e7389. [Google Scholar] [CrossRef]
- He, J.; Huang, B.; Zhang, K.; Liu, M.; Xu, T. Long non-coding RNA in cervical cancer: From biology to therapeutic opportunity. Biomed. Pharmacother. 2020, 127, 110209. [Google Scholar] [CrossRef]
- Ye, Z.; Zhang, Y.; Liang, Y.; Lang, J.; Zhang, X.; Zang, G.; Yuan, D.; Tian, G.; Xiao, M.; Yang, J. Cervical Cancer Metastasis and Recurrence Risk Prediction Based on Deep Convolutional Neural Network. Curr. Bioinform. 2022, 17, 164–173. [Google Scholar] [CrossRef]
- Men, L.; Ilk, N.; Tang, X.; Liu, Y. Multi-disease prediction using LSTM recurrent neural networks. Expert. Syst. Appl. 2021, 177, 114905. [Google Scholar] [CrossRef]
- Gholami, H.; Mohammadifar, A.; Golzari, S.; Song, Y.; Pradhan, B. Interpretability of simple RNN and GRU deep learning models used to map land susceptibility to gully erosion. Sci. Total Environ. 2023, 904, 166960. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, Z.; Dong, Y.; Tu, J. An interpretable LSTM deep learning model predicts the time-dependent swelling behaviour in CERCER composite fuels. Mater. Today Commun. 2023, 37, 106998. [Google Scholar] [CrossRef]
- Amendolara, A.B.; Sant, D.; Rotstein, H.G.; Fortune, E. LSTM-based recurrent neural network provides effective short-term flu forecasting. BMC Public Health 2023, 23, 1788. [Google Scholar] [CrossRef]
