Interplay Between Type 2 Diabetes Susceptibility and Prostate Cancer Progression: Functional Insights into C2CD4A
Abstract
1. Introduction
2. Patients and Methods
2.1. Patient and Response Assessment
2.2. SNP Selection and Genotyping
2.3. Bioinformatic Analyses
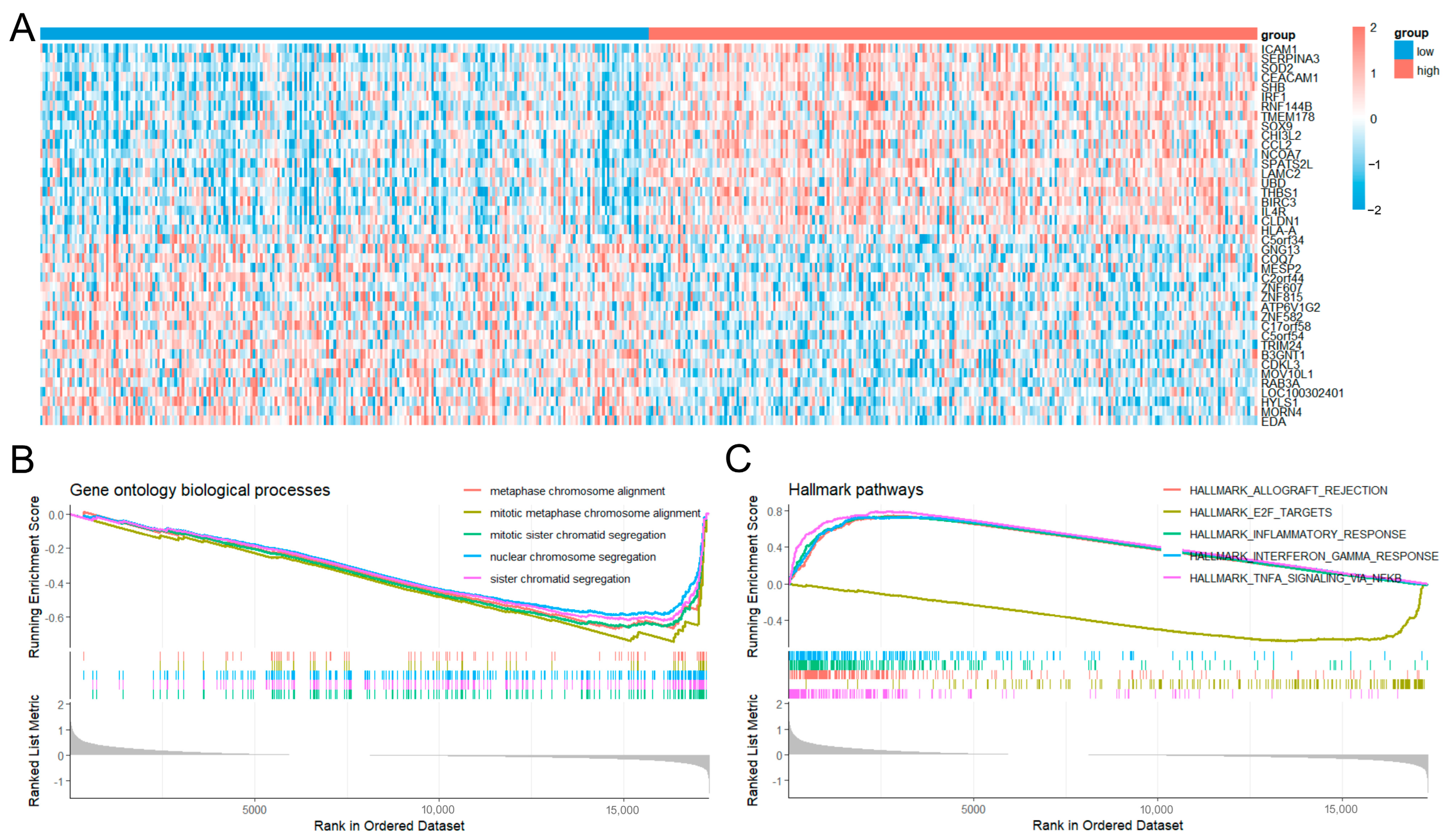
2.4. Differential Gene Expression and Gene Enrichment Analyses
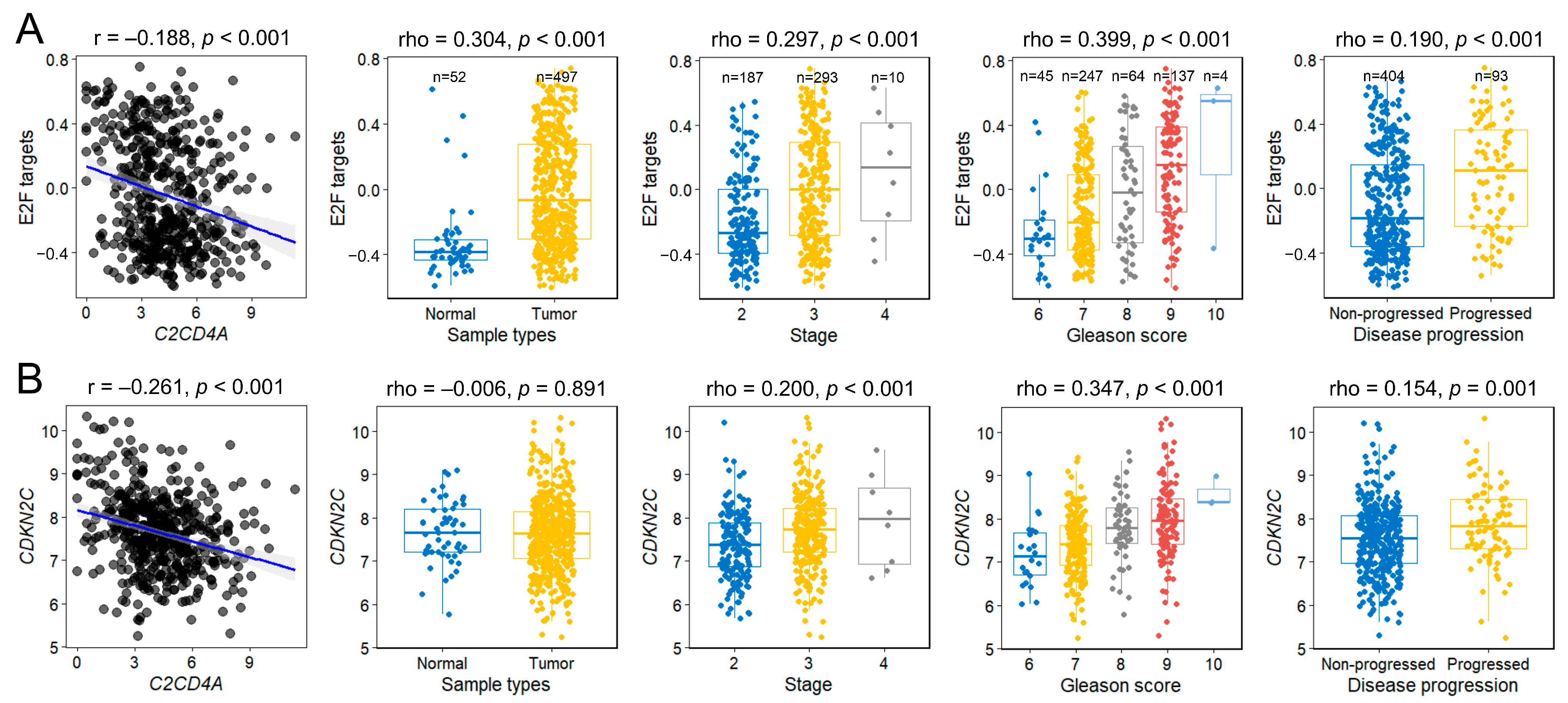
2.5. Gene Set Variation Analysis
2.6. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Eastham, J.A.; Auffenberg, G.B.; Barocas, D.A.; Chou, R.; Crispino, T.; Davis, J.W.; Eggener, S.; Horwitz, E.M.; Kane, C.J.; Kirkby, E.; et al. Clinically Localized Prostate Cancer: AUA/ASTRO Guideline, Part I: Introduction, Risk Assessment, Staging, and Risk-Based Management. J. Urol. 2022, 208, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.M.; Daignault-Newton, S.; Spratt, D.E.; Dunn, R.L.; Singhal, U.; Okoth, L.A.; Feng, F.Y.; Johnson, A.M.; Lane, B.R.; Linsell, S.; et al. Impact of Gene Expression Classifier Testing on Adjuvant Treatment Following Radical Prostatectomy: The G-MINOR Prospective Randomized Cluster-crossover Trial. Eur. Urol. 2025, 87, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016, 375, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.C.; Shen, W.C.; Shih, T.C.; Tsai, C.W.; Chang, W.S.; Cho, Y.; Tsai, C.H.; Bau, D.T. The current progress and future prospects of personalized radiogenomic cancer study. Biomedicine 2015, 5, 2. [Google Scholar] [CrossRef]
- Bansal, D.; Bhansali, A.; Kapil, G.; Undela, K.; Tiwari, P. Type 2 diabetes and risk of prostate cancer: A meta-analysis of observational studies. Prostate Cancer Prostatic Dis. 2013, 16, 151–158. [Google Scholar] [CrossRef]
- Pagano, A.P.; da Silva, B.R.; Vieira, F.T.; Meira Filho, L.F.; Purcell, S.A.; Lewis, J.D.; Mackenzie, M.L.; Robson, P.J.; Vena, J.E.; Silva, F.M.; et al. Association Between Diabetes and Risk of Prostate Cancer: A Systematic Review and Meta-Analysis of Observational Studies. World J. Mens. Health 2025, 43, 304–320. [Google Scholar] [CrossRef]
- Cai, H.; Xu, Z.; Xu, T.; Yu, B.; Zou, Q. Diabetes mellitus is associated with elevated risk of mortality amongst patients with prostate cancer: A meta-analysis of 11 cohort studies. Diabetes Metab. Res. Rev. 2015, 31, 336–343. [Google Scholar] [CrossRef]
- Kelkar, S.; Oyekunle, T.; Eisenberg, A.; Howard, L.; Aronson, W.J.; Kane, C.J.; Amling, C.L.; Cooperberg, M.R.; Klaassen, Z.; Terris, M.K.; et al. Diabetes and Prostate Cancer Outcomes in Obese and Nonobese Men After Radical Prostatectomy. JNCI Cancer Spectr. 2021, 5, pkab023. [Google Scholar] [CrossRef]
- Gallagher, E.J.; LeRoith, D. Obesity and Diabetes: The Increased Risk of Cancer and Cancer-Related Mortality. Physiol. Rev. 2015, 95, 727–748. [Google Scholar] [CrossRef]
- Elemam, N.M.; Hotait, H.Y.; Saleh, M.A.; El-Huneidi, W.; Talaat, I.M. Insulin-like growth factor family and prostate cancer: New insights and emerging opportunities. Front. Endocrinol. 2024, 15, 1396192. [Google Scholar] [CrossRef]
