Clinical and Genetic Characterization of Noonan Syndrome in a Romanian Cohort from Transylvania: Details on PTPN11 c.922A>G Variant and Phenotypic Spectrum
Abstract
1. Introduction
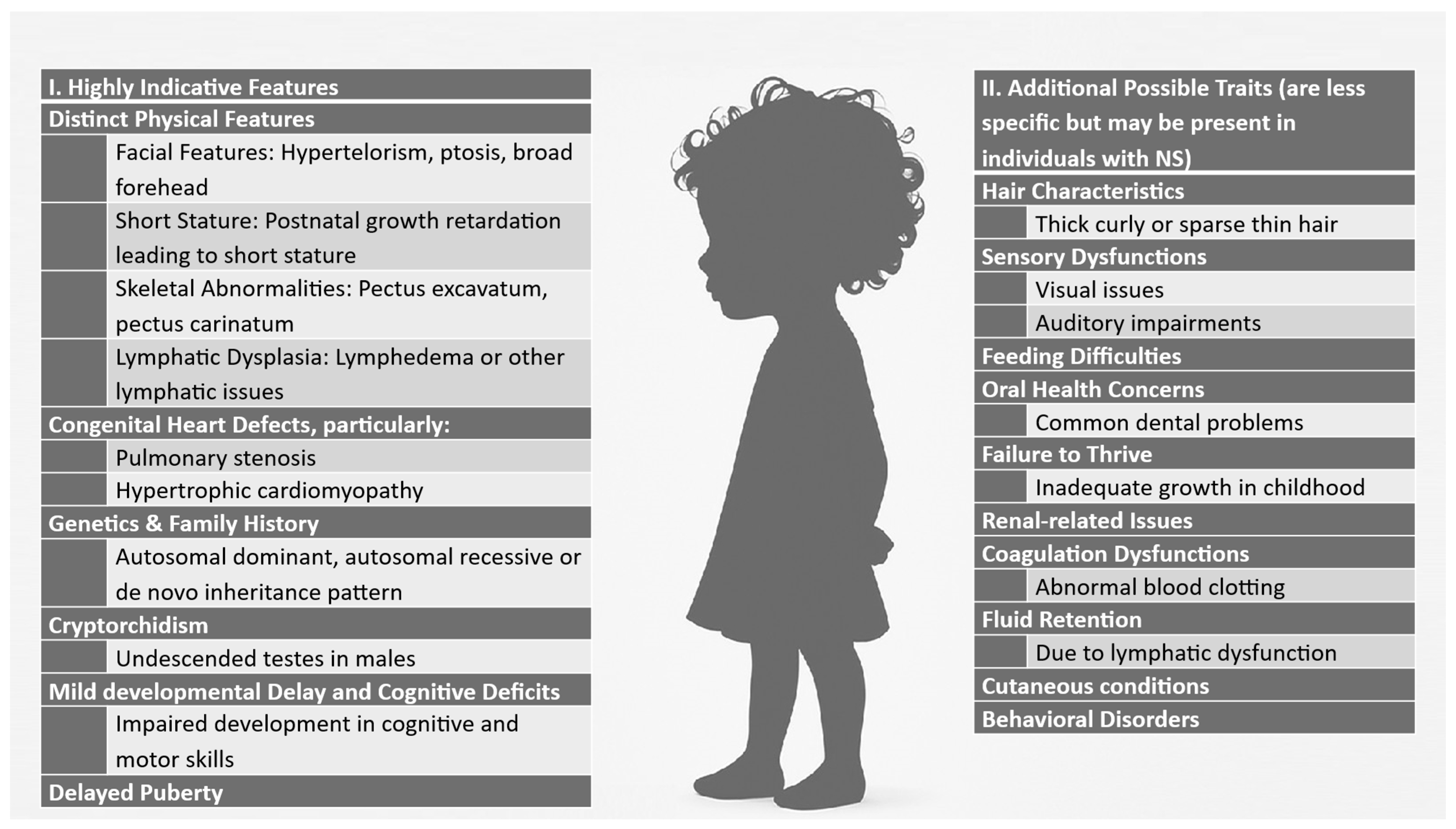
2. Materials and Methods
2.1. Patients
- A typical facial appearance together with at least one additional major criterion or two minor criteria, or
- Suggestive facial dysmorphology plus two major or three minor signs.