- Perkins, R.B.; Guido, R.L.; Saraiya, M.; Sawaya, G.F.; Wentzensen, N.; Schiffman, M.; Feldman, S. Summary of Current Guidelines for Cervical Cancer Screening and Management of Abnormal Test Results: 2016–2020. J. Womens Health 2021, 30, 5–13. [Google Scholar] [CrossRef]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Li, X.; Jin, F.; Li, Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J. Cell. Mol. Med. 2021, 25, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Wang, T.; Seklouli, A.S.; Zhang, H.; Bouras, A. A review on missing values for main challenges and methods. Inf. Syst. 2023, 119, 102268. [Google Scholar] [CrossRef]
- Alghamdi, T.A.; Javaid, N. A Survey of Preprocessing Methods Used for Analysis of Big Data Originated from Smart Grids. IEEE Access 2022, 10, 29149–29171. [Google Scholar] [CrossRef]
- Gao, C.; Yan, J.; Zhou, S.; Chen, B.; Varshney, K.P.; Liu, H. Long short-term memory-based recurrent neural networks for nonlinear target tracking. Signal Process. 2019, 164, 67–73. [Google Scholar] [CrossRef]
- Belagoune, S.; Bali, N.; Bakdi, A.; Baadji, B.; Atif, K. Deep learning through LSTM classification and regression for transmission line fault detection, diagnosis, and location in large-scale multi-machine power systems. Measurement 2021, 177, 109330. [Google Scholar] [CrossRef]