- Janssen, J. Hyperinsulinemia and Its Pivotal Role in Aging, Obesity, Type 2 Diabetes, Cardiovascular Disease and Cancer. Int. J. Mol. Sci. 2021, 22, 7797. [Google Scholar] [CrossRef]
- Sharma, U.; Sahu, A.; Shekhar, H.; Sharma, B.; Haque, S.; Kaur, D.; Tuli, H.S.; Mishra, A.; Ahmad, F. The heat of the battle: Inflammation’s role in prostate cancer development and inflammation-targeted therapies. Discov. Oncol. 2025, 16, 108. [Google Scholar] [CrossRef]
- Tsalamandris, S.; Antonopoulos, A.S.; Oikonomou, E.; Papamikroulis, G.A.; Vogiatzi, G.; Papaioannou, S.; Deftereos, S.; Tousoulis, D. The Role of Inflammation in Diabetes: Current Concepts and Future Perspectives. Eur. Cardiol. 2019, 14, 50–59. [Google Scholar] [CrossRef]
- Pungsrinont, T.; Kallenbach, J.; Baniahmad, A. Role of PI3K-AKT-mTOR Pathway as a Pro-Survival Signaling and Resistance-Mediating Mechanism to Therapy of Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 11088. [Google Scholar] [CrossRef]
- Yoon, M.S. The Role of Mammalian Target of Rapamycin (mTOR) in Insulin Signaling. Nutrients 2017, 9, 1176. [Google Scholar] [CrossRef] [PubMed]
- Imamura, M.; Maeda, S. Perspectives on genetic studies of type 2 diabetes from the genome-wide association studies era to precision medicine. J. Diabetes Investig. 2024, 15, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Raynor, L.A.; Pankow, J.S.; Rasmussen-Torvik, L.J.; Tang, W.; Prizment, A.; Couper, D.J. Pleiotropy and pathway analyses of genetic variants associated with both type 2 diabetes and prostate cancer. Int. J. Mol. Epidemiol. Genet. 2013, 4, 49–60. [Google Scholar] [PubMed]
- Yu, O.H.; Foulkes, W.D.; Dastani, Z.; Martin, R.M.; Eeles, R.; Consortium, P.; Investigators, C.G.; Richards, J.B. An assessment of the shared allelic architecture between type II diabetes and prostate cancer. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1473–1475. [Google Scholar] [CrossRef]
- significance of prostate cancer susceptibility variants on prostate-specific antigen recurrence after radical prostatectomy. Cancer Epidemiol. Biomark. Prev. 2009, 18, 3068–3074. [CrossRef]
- Huang, S.P.; Lan, Y.H.; Lu, T.L.; Pao, J.B.; Chang, T.Y.; Lee, H.Z.; Yang, W.H.; Hsieh, C.J.; Chen, L.M.; Huang, L.C.; et al. Clinical significance of runt-related transcription factor 1 polymorphism in prostate cancer. BJU Int. 2011, 107, 486–492. [Google Scholar] [CrossRef]
- Bao, B.Y.; Pao, J.B.; Lin, V.C.; Huang, C.N.; Chang, T.Y.; Lan, Y.H.; Lu, T.L.; Lee, H.Z.; Chen, L.M.; Ting, W.C.; et al. Individual and cumulative association of prostate cancer susceptibility variants with clinicopathologic characteristics of the disease. Clin. Chim. Acta 2010, 411, 1232–1237. [Google Scholar] [CrossRef]