2.2. DNA Extraction and PCR-RFLP Analyses
3. Results
3.1. Clinical Characteristics
3.2. Genetic Results
3.3. Details for Case No. 1 Positive for PTPN11 c.922A>G
3.4. Details for Case No. 2 Positive for PTPN11 c.922A>G
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Autosomal dominant |
| AR | Autosomal recessive |
| ASD | Autism spectrum disorder |
| CGH | Comparative genomic hybridization |
| CMA/AVM | Capillary malformation/arteriovenous malformation |
| ECG | Electrocardiogram |
| HCM | Hypertrophic cardiomyopathy |
| HT | Hypertension |
| MIM/ OMIM | Mendelian Inheritance in Man/ Online Mendelian Inheritance in Man |
| MLPA | Multiplex Ligation-dependent Probe Amplification |
| NGS | Next-generation sequencing |
| NS | Noonan syndrome |
| PCR-RFLP | Polymerase Chain Reaction–Restriction Fragment Length Polymorphism |
| PTPN11 | Protein Tyrosine Phosphatase, Non-Receptor Type 11 |
| PVS | Pulmonary valve stenosis |
| RAS/MAPK | Rat Sarcoma/Mitogen-Activated Protein Kinase |
| SDS | Standard Deviation Score |
| WBS | Williams–Beuren syndrome |
References
- Tidyman, W.E.; Rauen, K.A. The RASopathies: Developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev. 2009, 19, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Rauen, K.A. The RASopathies. Annu. Rev. Genom. Hum. Genet. 2013, 14, 355–369. [Google Scholar] [CrossRef]
- Zenker, M.; Binder, G.; Koch, A.; Niemeyer, C.M.; Rauch, A. Noonan syndrome: Improving recognition and diagnosis. Arch. Dis. Child. 2022, 107, 1073–1078. [Google Scholar] [CrossRef]
- Yılmaz Uzman, C.; Gursoy, S.; Ozkan, B.; Vuran, G.; Ayyildiz Emecen, D.; Koprulu, O.; Bilen, M.M.; Hazan, F. Clinical features and molecular genetics of patients with RASopathies: Expanding the phenotype with rare genes and novel variants. Eur. J. Pediatr. 2024, 184, 108. [Google Scholar] [CrossRef] [PubMed]
- Musante, L.; Kehl, H.; Majewski, F.; Meinecke, P.; Schweiger, S.; Gillessen-Kaesbach, G.; Hinkel, G.K.; Tinschert, S.; Hoeltzenbein, M.; Reis, A.; et al. Spectrum of mutations in PTPN11 and genotype–phenotype correlation in 96 patients with Noonan syndrome and five patients with cardio-facio-cutaneous syndrome. Eur. J. Hum. Genet. 2003, 11, 201–206. [Google Scholar] [CrossRef]
- Johnston, J.J.; van der Smagt, J.J.; Rosenfeld, J.A.; Pagnamenta, A.T.; Alswaid, A.; Baker, E.H.; Blair, E.; Borck, G.; Brinkmann, J.; Craigen, W.; et al. Autosomal recessive Noonan syndrome associated with biallelic LZTR1 variants. Genet. Med. 2018, 20, 1175–1185. [Google Scholar] [CrossRef]
- Tartaglia, M.; Gelb, B.D.; Zenker, M. Noonan syndrome and clinically related disorders. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 161–179. [Google Scholar] [CrossRef]
- Sharland, M.; Burch, M.; McKenna, W.M.; Paton, M.A. A clinical study of Noonan syndrome. Arch. Dis. Child. 1992, 67, 178–183. [Google Scholar] [CrossRef]
- Baldo, F.; Fachin, A.; Da Re, B.; Rubinato, E.; Bobbo, M.; Barbi, E. New insights on Noonan syndrome’s clinical phenotype: A single center retrospective study. BMC Pediatr. 2022, 22, 734. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.A.; Allanson, J.E.; Dahlgren, J.; Gelb, B.D.; Hall, B.; Pierpont, M.E.; Roberts, A.E.; Robinson, W.; Takemoto, C.M.; Noonan, J.A. Noonan syndrome: Clinical features, diagnosis, and management guidelines. Pediatrics 2010, 126, 746–759. [Google Scholar] [CrossRef]