- Ding, Y.; Zhu, Y.; Feng, J.; Zhang, P.; Cheng, Z. Interpretable spatio-temporal attention LSTM model for flood forecasting. Neurocomputing 2020, 403, 348–359. [Google Scholar] [CrossRef]
- Zou, J.; Lin, Z.; Jiao, W.; Chen, J.; Lin, L.; Zhang, F.; Zhang, X.; Zhao, J. A multi-omics-based investigation of the limitation and immunological impact of necroptosis-related mRNA in patients with cervical squamous carcinoma and adenocarcinoma. Sci. Rep. 2022, 12, 16773. [Google Scholar] [CrossRef]
- Li, N.; Yu, K.; Lin, Z.; Zeng, D. Identifying a cervical cancer survival signature based on mRNA expression and genome-wide copy number variations. Exp. Biol. Med. 2022, 247, 207–220. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Ge, X.; Lei, S.; Wang, P.; Wang, W.; Wang, W. The metabolism-related lncRNA signature predicts the prognosis of breast cancer patients. Sci. Rep. 2024, 14, 3500. [Google Scholar] [CrossRef]
- Allgaier, J.; Pryss, R. Cross-Validation Visualized: A Narrative Guide to Advanced Methods. Mach. Learn. Knowl. Extr. 2024, 6, 1378–1388. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, X.; Han, X.; Pandey, V.; Lobie, P.E.; Zhu, T. The potential of long noncoding RNAs for precision medicine in human cancer. Cancer Lett. 2021, 50, 12–19. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, J.; Li, L.; Han, M.; Dong, J.; Wang, X. Comprehensive assessment of postoperative recurrence and survival in patients with cervical cancer. Eur. J. Surg. Oncol. 2024, 50, 108583. [Google Scholar] [CrossRef]
- Burmeister, C.A.; Khan, S.F.; Schäfer, G.; Mbatani, N.; Adams, T.; Moodley, J.; Prince, S. Cervical cancer therapies: Current challenges and future perspectives. Tumour Virus Res. 2022, 13, 200238. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Hu, L.; Huang, D.; Chen, K.; Qiu, X.; Qiu, B. Six-lncRNA Immune Prognostic Signature for Cervical Cancer. Front. Genet. 2020, 11, 533628. [Google Scholar]
- Zhou, Y.; Wang, Y.; Lin, M.; Wu, D.; Zhao, M. LncRNA HOTAIR promotes proliferation and inhibits apoptosis by sponging miR-214-3p in HPV16-positive cervical cancer cells. Cancer Cell Int. 2021, 21, 400. [Google Scholar] [CrossRef]
- Sun, M.; Liu, X.; Xia, L.; Chen, Y.; Kuang, L.; Gu, X.; Li, T. A nine-lncRNA signature predicts distant relapse-free survival of HER2-negative breast cancer patients receiving taxane and anthracycline-based neoadjuvant chemotherapy. Biochem. Pharmacol. 2021, 189, 114285. [Google Scholar] [CrossRef] [PubMed]