- Freedland, S.J.; Sutter, M.E.; Dorey, F.; Aronson, W.J. Defining the ideal cutpoint for determining PSA recurrence after radical prostatectomy. Prostate-specific antigen. Urology 2003, 61, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Huang, S.P.; Chen, Y.T.; Wu, H.E.; Cheng, W.C.; Huang, C.Y.; Yu, C.C.; Lin, V.C.; Geng, J.H.; Lu, T.L.; et al. TNFRSF13B is a potential contributor to prostate cancer. Cancer Cell Int. 2022, 22, 180. [Google Scholar] [CrossRef]
- Xue, A.; Wu, Y.; Zhu, Z.; Zhang, F.; Kemper, K.E.; Zheng, Z.; Yengo, L.; Lloyd-Jones, L.R.; Sidorenko, J.; Wu, Y.; et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 2018, 9, 2941. [Google Scholar] [CrossRef]
- Huang, S.P.; Chen, L.C.; Chen, Y.T.; Lee, C.H.; Huang, C.Y.; Yu, C.C.; Lin, V.C.; Lu, T.L.; Bao, B.Y. PTBP1 Genetic Variants Affect the Clinical Response to Androgen-deprivation Therapy in Patients With Prostate Cancer. Cancer Genom. Proteom. 2021, 18, 325–334. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
- Kwong, A.; Boughton, A.P.; Wang, M.; VandeHaar, P.; Boehnke, M.; Abecasis, G.; Kang, H.M. FIVEx: An interactive eQTL browser across public datasets. Bioinformatics 2022, 38, 559–561. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Chang, C.W.; Shen, Y.C.; Yan, S.J. HP1a-mediated heterochromatin formation inhibits high dietary sugar-induced tumor progression. Cell Death Dis. 2021, 12, 1130. [Google Scholar] [CrossRef]
- Jeon, Y.H.; Kim, G.W.; Kim, S.Y.; Yi, S.A.; Yoo, J.; Kim, J.Y.; Lee, S.W.; Kwon, S.H. Heterochromatin Protein 1: A Multiplayer in Cancer Progression. Cancers 2022, 14, 763. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.S.; Jun, H.S. Effects of Glucagon-Like Peptide-1 on Oxidative Stress and Nrf2 Signaling. Int. J. Mol. Sci. 2017, 19, 26. [Google Scholar] [CrossRef] [PubMed]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Pradas-Juni, M.; Hansmeier, N.R.; Link, J.C.; Schmidt, E.; Larsen, B.D.; Klemm, P.; Meola, N.; Topel, H.; Loureiro, R.; Dhaouadi, I.; et al. A MAFG-lncRNA axis links systemic nutrient abundance to hepatic glucose metabolism. Nat. Commun. 2020, 11, 644. [Google Scholar] [CrossRef]
- Wang, M.; Ren, Y.; Hu, S.; Liu, K.; Qiu, L.; Zhang, Y. TCF11 Has a Potent Tumor-Repressing Effect Than Its Prototypic Nrf1alpha by Definition of Both Similar Yet Different Regulatory Profiles, With a Striking Disparity from Nrf2. Front. Oncol. 2021, 11, 707032. [Google Scholar] [CrossRef]
- Kuo, T.; Kraakman, M.J.; Damle, M.; Gill, R.; Lazar, M.A.; Accili, D. Identification of C2CD4A as a human diabetes susceptibility gene with a role in beta cell insulin secretion. Proc. Natl. Acad. Sci. USA 2019, 116, 20033–20042. [Google Scholar] [CrossRef]
- Lu, J.; Chen, Q. Transcriptome-based identification of molecular markers related to the development and prognosis of Colon cancer. Nucleosides Nucleotides Nucleic Acids 2021, 40, 1114–1124. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhang, W.; Shang, W. Identification and validation of a prognostic risk-scoring model based on sphingolipid metabolism-associated cluster in colon adenocarcinoma. Front. Endocrinol. 2022, 13, 1045167. [Google Scholar] [CrossRef]