- Tafazoli, A.; Ahmadi, M.; Fekri, S.; Vahidi Mehrjardi, M.Y.; Poursadegh Zonouzi, A.; Saliminejad, K.; Kamali, K.; Kamali, M.; Taghdiri, M.; Najmabadi, H. Noonan syndrome—A new survey. Arch. Med. Sci. 2017, 13, 215–222. [Google Scholar] [CrossRef]
- Orlova, A.; Guseva, D.; Demina, N.; Polyakov, A.; Ryzhkova, O. Spectrum of Mutations in PTPN11 in Russian Cohort. Genes 2024, 15, 345. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Allanson, J.; Tartaglia, M.; Gelb, B. Noonan syndrome. Lancet 2013, 381, 333–342. [Google Scholar] [CrossRef]
- Dodi, G.; Marini, R.; Virdis, R.; Giordano, M.; De Angelis, C.; Catassi, C. Novel insights in Noonan syndrome. Pediatr. Dev. Pathol. 2020, 5, 1–6. [Google Scholar] [CrossRef]
- Van der Burgt, I.; Berends, E.; Lommen, E.; Van Beersum, S.; Hamel, B.; Mariman, E. Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am. J. Med. Genet. 1994, 53, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, M.; Mehler, E.L.; Goldberg, R.; Zampino, G.; Brunner, H.G.; Kremer, H.; van der Burgt, I.; Crosby, A.H.; Ion, A.; Jeffery, S.; et al. PTPN11 mutations in Noonan syndrome: Molecular spectrum, genotype–phenotype correlation, and phenotypic heterogeneity. Am. J. Hum. Genet. 2002, 70, 1555–1563. [Google Scholar] [CrossRef]
- Roberts, A.E.; Allanson, J.E.; Tartaglia, M.; Gelb, B.D. Noonan Syndrome. In GeneReviews® [Internet]; Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J., Gripp, K.W., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 2001; Revised in June 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1124/ (accessed on 26 October 2025).
- Narayanan, D.L.; Pandey, H.; Moirangthem, A.; Mandal, K.; Gupta, R.; Puri, R.D.; Patil, S.J.; Phadke, S.R. Hotspots in PTPN11 gene among Indian children with Noonan syndrome. Indian Pediatr. 2017, 54, 638–643. [Google Scholar] [CrossRef]
- Ilic, N.; Krasic, S.; Maric, N.; Gasic, V.; Krstic, J.; Cvetkovic, D.; Miljkovic, V.; Zec, B.; Maver, A.; Vukomanovic, V.; et al. Noonan syndrome: Relation of genotype to cardiovascular phenotype—A multi-center retrospective study. Genes 2024, 15, 1463. [Google Scholar] [CrossRef]
- Pierpont, M.E.; Digilio, M.C. Cardiovascular disease in Noonan syndrome. Curr. Opin. Pediatr. 2018, 30, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Athota, J.P.; Bhat, M.; Nampoothiri, S.; Gowrishankar, K.; Narayanachar, S.G.; Puttamallesh, V.; Jayaram, S.; Srivastava, A.; Girisha, K.M. Molecular and clinical studies in 107 Noonan syndrome affected individuals with PTPN11 mutations. BMC Med. Genet. 2020, 21, 50. [Google Scholar] [CrossRef]
- Rojnueangnit, K.; Xie, J.; Gomes, A.; Sharp, A.; Callens, T.; Chen, Y.; Liu, Y.; Cochran, M.; Abbott, M.A.; Atkin, J.; et al. High Incidence of Noonan Syndrome Features Including Short Stature and Pulmonic Stenosis in Patients Carrying NF1 Missense Mutations Affecting p.Arg1809: Genotype-Phenotype Correlation. Hum. Mutat. 2015, 36, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Peduto, C.; Zanobio, M.; Nigro, V.; Perrotta, S.; Piluso, G.; Santoro, C. Neurofibromatosis type 1: Pediatric aspects and review of genotype–phenotype correlations. Cancers 2023, 15, 1217. [Google Scholar] [CrossRef]
- Online Mendelian Inheritance in Man (OMIM). Available online: https://omim.org/ (accessed on 11 August 2025).