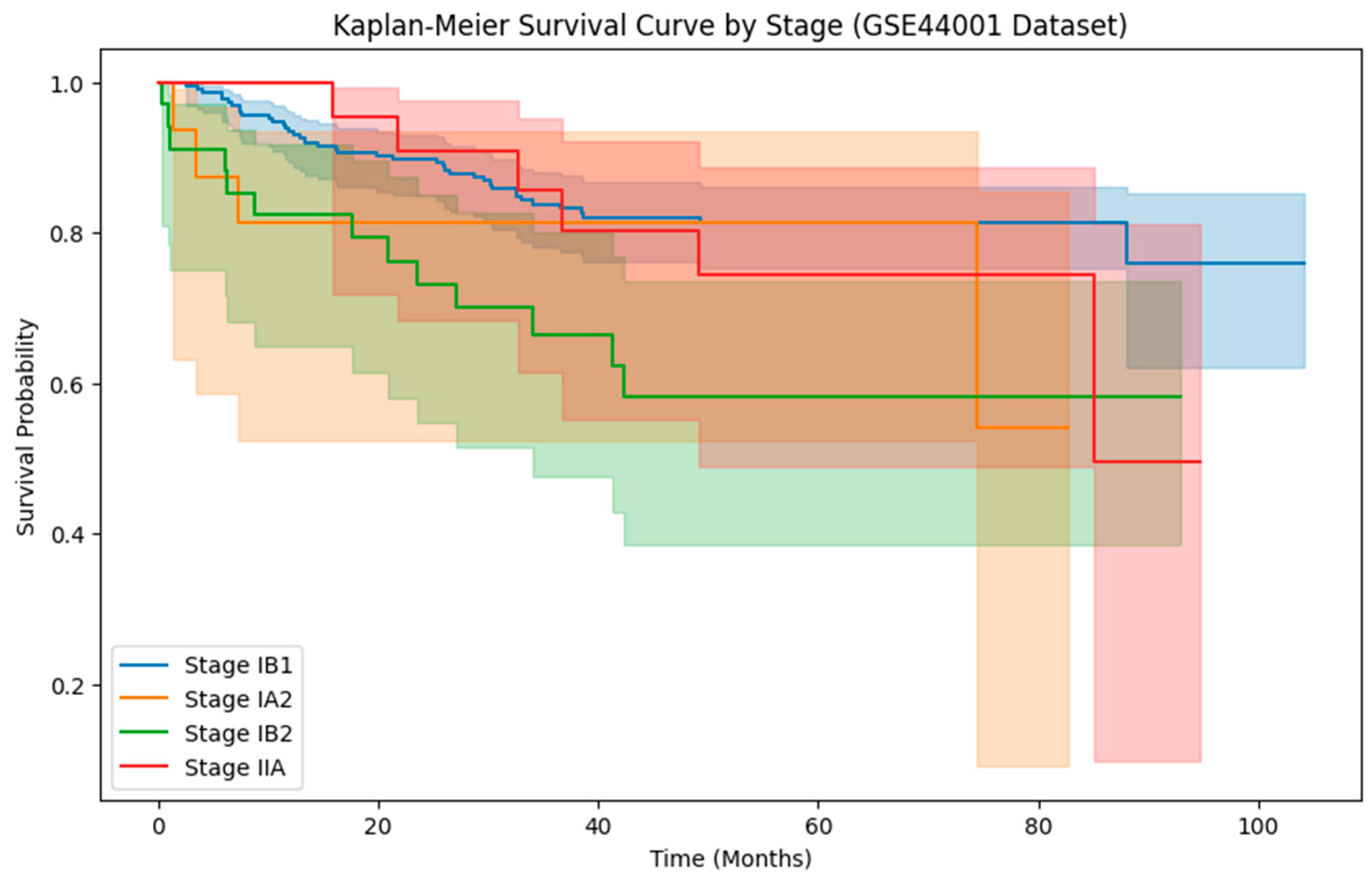
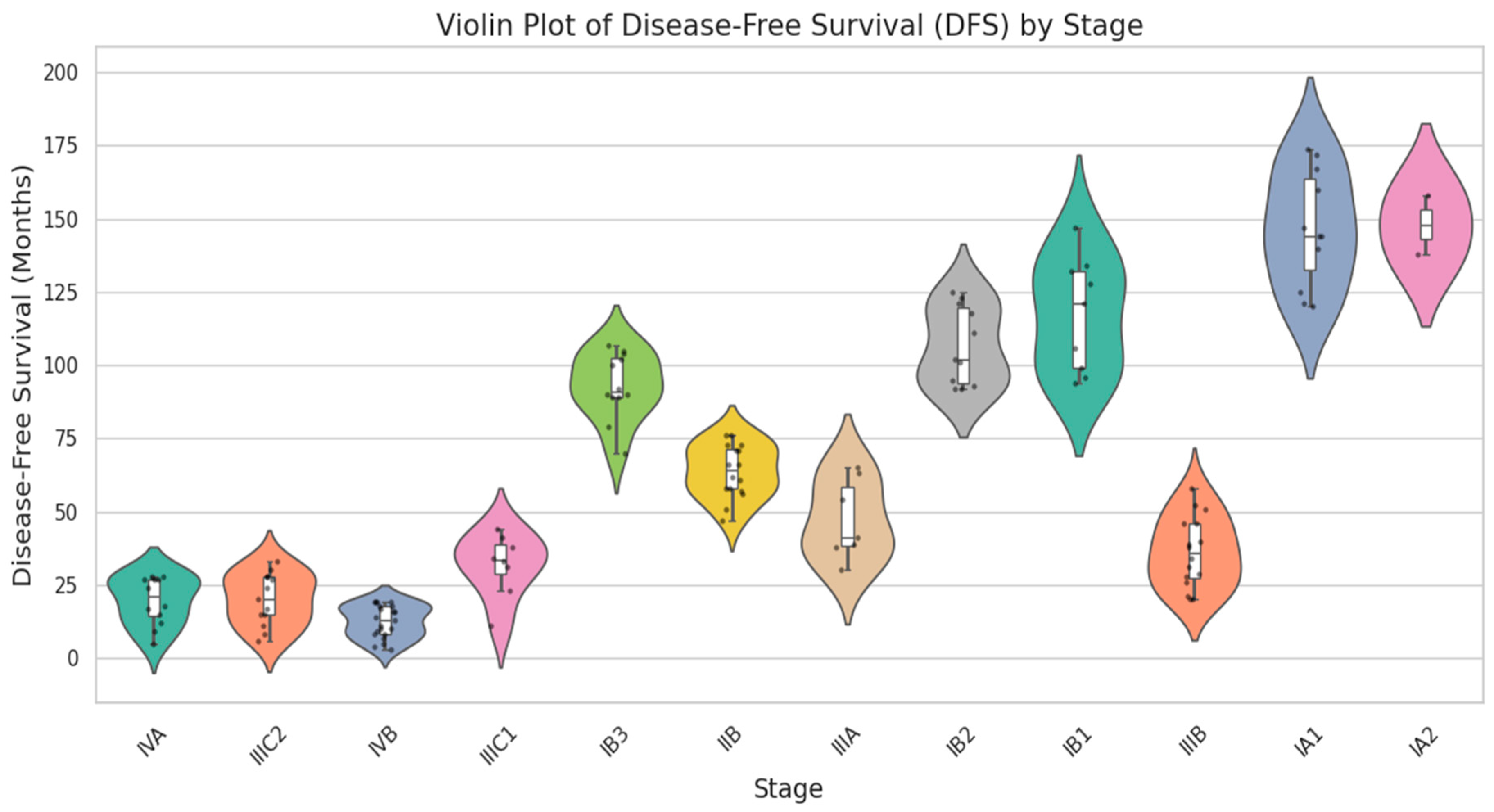
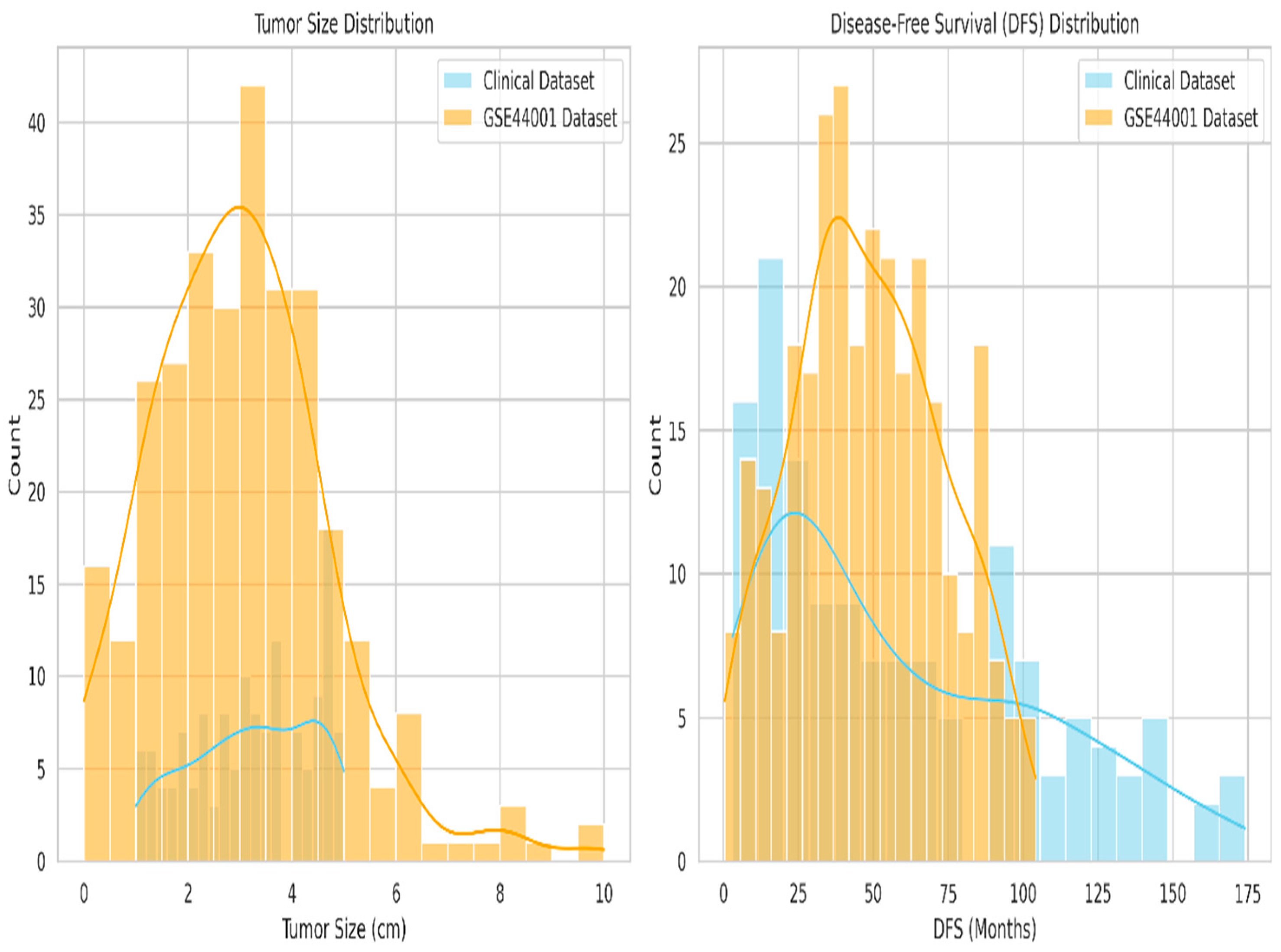
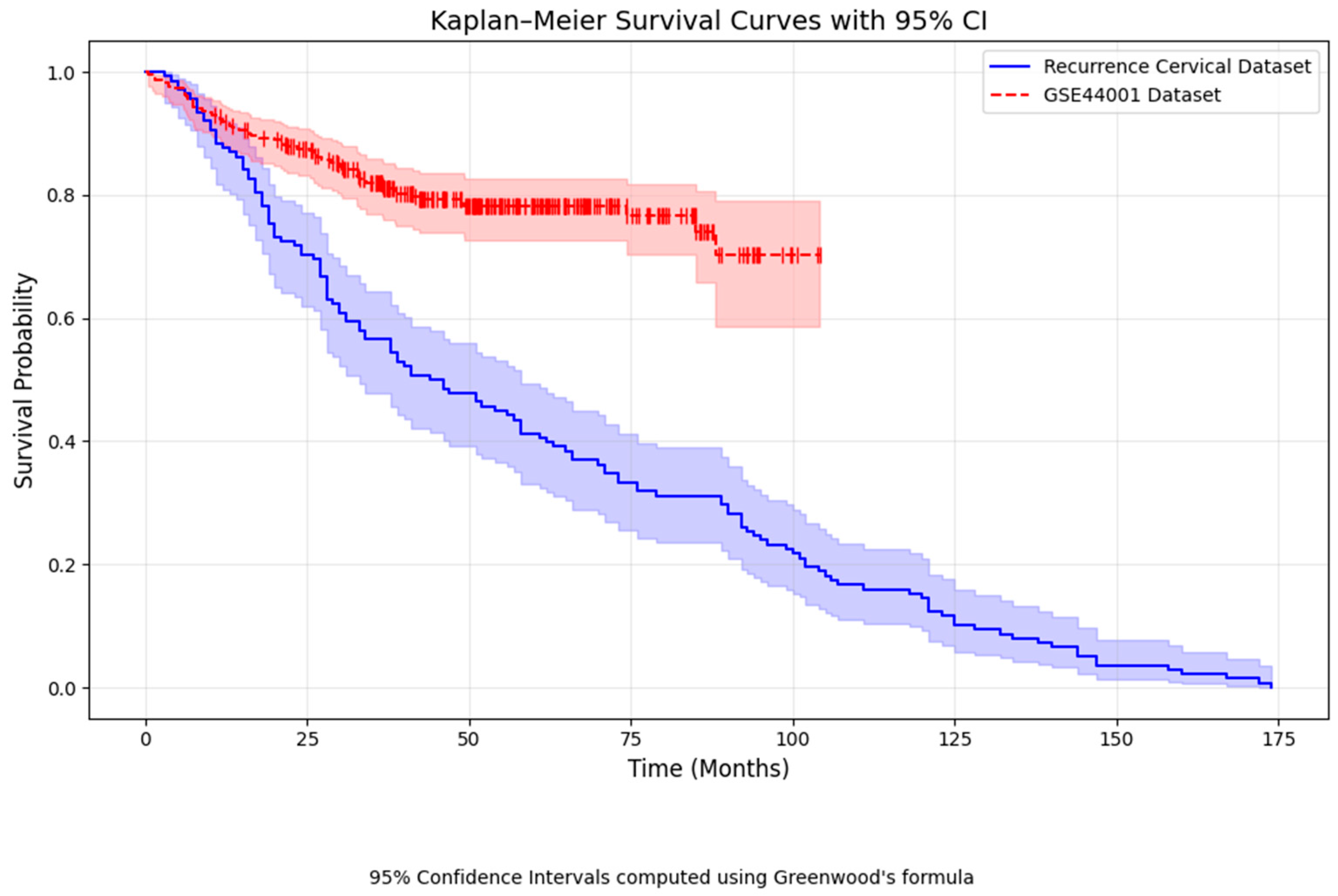
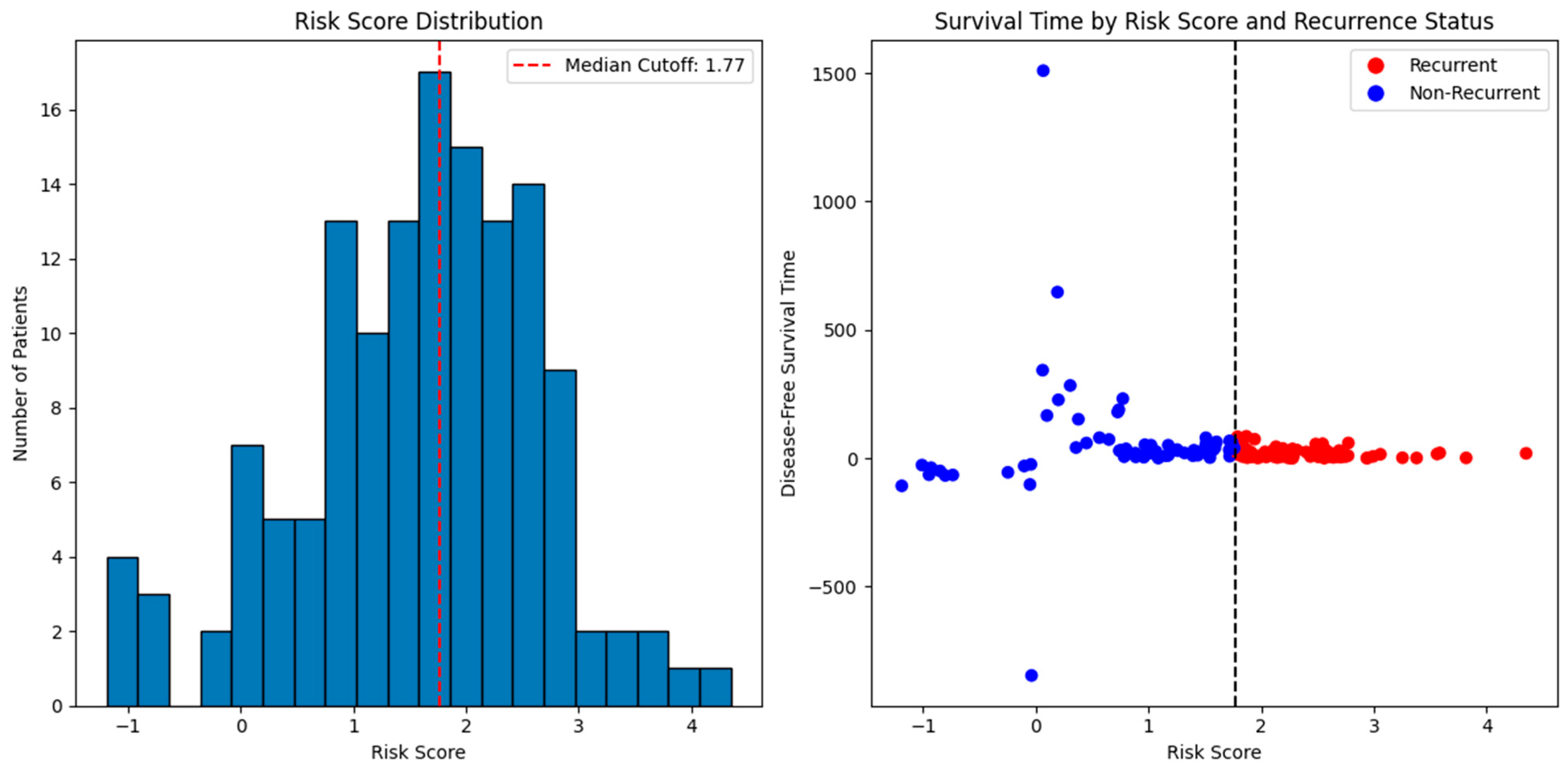
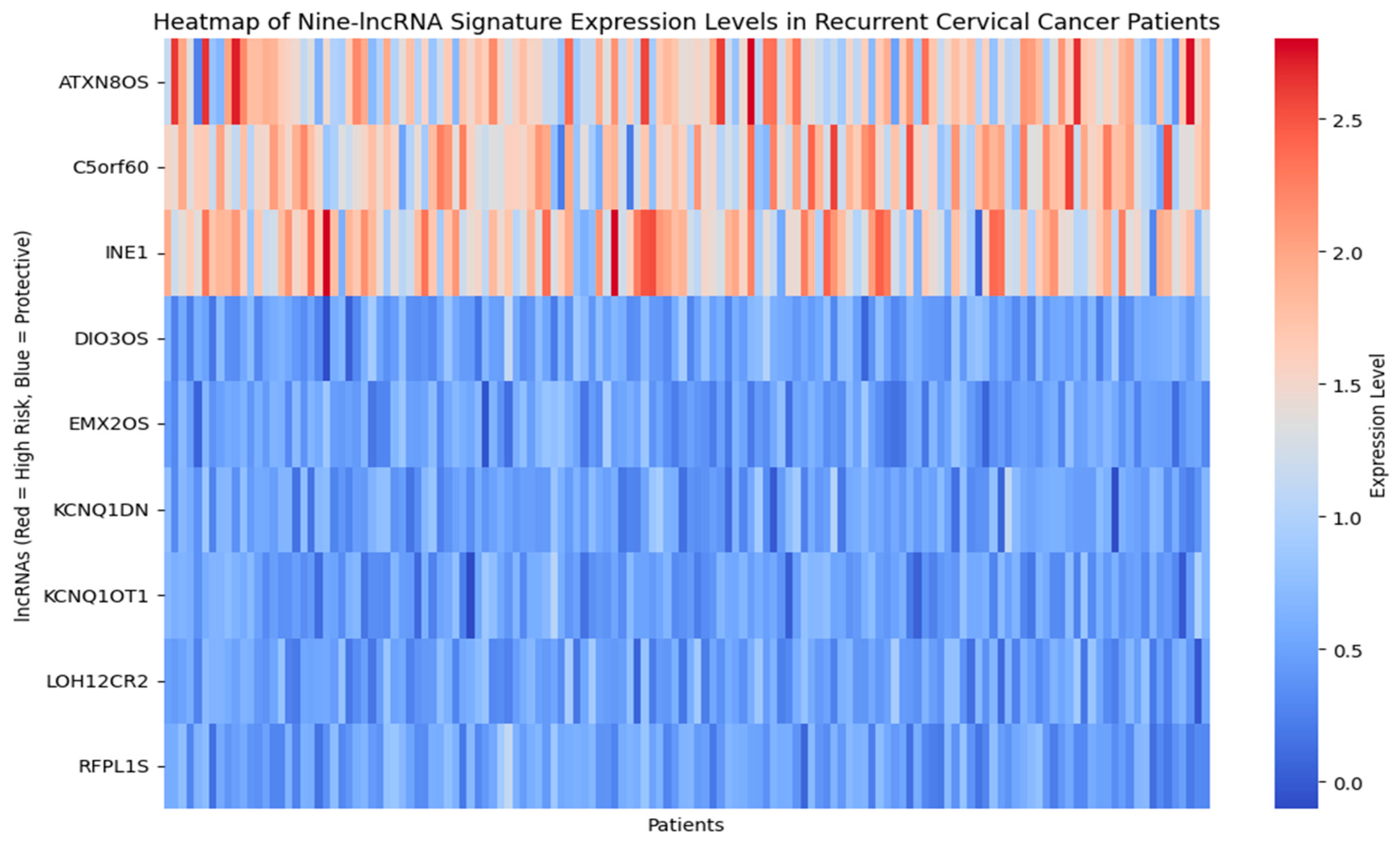
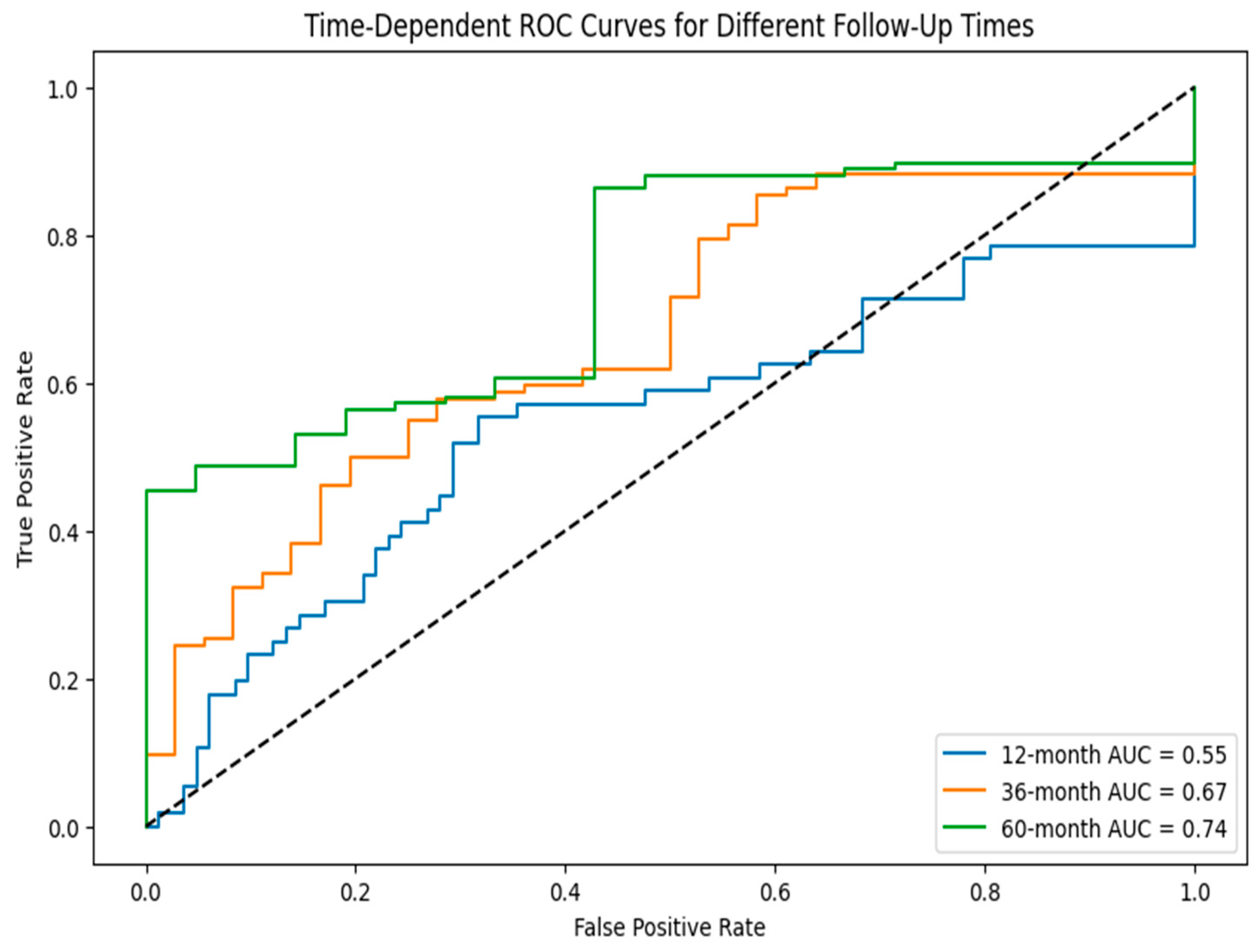
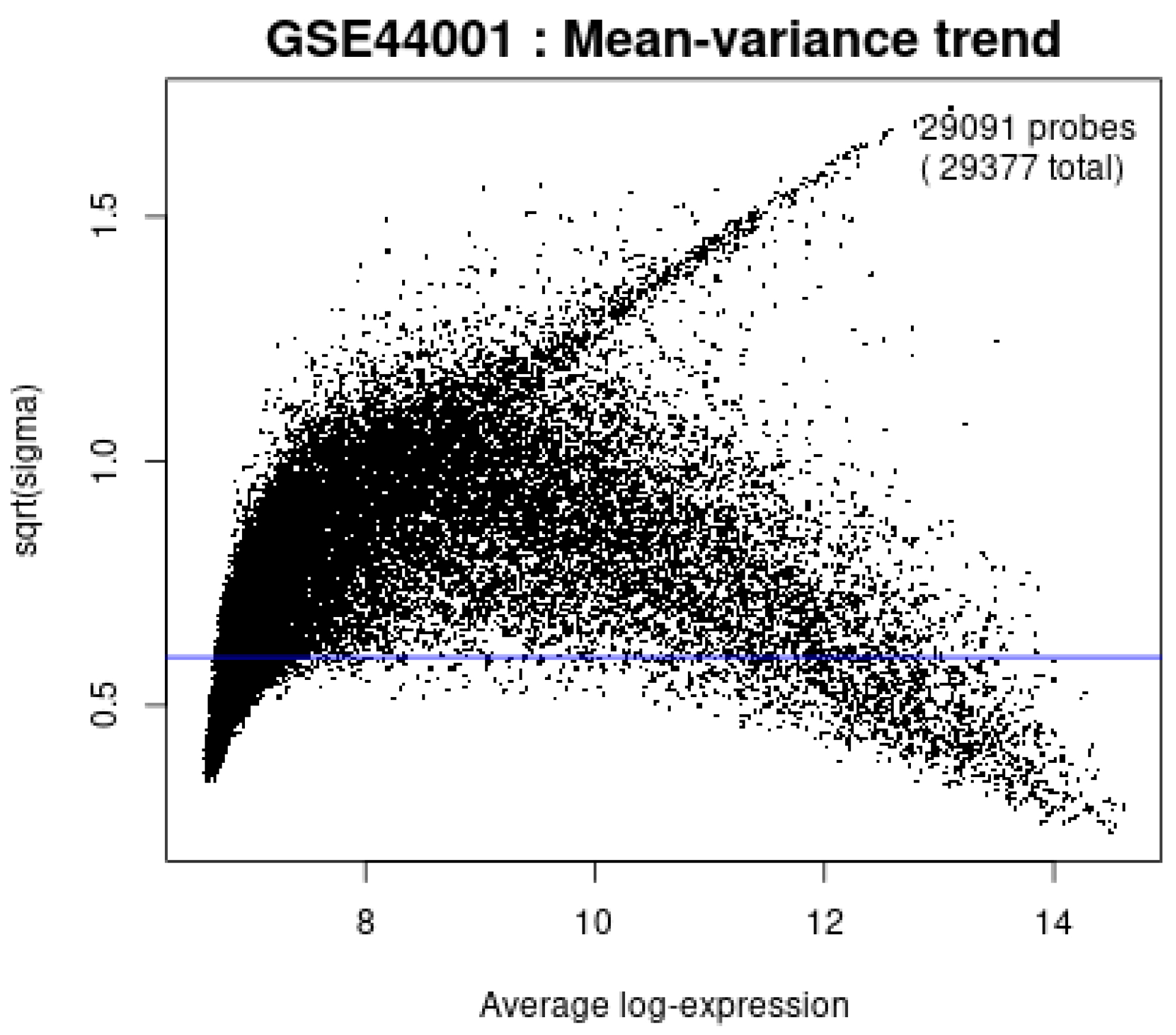
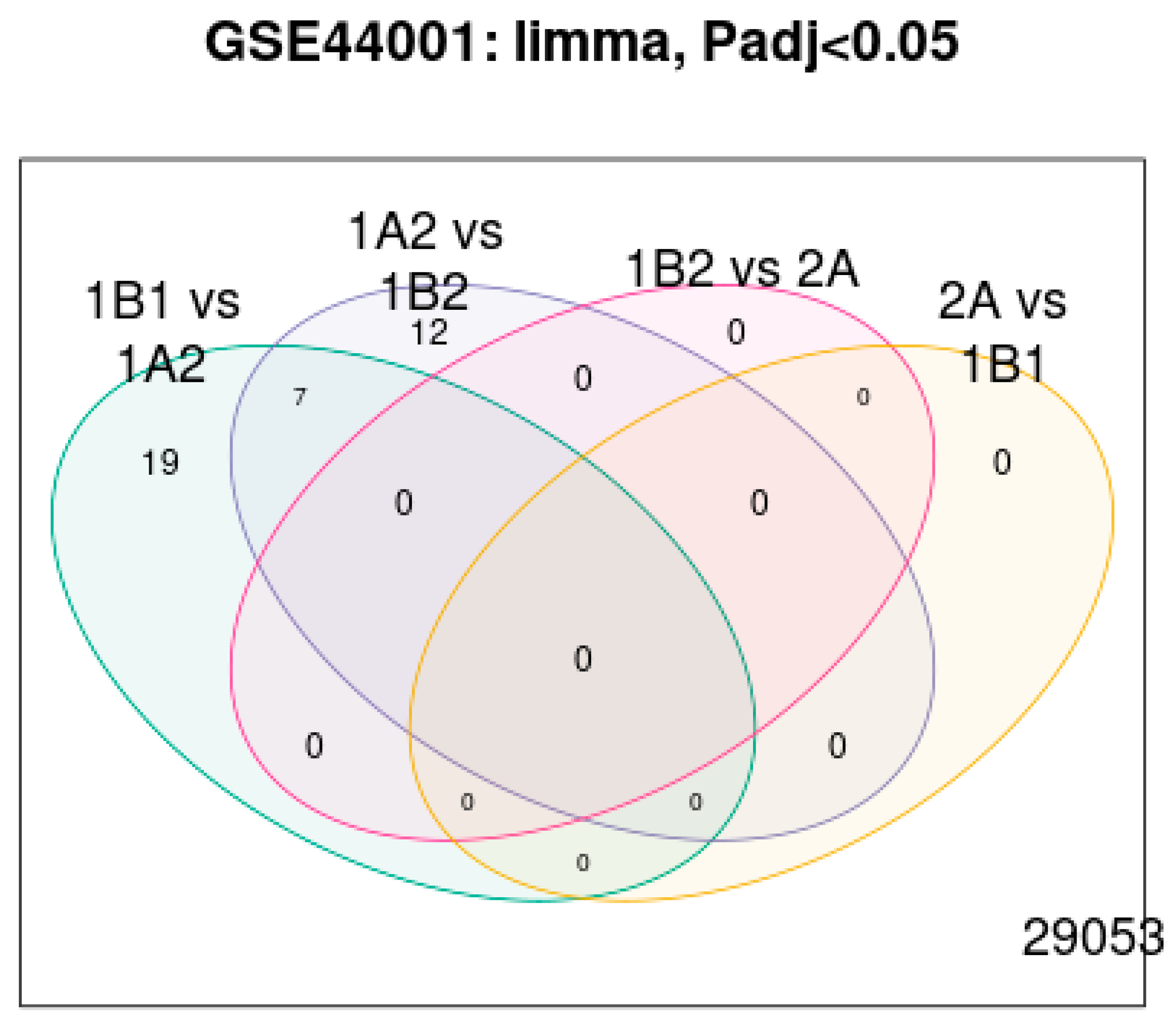
| Category | Details |
|---|---|
| Total patients with recurrence (n) | 138 |
| Age (years), mean ± SD | 49.7 ± 14.6 |
| FIGO 2009 staging | n (%) |
| IVB | 21 (15.2%) |
| IVA | 12 (8.7%) |
| IIIC-2 | 13 (9.4%) |
| IIIC-1 | 8 (5.8%) |
| IIIB | 16 (11.6%) |
| IIIA | 7 (5.1%) |
| IIB | 16 (11.6%) |
| IB3 | 12 (8.7%) |
| IB2 | 11 (8.0%) |
| IB-1 | 9 (6.5%) |
| IA-2 | 2 (1.4%) |
| IA-1 | 11 (8.0%) |
| Stages, n (%) | |
| Early | 22 (15.9%) |
| Locally advanced | 67 (48.6%) |
| Advanced | 49 (35.5%) |
| Histological subtypes, n (%) | |
| Squamous cell carcinoma (SCC) | 51 (37.0%) |
| Adenocarcinoma (ADC) | 53 (38.4%) |
| Other | 34 (24.6%) |
| Features | Mean | Median | Standard Deviation |
|---|---|---|---|
| Age | 49.65 | 50.0 | 14.57 |
| Age of Initial Diagnosis | 50.35 | 52.0 | 14.89 |
| Post Menopause(years) | 7.60 | 6.0 | 7.6 |
| Tumor size | 3.21 | 3.34 | 1.183 |
| DFS months | 59.45 | 45.0 | 45.75 |