- Rong, Z.; Luo, Z.; Fu, Z.; Zhang, P.; Li, T.; Zhang, J.; Zhu, Z.; Yu, Z.; Li, Q.; Qiu, Z.; et al. The novel circSLC6A6/miR-1265/C2CD4A axis promotes colorectal cancer growth by suppressing p53 signaling pathway. J. Exp. Clin. Cancer Res. 2021, 40, 324. [Google Scholar] [CrossRef] [PubMed]
- Li, A.H.; Park, S.Y.; Li, P.; Zhou, C.; Kluz, T.; Li, J.; Costa, M.; Sun, H. Transcriptome Analysis Reveals Anti-Cancer Effects of Isorhapontigenin (ISO) on Highly Invasive Human T24 Bladder Cancer Cells. Int. J. Mol. Sci. 2024, 25, 1783. [Google Scholar] [CrossRef]
- Han, F.; Qian, L.; Zhang, Y.; Liu, P.; Li, R.; Chen, M. C2CD4A/B variants in the predisposition of lung cancer in the Chinese Han population. Funct. Integr. Genom. 2022, 22, 331–340. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jiang, B.; Guo, J.; Shao, H.; Del Priore, I.S.; Chang, Q.; Kudo, R.; Li, Z.; Razavi, P.; Liu, B.; et al. INK4 Tumor Suppressor Proteins Mediate Resistance to CDK4/6 Kinase Inhibitors. Cancer Discov. 2022, 12, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Li, G.S.; Chen, G.; Liu, J.; Tang, D.; Zheng, J.H.; Luo, J.; Jin, M.H.; Lu, H.S.; Bao, C.X.; Tian, J.; et al. Clinical significance of cyclin-dependent kinase inhibitor 2C expression in cancers: From small cell lung carcinoma to pan-cancers. BMC Pulm. Med. 2022, 22, 246. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Chen, L.; Niu, Q.; Ge, Q.; Zhang, J.; Tao, J.; Zhou, J.; Liang, C. Identification and validation of an E2F-related gene signature for predicting recurrence-free survival in human prostate cancer. Cancer Cell Int. 2022, 22, 382. [Google Scholar] [CrossRef]
- Vishnoi, K.; Viswakarma, N.; Rana, A.; Rana, B. Transcription Factors in Cancer Development and Therapy. Cancers 2020, 12, 2296. [Google Scholar] [CrossRef]
- Ankers, J.M.; Awais, R.; Jones, N.A.; Boyd, J.; Ryan, S.; Adamson, A.D.; Harper, C.V.; Bridge, L.; Spiller, D.G.; Jackson, D.A.; et al. Dynamic NF-kappaB and E2F interactions control the priority and timing of inflammatory signalling and cell proliferation. eLife 2016, 5, e10473. [Google Scholar] [CrossRef]
- Klein, K.; Witalisz-Siepracka, A.; Gotthardt, D.; Agerer, B.; Locker, F.; Grausenburger, R.; Knab, V.M.; Bergthaler, A.; Sexl, V. T Cell-Intrinsic CDK6 Is Dispensable for Anti-Viral and Anti-Tumor Responses In Vivo. Front. Immunol. 2021, 12, 650977. [Google Scholar] [CrossRef]


| Characteristics | No BCR | BCR | p |
| No. of patients, n (%) | 415 (64.4) | 229 (35.6) | |
| Median age at diagnosis, years (IQR) | 66 (62–70) | 66 (61–71) | 0.299 |
| Median PSA at diagnosis, ng/mL (IQR) | 9.3 (6.2–15.0) | 14.7 (8.4–25.9) | <0.001 |
| Pathologic stage, n (%) | |||
| T1/T2 | 275 (66.4) | 89 (39.6) | <0.001 |
| T3/T4/N1 | 139 (33.6) | 136 (60.4) | |
| Pathologic Gleason score, n (%) | |||
| 2–7 | 369 (88.4) | 164 (71.6) | <0.001 |