- Van Der Burgt, I.; Brunner, H. Genetic heterogeneity in Noonan syndrome: Evidence for an autosomal recessive form. Am. J. Med. Genet. 2000, 94, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Porras, A.R.; Summar, M.; Linguraru, M.G. Objective differential diagnosis of Noonan and Williams–Beuren syndromes in diverse populations using quantitative facial phenotyping. Mol. Genet. Genom. Med. 2021, 9, e1636. [Google Scholar] [CrossRef]
- Kehrer-Sawatzki, H.; Mautner, V.F.; Cooper, D.N. Emerging genotype–phenotype relationships in patients with large NF1 deletions. Hum. Genet. 2017, 136, 349–376. [Google Scholar] [CrossRef]
- Pacot, L.; Girish, M.; Knight, S.; Spurlock, G.; Varghese, V.; Ye, M.; Thomas, N.; Pasmant, E.; Upadhyaya, M. Correlation between large rearrangements and patient phenotypes in NF1 deletion syndrome: An update and review. BMC Med. Genom. 2024, 17, 73. [Google Scholar] [CrossRef]
- Perrino, F.; Licchelli, S.; Serra, G.; Piccini, G.; Caciolo, C.; Pasqualetti, P.; Cirillo, F.; Leoni, C.; Digilio, M.; Zampino, G.; et al. Psychopathological features in Noonan syndrome. Eur. J. Paediatr. Neurol. 2018, 22, 170–177. [Google Scholar] [CrossRef]
- Orphanet: An Online Rare Disease and Orphan Drug Data Base. Available online: https://www.orpha.net (accessed on 14 August 2025).
- Xie, B.; Fan, X.; Lei, Y.; Chen, R.; Wang, J.; Fu, C.; Yi, S.; Luo, J.; Zhang, S.; Yang, Q.; et al. A novel de novo microdeletion at 17q11.2 adjacent to NF1 gene associated with developmental delay, short stature, microcephaly and dysmorphic features. Mol. Cytogenet. 2016, 9, 64. [Google Scholar] [CrossRef]
- Bertola, D.R.; Pereira, A.C.; Kim, C.A.; Albano, L.M.J.; Takahashi, T.N.; Passos-Bueno, M.R.; Krieger, J.E. PTPN11 gene analysis in 74 Brazilian patients with Noonan syndrome or Noonan-like phenotype. Genet. Test. 2006, 10, 186–191. [Google Scholar] [CrossRef]
- Sznajer, Y.; Keren, B.; Baumann, C.; Pereira, S.; Alberti, C.; Elion, J.; Cavé, H.; Verloes, A. The spectrum of cardiac anomalies in Noonan syndrome as a result of mutations in the PTPN11 gene. Pediatrics 2007, 119, e1325–e1331. [Google Scholar] [CrossRef] [PubMed]
- Brasil, A.S.; Pereira, A.C.; Wanderley, L.T.; Kim, C.A.; Malaquias, A.C.; Jorge, A.A.L.; Bertola, D.R.; Krieger, J.E. PTPN11 and KRAS gene analysis in patients with Noonan and Noonan-like syndromes. Genet. Test. Mol. Biomark. 2010, 14, 425–432. [Google Scholar] [CrossRef]
- Zepeda-Olmos, P.M.; Esparza-García, E.; Robles-Espinoza, K.; González-García, J.R.; Rodríguez Gutiérrez, P.G.; Magaña-Torres, M.T. Variants of the PTPN11 gene in Mexican patients with Noonan syndrome. Genes 2024, 15, 1379. [Google Scholar] [CrossRef]