| Feature | Missing (n) | Missing (%) |
|---|---|---|
| Age | 5 | 0.7% |
| Age of Initial Diagnosis | 6 | 0.8% |
| Post Menopause in Years | 28 | 3.8% |
| Symptoms | 10 | 1.4% |
| Duration of Symptoms | 35 | 4.7% |
| Comorbidities | 12 | 1.6% |
| Comorbidities Details | 45 | 6.1% |
| Addictive Habits | 60 | 8.1% |
| PV Examination | 15 | 2.0% |
| PR Examination | 18 | 2.4% |
| Primary Lesion—MRI | 55 | 7.4% |
| Primary Lesion—CT Scan | 110 | 14.9% |
| HPV Infection | 95 | 12.9% |
| HPV Vaccination Status | 140 | 18.9% |
| Smoking | 80 | 10.8% |
| Chlamydia Infection | 65 | 8.8% |
| BMI | 50 | 6.8% |
| Oral Contraceptives Use | 90 | 12.2% |
| Number of Full-term Pregnancies | 25 | 3.4% |
| Age at First Full-term Pregnancy | 60 | 8.1% |
| History | 75 | 10.1% |
| Tumor Size (cm) | 30 | 4.1% |
| Lymph Node Metastasis | 40 | 5.4% |
| Histological Type | 15 | 2.0% |
| Treatment Type | 5 | 0.7% |
| FIGO Stage | 0 | 0.0% |
| Imaging | 12 | 1.6% |
| DFS (Months) | 0 | 0.0% |
| Metric | Value | Interpretation |
|---|---|---|
| Number of matched records | 138 | Successfully matched patients using tumor size and DFS |
| GSE 44001 Data | 299 | Full public dataset size |
| Tumor size difference in average (cm) | 0.105 | Very close alignment in tumor size |