| 8–10 | 48 (11.6) | 65 (28.4) | |
| ISUP grade group | |||
| 1 | 117 (28.2) | 36 (15.7) | <0.001 |
| 2 and 3 | 250 (60.2) | 128 (55.9) | |
| 4 | 22 (5.3) | 20 (8.7) | |
| 5 | 26 (6.3) | 45 (19.7) | |
| D’Amico risk classification, n (%) | |||
| Low | 65 (15.7) | 12 (5.3) | <0.001 |
| Intermediate | 166 (40.1) | 50 (21.9) | |
| High | 183 (44.2) | 166 (72.8) | |
| Surgical margin, n (%) | |||
| Negative | 320 (77.1) | 140 (61.1) | <0.001 |
| Positive | 95 (22.9) | 89 (38.9) | |
| Lymph node metastasis, n (%) | |||
| Negative | 323 (95.8) | 182 (95.3) | 0.763 |
| Positive | 14 (4.2) | 9 (4.7) |
| SNP ID | Chromosome | Position | Mapped Gene | Allele | MAF | HWE | Risk Allele for T2D | Risk Allele for BCR | HR | p |
| rs2296173 | 1 | 39913351 | MACF1 | A>G | 0.166 | 1.000 | G | A | 0.876 | 0.317 |
| rs12088739 | 1 | 51506886 | MIR4421 | A>G | 0.100 | 0.664 | A | A | 0.804 | 0.197 |
| rs1127655 | 1 | 117530507 | PTGFRN | C>T | 0.438 | 0.080 | C | T | 1.161 | 0.093 |
| rs340874 | 1 | 214159256 | PROX1-AS1 | T>C | 0.387 | 0.282 | C | C | 1.018 | 0.859 |
| rs2820426 | 1 | 219660535 | LOC102723886 (LYPLAL1) | A>G | 0.330 | 0.426 | G | A | 0.898 | 0.299 |
| rs348330 | 1 | 229672955 | ABCB10 | G>A | 0.435 | 0.130 | G | G | 0.883 | 0.212 |
| rs2867125 | 2 | 622827 | TMEM18 | C>T | 0.078 | 0.165 | C | T | 1.028 | 0.875 |
| rs780094 | 2 | 27741237 | GCKR | T>C | 0.492 | 0.814 | C | T | 0.969 | 0.737 |
| rs243019 | 2 | 60585806 | MIR4432HG | C>T | 0.345 | 0.603 | C | C | 0.853 | 0.109 |
| rs1009358 | 2 | 65276452 | CEP68 | T>C | 0.284 | 0.441 | T | T | 0.976 | 0.822 |
| rs10169613 | 2 | 111934977 | BCL2L11 | C>T | 0.455 | 0.692 | C | T | 1.038 | 0.685 |
| rs12617659 | 2 | 121309759 | LOC105373585 (GLI2) | C>T | 0.184 | 0.192 | C | C | 0.918 | 0.480 |
| rs7572970 | 2 | 161136656 | RBMS1 | G>A | 0.185 | 0.605 | G | G | 0.995 | 0.968 |
| rs13389219 | 2 | 165528876 | COBLL1 | C>T | 0.098 | 0.501 | C | C | 0.830 | 0.286 |
| rs2972144 | 2 | 227101411 | MIR5702 | G>A | 0.064 | 1.000 | G | A | 1.129 | 0.518 |
| rs7561798 | 2 | 228973660 | SPHKAP | G>A | 0.325 | 0.858 | G | A | 1.075 | 0.480 |
| rs1899951 | 3 | 12394840 | PPARG | C>T | 0.053 | 0.701 | C | T | 1.081 | 0.710 |
| rs1496653 | 3 | 23454790 | UBE2E2 | A>G | 0.213 | 1.000 | A | G | 1.132 | 0.276 |
| rs11926707 | 3 | 46925539 | PTH1R | C>T | 0.359 | 0.932 | C | C | 0.961 | 0.684 |
| rs2292662 | 3 | 63897215 | ATXN7 | C>T | 0.424 | 0.520 | C | T | 1.091 | 0.354 |
| rs6795735 | 3 | 64705365 | ADAMTS9-AS2 | T>C | 0.242 | 0.915 | C | T | 0.812 | 0.067 |
| rs4472028 | 3 | 152053250 | MBNL1 | C>T | 0.438 | 1.000 | T | T | 1.107 | 0.285 |
| rs11925227 | 3 | 170766618 | TNIK | G>A | 0.169 | 0.576 | G | G | 0.901 | 0.423 |
| rs7651090 | 3 | 185513392 | IGF2BP2 | A>G | 0.249 | 0.599 | G | A | 1.000 | 0.999 |
| rs3887925 | 3 | 186665645 | ST6GAL1 | C>T | 0.475 | 0.157 | T | C | 0.952 | 0.591 |
| rs1801214 | 4 | 6303022 | WFS1 | T>C | 0.077 | 0.045 | T | C | 1.113 | 0.490 |