- Yoshida, R.; Hasegawa, T.; Hasegawa, Y.; Nagai, T.; Kinoshita, E.; Tanaka, Y.; Kameyama, J.; Nagashima, T.; Hasegawa, S.; Ohashi, H.; et al. Protein-tyrosine phosphatase, nonreceptor type 11 mutation analysis and clinical assessment in 45 patients with Noonan syndrome. J. Clin. Endocrinol. Metab. 2004, 89, 3359–3364. [Google Scholar] [CrossRef]
- Ferreira, L.V.; Souza, S.A.; Montenegro, L.R.; Arnhold, I.J.; Pasqualini, T.; Heinrich, J.J.; Keselman, A.C.; Mendonça, B.B.; Jorge, A.A. Variabilidade do fenótipo de pacientes com síndrome de Noonan com e sem mutações no gene PTPN11. Arq. Bras. Endocrinol. Metabol. 2007, 51, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Kosaki, K.; Suzuki, T.; Muroya, K.; Hasegawa, T.; Sato, S.; Matsuo, N.; Kosaki, R.; Nagai, T.; Hasegawa, Y.; Ogata, T. PTPN11 (protein-tyrosine phosphatase, nonreceptor-type 11) mutations in seven Japanese patients with Noonan syndrome. J. Clin. Endocrinol. Metab. 2002, 87, 3529–3533. [Google Scholar] [CrossRef]
- Ouboukss, F.; Adadi, N.; Amasdl, S.; Smaili, W.; Laarabi, F.Z.; Lyahyai, J.; Sefiani, A.; Ratbi, I. High frequency of hotspot mutation in PTPN11 gene among Moroccan patients with Noonan syndrome. J. Appl. Genet. 2024, 65, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Allanson, J. Noonan syndrome. J. Med. Genet. 1987, 24, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, T. Noonan syndrome: Introduction and basic clinical features. Horm. Res. Paediatr. 2009, 72, 3–7. [Google Scholar] [CrossRef]
- Mastromoro, G.; De Luca, A.; Marchionni, E.; Spagnuolo, A.; Ventriglia, F.; Manganaro, L.; Pizzuti, A. External hydrocephalus as a prenatal feature of Noonan syndrome. Ann. Hum. Genet. 2021, 85, 249–252. [Google Scholar] [CrossRef]
- Ejarque, I.; Millán-Salvador, J.M.; Oltra, S.; Pesudo-Martínez, J.V.; Beneyto, M.; Pérez-Aytés, A. Malformación de Arnold–Chiari en el síndrome de Noonan y otros síndromes de la vía RAS/MAPK [Arnold–Chiari malformation in Noonan syndrome and other syndromes of the RAS/MAPK pathway]. Rev. Neurol. 2015, 60, 408–412. [Google Scholar] [CrossRef]
- Făgărășanu, A.; Al Hussein, H.; Ghiragosian Rusu, S.E. RAF-1 mutation associated with a risk for ventricular arrhythmias in a child with Noonan syndrome and cardiovascular pathology. J. Crit. Care Med. 2022, 8, 126–130. [Google Scholar] [CrossRef]
- Harpa, M.M.; Anitei, E.-D.; Ghiragosian, C.; Calburean, P.; Opris, D.R.; Banceu, M.C.; Arbanasi, E.M.; Suciu, H.; Al Hussein, H. Endoscopic mitral surgery in Noonan syndrome—Case report and considerations. J. Clin. Med. 2025, 14, 583. [Google Scholar] [CrossRef] [PubMed]
- Nicola, A.A.; Greere, M.; Birceanu, A.L.; Balanescu, D.V.; Duna, M.; Predeteanu, D. A rare association: Ankylosing spondylitis and a genetic disease. Rom. J. Rheumatol. 2019, 28, 111–116. [Google Scholar] [CrossRef]