| DFS difference average(months) | 8.457 | Acceptable difference considering real-world variability |
| t-test p-value (Tumor Size) | 0.1194 | No significant difference in tumor size distributions |
| KS-test p-value (Tumor Size) | 0.0290 | Mild distributional shift detected |
| Cohen’s d (Tumor Size) | 0.161 | distributions are broadly similar |
| Variable | Coef | exp(Coef) | Z | p-Value | 95% Confidence Interval |
|---|---|---|---|---|---|
| Stage | 0.39 | 1.48 | 13.92 | <0.005 | (1.40–1.56) |
| Largest Diameter (cm) | 0.17 | 1.19 | 2.93 | <0.005 | (1.06–1.33) |
| Biomarker | Prognostic Value | Classification | Estimated Prognosis |
|---|---|---|---|
| ATXN8OS Marker | 28.45 | Elevated Risk | Decreased progression-free period |
| C5orf60 Indicator | 32.18 | Elevated Risk | Decreased progression-free period |
| INE1 Index | 75.93 | Elevated Risk | Decreased progression-free period |
| DIO3OS Metric | −45.27 | Reduced Risk | Prolonged progression-free period |
| EMX2OS Score | −1.25 | Moderate Risk | Medium progression-free period |
| KCNQ1DN Marker | −4.87 | Low Risk | Prolonged progression-free period |
| LOH12CR2 Gauge | −0.85 | Low Risk | Prolonged progression-free period |
| RFPL1S Value | −0.62 | Low Risk | Prolonged progression-free period |