| rs17086692 | 4 | 53134293 | SPATA18 | G>T | 0.257 | 0.918 | G | T | 1.145 | 0.196 |
| rs993380 | 4 | 83584496 | SCD5 | G>A | 0.364 | 0.735 | A | A | 1.120 | 0.233 |
| rs7674212 | 4 | 103988899 | SLC9B2 | G>T | 0.382 | 0.868 | G | T | 1.130 | 0.209 |
| rs11098676 | 4 | 123833154 | NUDT6 | C>T | 0.039 | 1.000 | C | C | 0.558 | 0.047 |
| rs7685296 | 4 | 153254121 | TMEM154 | C>T | 0.442 | 1.000 | C | T | 1.035 | 0.713 |
| rs1061813 | 5 | 14847331 | ANKH | A>G | 0.182 | 0.012 | G | A | 0.854 | 0.225 |
| rs4865796 | 5 | 53272664 | ARL15 | A>G | 0.117 | 0.124 | A | G | 1.024 | 0.867 |
| rs459193 | 5 | 55806751 | C5orf67 | G>A | 0.499 | 0.387 | G | A | 1.122 | 0.235 |
| rs2307111 | 5 | 75003678 | POC5 | C>T | 0.431 | 0.576 | T | T | 1.175 | 0.089 |
| rs6878122 | 5 | 76427311 | ZBED3-AS1 | A>G | 0.060 | 0.499 | G | A | 0.975 | 0.898 |
| rs10077431 | 5 | 112927686 | YTHDC2 | C>A | 0.101 | 0.829 | C | A | 1.296 | 0.064 |
| rs1050226 | 6 | 7281654 | RREB1 | A>G | 0.414 | 0.808 | A | G | 1.046 | 0.623 |
| rs7756992 | 6 | 20679709 | CDKAL1 | A>G | 0.482 | 0.432 | G | A | 0.992 | 0.934 |
| rs2857605 | 6 | 31524851 | NFKBIL1 | T>C | 0.156 | 0.072 | T | C | 1.250 | 0.066 |
| rs1063355 | 6 | 32627714 | HLA-DQB1 | G>T | 0.329 | 0.656 | G | G | 0.840 | 0.089 |
| rs9369425 | 6 | 43810974 | LOC107986598 (VEGFA) | A>G | 0.144 | 0.265 | G | G | 1.191 | 0.177 |
| rs72892910 | 6 | 50816887 | TFAP2B | G>T | 0.134 | 0.173 | T | T | 1.252 | 0.074 |
| rs853974 | 6 | 127068983 | RPS4XP9 | C>T | 0.468 | 1.000 | T | T | 1.014 | 0.883 |
| rs2246012 | 6 | 131898208 | ARG1, MED23 | T>C | 0.402 | 0.327 | C | T | 0.967 | 0.722 |
| rs622217 | 6 | 160766770 | SLC22A3 | T>C | 0.296 | 0.258 | T | T | 0.918 | 0.410 |
| rs17168486 | 7 | 14898282 | DGKB | C>T | 0.488 | 0.529 | T | C | 0.940 | 0.526 |
| rs2191348 | 7 | 15064255 | AGMO | T>G | 0.311 | 0.000 | T | T | 0.836 | 0.107 |
| rs2908282 | 7 | 44248828 | YKT6 | G>A | 0.194 | 1.000 | A | G | 0.933 | 0.569 |
| rs2299383 | 7 | 103418846 | RELN | C>T | 0.404 | 0.807 | T | C | 0.942 | 0.524 |
| rs13239186 | 7 | 117510621 | CTTNBP2 | C>T | 0.188 | 0.438 | T | T | 1.060 | 0.617 |
| rs13234269 | 7 | 130429186 | LOC105375508 (KLF14) | T>A | 0.327 | 0.789 | T | A | 1.033 | 0.750 |
| rs11774915 | 8 | 9188762 | LOC157273(TNKS) | C>T | 0.316 | 0.364 | T | T | 1.040 | 0.684 |
| rs10100265 | 8 | 10633159 | PINX1 | A>C | 0.357 | 0.494 | A | A | 0.900 | 0.296 |
| rs17411031 | 8 | 19852310 | LPL | C>G | 0.187 | 1.000 | C | C | 0.848 | 0.198 |
| rs10087241 | 8 | 30863722 | PURG | A>G | 0.033 | 1.000 | G | G | 1.108 | 0.691 |
| rs12681990 | 8 | 36859186 | KCNU1 | T>C | 0.343 | 0.029 | C | T | 0.961 | 0.675 |
| rs516946 | 8 | 41519248 | ANK1 | C>T | 0.130 | 0.484 | C | T | 1.215 | 0.139 |
| rs7845219 | 8 | 95937502 | TP53INP1 | C>T | 0.281 | 0.923 | T | C | 0.960 | 0.699 |
| rs3802177 | 8 | 118185025 | SLC30A8 | G>A | 0.461 | 0.134 | G | A | 1.013 | 0.890 |
| rs2294120 | 8 | 146003567 | ZNF34 | G>A | 0.286 | 0.923 | A | A | 1.138 | 0.198 |