- Radu, D.-N.; Șuteu, C.; Popa, M.; Bănescu, C. Neurofibromatosis 1–Noonan syndrome associated with pulmonary stenosis and hypertrophic cardiomyopathy. Acta Med. Marisiensis 2020, 66, 128–131. [Google Scholar] [CrossRef]
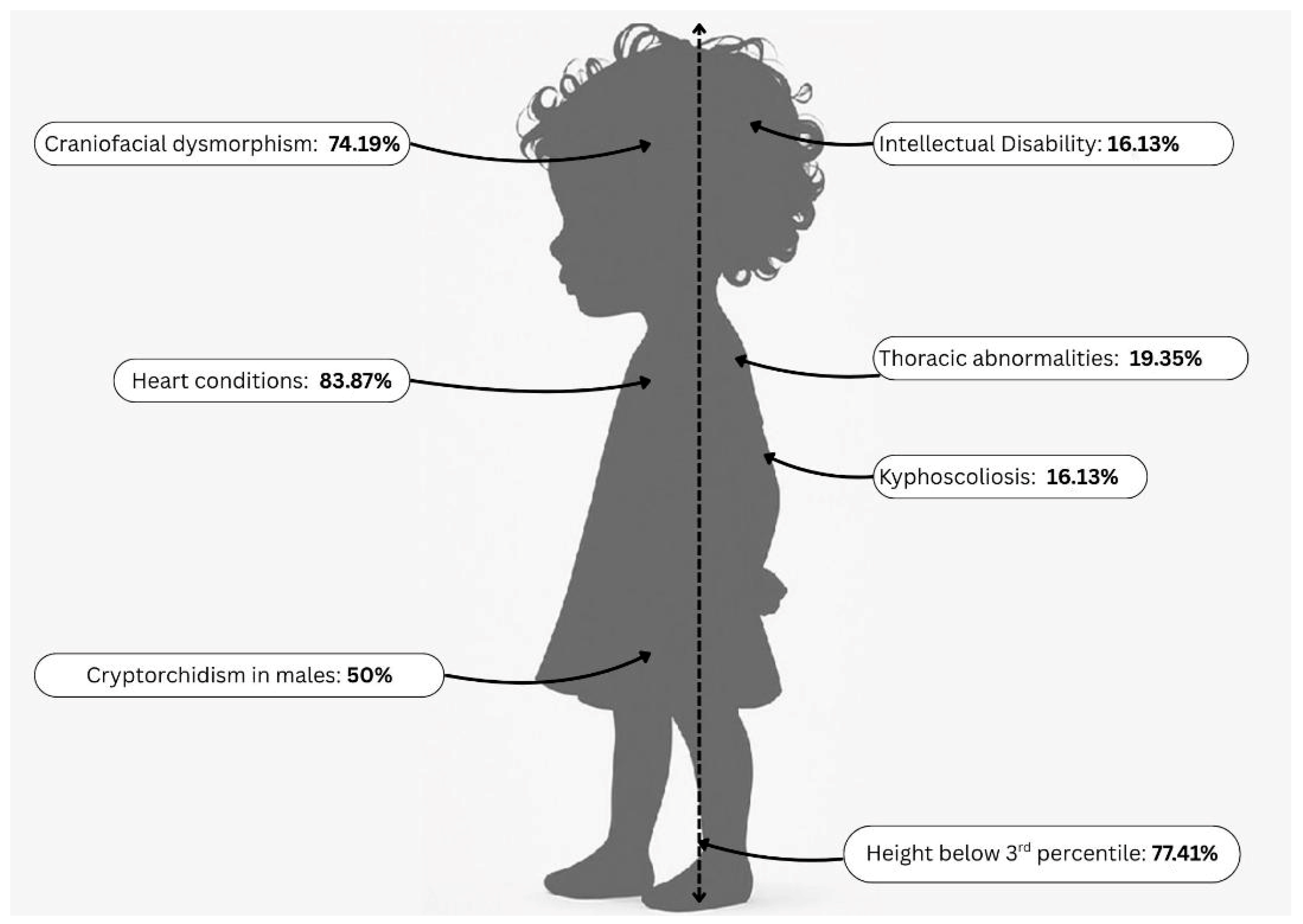
| Key Clinical Features in the Study Cohort | Number of Patients, n (%) |
|---|---|
| Height below third percentile | 24 (77.41%) |
| Height between percentile 3 and 10 | 1 (3.22%) |
| Heart conditions | 26 (83.87%) |
| Craniofacial dysmorphism | 23 (74.19%) |
| Triangular face | 5 (16.66%) |
| Hypertelorism | 10 (33.33%) |
| Palpebral ptosis | 2 (6.63%) |
| Downslanting palpebral fissures | 14 (46.66%) |
| Low set ears | 13 (43.33%) |
| Pterygium colli | 3 (9.67%) |
| Thoracic abnormalities | 6 (19.35%) |
| Broad thorax | 1 (3.33%) |
| Pectus carinatum | 2 (6.45%) |
| Pectus excavatum | 3 (9.67%) |
| Wide-spaced nipples | 5 (16.13%) |
| Kyphoscoliosis | 5 (16.13%) |
| Intellectual disability | 5 (16.13%) |
| Cryptorchidism in males | 11 (50%) |
| Congenital Heart Defect | Number of Patients, n (%) |
|---|---|
| PVS | 16 (51.61%) |
| PVS and HCM * | 3 (9.68%) |
| Pulmonary valve dysplasia | 1 (3.23%) |
| Left HCM | 1 (3.23%) |
| Atrial sept defect operated | 1 (3.23%) |
| Atrial septal aneurysm with left–right shunt | 1 (3.23%) |
| Small muscular sept defect | 1 (3.23%) |
| Patent foramen ovale | 2 (6.45%) |
| No heart condition | 5 (16.13%) |
| Features | Noonan Syndrome NF1 | Williams–Beuren Syndrome | Neurofibromatosis Type 1 Microdeletion Syndrome | References |
|---|---|---|---|---|
| Genetic Causes | Variants in BRAF, KRAS, MAP2K1, MRAS, NRAS, PTPN11, RAF1, RASA2, RIT1, RRAS2, SOS1, SOS2, LZTR1 (RAS/MAPK pathway); several additional genes, each linked to an NS-like phenotype, were identified in fewer than ten individuals | Hemizygous deletion of 1.5 to 1.8 Mb, approx. 28 genes from 7q11.23; is a contiguous gene deletion syndrome | Deletion of 17q11.2 (including NF1 gene) | [3,17,22,23,24] |
| Alternative name |
|
|
| [24] |
| Gene/Locus MIM number | *176876 | *130160 | *613113 | [24] |
| Phenotype MIM number | #163950 | #194050 | #601321 | [24] |
| Inheritance pattern | Mostly de novo or AD variants; AR forms of NS include NS2, caused by variants in the LZTR1 gene, and NS14, caused by variants in the SPRED2 gene | Mostly de novo, AD | Mostly de novo but can be inherited AD | [6,25] |
| Facial features | Triangular face, hypertelorism, ptosis, low-set ears: similar facial phenotypes to WBS modulated by ethnic background | “Elfin-like” face, broad forehead, full cheeks, wide mouth | Coarse facial features, hypertelorism, down-slanting palpebral fissures | [22,23,26,27] |