| KCNQ1OT1 Indicator | −0.95 | Low Risk | Prolonged progression-free period |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Senthilkumar, G.; Pitchaimuthu, R.; Panneerselvam, P.S.; Alagarswamy, R.P.; Dhanasekaran, S. Integrative Long Non-Coding RNA Analysis and Recurrence Prediction in Cervical Cancer Using a Recurrent Neural Network. Diagnostics 2025, 15, 2848. https://doi.org/10.3390/diagnostics15222848
Senthilkumar G, Pitchaimuthu R, Panneerselvam PS, Alagarswamy RP, Dhanasekaran S. Integrative Long Non-Coding RNA Analysis and Recurrence Prediction in Cervical Cancer Using a Recurrent Neural Network. Diagnostics. 2025; 15(22):2848. https://doi.org/10.3390/diagnostics15222848
Chicago/Turabian StyleSenthilkumar, Geeitha, Renuka Pitchaimuthu, Prabu Sankar Panneerselvam, Rama Prasath Alagarswamy, and Seshathiri Dhanasekaran. 2025. "Integrative Long Non-Coding RNA Analysis and Recurrence Prediction in Cervical Cancer Using a Recurrent Neural Network" Diagnostics 15, no. 22: 2848. https://doi.org/10.3390/diagnostics15222848
APA StyleSenthilkumar, G., Pitchaimuthu, R., Panneerselvam, P. S., Alagarswamy, R. P., & Dhanasekaran, S. (2025). Integrative Long Non-Coding RNA Analysis and Recurrence Prediction in Cervical Cancer Using a Recurrent Neural Network. Diagnostics, 15(22), 2848. https://doi.org/10.3390/diagnostics15222848