| rs10974438 | 9 | 4291928 | GLIS3 | A>C | 0.378 | 0.616 | C | A | 0.950 | 0.596 |
| rs1063192 | 9 | 22003367 | CDKN2B-AS1/CDKN2B | A>G | 0.180 | 0.351 | A | G | 1.046 | 0.715 |
| rs10811661 | 9 | 22134094 | CDKN2B-AS1 | T>C | 0.408 | 0.626 | T | C | 1.076 | 0.432 |
| rs1758632 | 9 | 34025640 | UBAP2 | G>C | 0.156 | 0.100 | G | C | 1.038 | 0.783 |
| rs17791483 | 9 | 81898980 | LOC101927450 (TLE1) | A>G | 0.052 | 1.000 | A | G | 1.180 | 0.424 |
| rs2796441 | 9 | 84308948 | LOC101927502 (TLE1) | A>G | 0.384 | 0.803 | G | A | 0.927 | 0.433 |
| rs10114341 | 9 | 96919182 | LOC107987099 (PTPDC1) | T>C | 0.090 | 0.338 | T | T | 0.767 | 0.148 |
| rs687621 | 9 | 136137065 | ABO | A>G | 0.340 | 0.541 | G | G | 1.028 | 0.787 |
| rs11257655 | 10 | 12307894 | CDC123 | T>C | 0.449 | 0.000 | T | T | 0.994 | 0.949 |
| rs2616132 | 10 | 71469514 | FAM241B | G>A | 0.484 | 0.814 | A | A | 1.075 | 0.436 |
| rs753270 | 10 | 80964975 | ZMIZ1 | T>C | 0.455 | 0.812 | C | C | 1.082 | 0.389 |
| rs11591741 | 10 | 101976501 | CHUK | G>C | 0.063 | 0.100 | G | G | 0.974 | 0.897 |
| rs2421016 | 10 | 124167512 | PLEKHA1 | C>T | 0.433 | 0.066 | C | C | 0.934 | 0.457 |
| rs2237892 | 11 | 2839751 | KCNQ1 | C>T | 0.340 | 0.485 | C | T | 1.003 | 0.974 |
| rs5215 | 11 | 17408630 | KCNJ11 | T>C | 0.374 | 0.276 | C | C | 1.084 | 0.414 |
| rs7929543 | 11 | 49351026 | TYRL | A>C | 0.148 | 0.061 | C | A | 0.843 | 0.223 |
| rs1552224 | 11 | 72433098 | ARAP1 | A>C | 0.064 | 1.000 | A | C | 1.325 | 0.095 |
| rs10830963 | 11 | 92708710 | MTNR1B | C>G | 0.431 | 0.690 | G | C | 0.919 | 0.366 |
| rs7931302 | 11 | 128236058 | ETS1 | A>C | 0.154 | 0.228 | C | A | 0.835 | 0.201 |
| rs67232546 | 11 | 128398938 | ETS1 | C>T | 0.172 | 0.679 | T | C | 0.811 | 0.114 |
| rs12299509 | 12 | 4406281 | CCND2 | G>A | 0.472 | 0.070 | G | A | 1.150 | 0.116 |
| rs11048456 | 12 | 26463082 | ITPR2 | C>T | 0.307 | 0.581 | C | C | 0.955 | 0.647 |
| rs10842994 | 12 | 27965150 | LOC105369709 (KLHL42) | C>T | 0.205 | 0.811 | C | T | 1.111 | 0.367 |
| rs2261181 | 12 | 66212318 | RPSAP52 | C>T | 0.115 | 0.334 | T | T | 1.277 | 0.064 |
| rs1480474 | 12 | 66326943 | HMGA2 | G>A | 0.085 | 0.610 | G | G | 0.927 | 0.671 |
| rs7138300 | 12 | 71439589 | TSPAN8 | C>T | 0.370 | 0.556 | C | T | 1.055 | 0.592 |
| rs11107116 | 12 | 93978504 | SOCS2 | G>T | 0.330 | 0.248 | T | G | 0.995 | 0.956 |
| rs940904 | 12 | 123491572 | PITPNM2 | A>G | 0.138 | 0.136 | A | A | 0.953 | 0.722 |
| rs825476 | 12 | 124568456 | ZNF664-FAM101A | T>C | 0.370 | 0.736 | T | T | 0.902 | 0.288 |
| rs576674 | 13 | 33554302 | KL | A>G | 0.212 | 0.906 | G | A | 0.953 | 0.681 |
| rs963740 | 13 | 51096095 | DLEU1 | T>A | 0.344 | 0.543 | A | A | 1.096 | 0.339 |
| rs1359790 | 13 | 80717156 | LOC105370275 (SPRY2) | G>A | 0.289 | 0.848 | G | A | 1.052 | 0.621 |
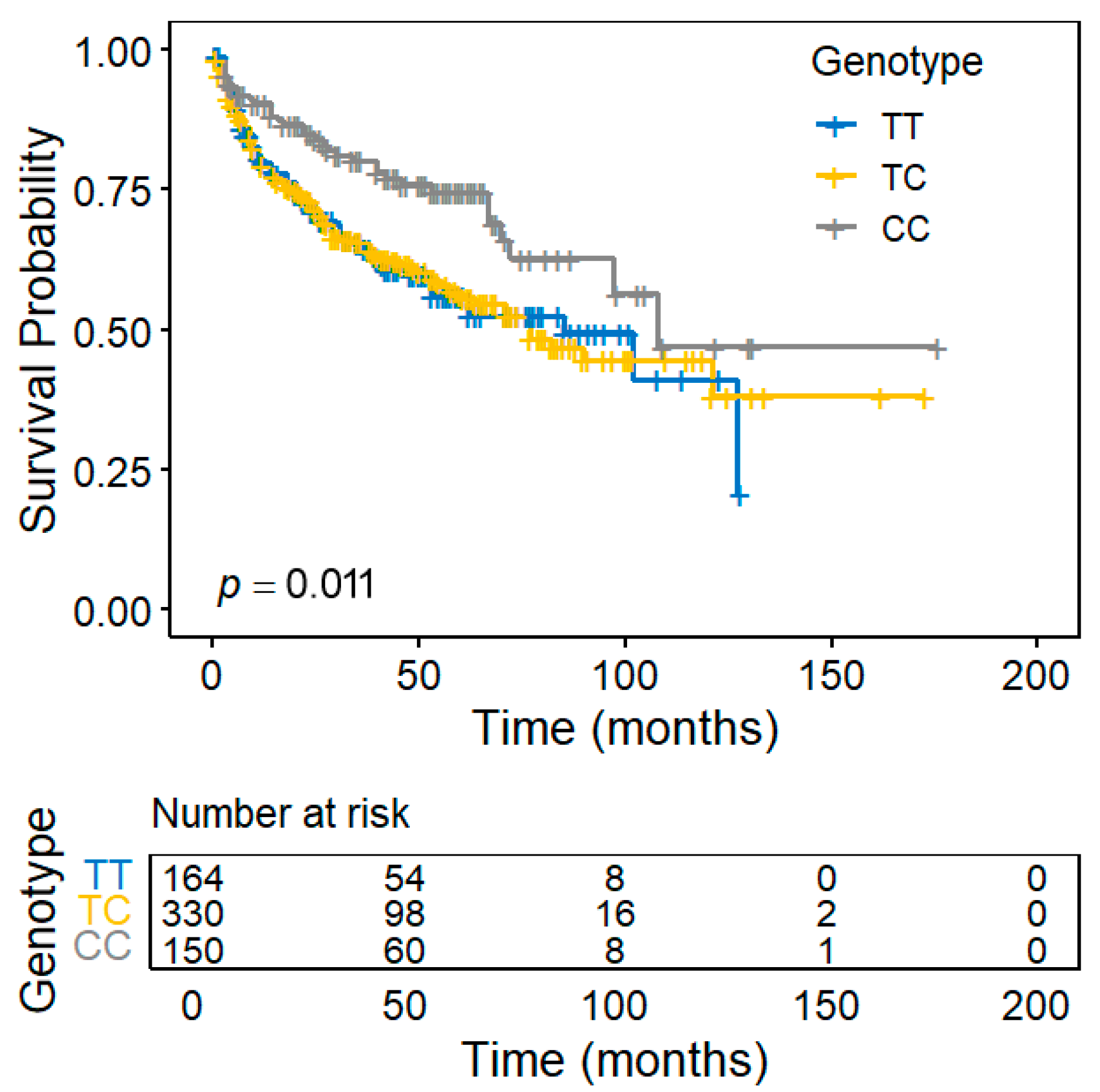
| rs4502156 | 15 | 62383155 | C2CD4A | T>C | 0.488 | 0.637 | T | T | 0.788 | 0.011 |
| rs982077 | 15 | 63823301 | USP3 | A>G | 0.056 | 0.713 | A | G | 1.377 | 0.087 |
| rs7177055 | 15 | 77832762 | LOC101929457 (HMG20A) | G>A | 0.333 | 0.002 | A | G | 0.913 | 0.343 |
| rs9940149 | 16 | 300641 | FAM234A | G>A | 0.429 | 0.689 | G | G | 0.951 | 0.598 |
| rs7185735 | 16 | 53822651 | FTO | A>G | 0.113 | 0.116 | G | G | 1.113 | 0.471 |
| rs244415 | 16 | 69666683 | NFAT5 | G>A | 0.105 | 1.000 | G | G | 0.925 | 0.603 |
| rs2925979 | 16 | 81534790 | CMIP | C>T | 0.426 | 0.378 | T | C | 0.953 | 0.606 |
| rs8068804 | 17 | 3985864 | ZZEF1 | G>A | 0.160 | 0.306 | A | G | 0.895 | 0.386 |
| rs12945601 | 17 | 17653411 | RAI1 | T>C | 0.095 | 0.822 | T | C | 1.114 | 0.487 |
| rs11651755 | 17 | 36099840 | HNF1B | T>C | 0.173 | 0.335 | C | T | 0.924 | 0.524 |
| rs9911983 | 17 | 45885756 | OSBPL7 | C>T | 0.157 | 0.767 | T | T | 1.056 | 0.665 |
| rs9894220 | 17 | 46989154 | UBE2Z | A>G | 0.251 | 0.834 | A | A | 0.850 | 0.137 |
| rs7240767 | 18 | 7070642 | LAMA1 | C>T | 0.306 | 0.927 | C | C | 0.995 | 0.958 |
| rs12970134 | 18 | 57884750 | MC4R | G>A | 0.156 | 0.765 | A | A | 1.253 | 0.068 |
| rs10401969 | 19 | 19407718 | SUGP1 | T>C | 0.105 | 0.529 | C | C | 1.221 | 0.172 |
| rs8108269 | 19 | 46158513 | GIPR | T>G | 0.438 | 0.750 | G | T | 0.945 | 0.550 |
| rs6515236 | 20 | 22435749 | LOC105372562 (FOXA2) | C>A | 0.333 | 0.185 | A | C | 0.883 | 0.232 |
| rs6059662 | 20 | 32675727 | EIF2S2 | G>A | 0.156 | 0.882 | G | G | 0.990 | 0.936 |
| rs4810426 | 20 | 43001721 | HNF4A | C>T | 0.413 | 0.686 | T | T | 1.128 | 0.199 |
| rs4823182 | 22 | 44377442 | SAMM50 | G>A | 0.492 | 0.388 | G | G | 0.974 | 0.772 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-T.; Chang, C.-F.; Chen, L.-C.; Huang, C.-Y.; Yu, C.-C.; Lin, V.C.-H.; Lu, T.-L.; Huang, S.-P.; Bao, B.-Y. Interplay Between Type 2 Diabetes Susceptibility and Prostate Cancer Progression: Functional Insights into C2CD4A. Diagnostics 2025, 15, 2767. https://doi.org/10.3390/diagnostics15212767
Chen Y-T, Chang C-F, Chen L-C, Huang C-Y, Yu C-C, Lin VC-H, Lu T-L, Huang S-P, Bao B-Y. Interplay Between Type 2 Diabetes Susceptibility and Prostate Cancer Progression: Functional Insights into C2CD4A. Diagnostics. 2025; 15(21):2767. https://doi.org/10.3390/diagnostics15212767
Chicago/Turabian StyleChen, Yei-Tsung, Chi-Fen Chang, Lih-Chyang Chen, Chao-Yuan Huang, Chia-Cheng Yu, Victor Chia-Hsiang Lin, Te-Ling Lu, Shu-Pin Huang, and Bo-Ying Bao. 2025. "Interplay Between Type 2 Diabetes Susceptibility and Prostate Cancer Progression: Functional Insights into C2CD4A" Diagnostics 15, no. 21: 2767. https://doi.org/10.3390/diagnostics15212767
APA StyleChen, Y.-T., Chang, C.-F., Chen, L.-C., Huang, C.-Y., Yu, C.-C., Lin, V. C.-H., Lu, T.-L., Huang, S.-P., & Bao, B.-Y. (2025). Interplay Between Type 2 Diabetes Susceptibility and Prostate Cancer Progression: Functional Insights into C2CD4A. Diagnostics, 15(21), 2767. https://doi.org/10.3390/diagnostics15212767