| Growth and stature | Short stature, postnatal growth retardation | Short stature, failure to thrive in infancy | Short stature, variable growth delay | [21,27] |
| Cardiac defects | In up to 90% of patients, PVS, HCM | Supravalvular aortic stenosis, pulmonary artery stenosis | Pulmonic stenosis, other congenital heart defects | [3,28] |
| Cognitive and developmental delay | Mild-to-moderate intellectual disability, learning difficulties | Intellectual disability, friendly/social personality | Developmental delay, speech delay, learning disabilities | [3,21,29] |
| Skeletal abnormalities | Pectus excavatum, pectus carinatum, scoliosis | Joint laxity, skeletal abnormalities | Pectus abnormalities, scoliosis | [3,21] |
| Skin manifestations | Generally normal | Soft skin, premature aging of the skin | Café-au-lait spots, axillary/inguinal freckling, multiple neurofibromas | [3] |
| Behavioral features | Varied; some social difficulties, verbal communication challenges | Overly friendly, anxiety, attention deficits, fear of loud noises | Attention deficits, possible ASD-like traits | [3,10] |
| Renal and urinary abnormalities | Sometimes present, cryptorchidism in males | Common, including renal artery stenosis | Genitourinary abnormalities, cryptorchidism in males; increased risk of HT | [3] |
| Cancer risk | Slightly increased risk of certain malignancies (e.g., leukemia, neuroblastoma) | No significant cancer predisposition | Increased risk of malignancies (due to NF1 gene deletion); e.g., plexiform neurofibromas, optic gliomas, malignant peripheral nerve sheath tumors | [3,10] |
| Sensory conditions vision issues, hearing changes | Strabismus, refractive errors, sensorineural hearing loss | Hyperacusis (sensitivity to sound), strabismus | Optic pathway gliomas, Lisch nodules (iris hamartomas), vision impairment | [3] |
| Distinctive features | Wide-spaced nipples, short/webbed neck, coagulation abnormalities | Outgoing personality, musical affinity, strong verbal skills | Café-au-lait macules, neurofibromas, and an increased risk of tumors | [3,10] |
| Diagnostic clues | Cardiac defects + short stature + facial dysmorphism | Supravalvular aortic stenosis + hypersociability + “elfin-like” face | Café-au-lait spots + neurofibromas | [3,26] |
| Prevalence | 1–5/10,000 | approx. 1/7500 | Not known; about 5% of NF1 cases are reported to have deletions of the entire NF1 gene | [30] |
| Study [Reference] | Total Number of Patients | Number of PTPN11 Variants | Percentage of PTPN11 Variants | Number of PTPN11 C.922A>G Variants | Percentage of PTPN11 C.922A>G Variants |
|---|---|---|---|---|---|
| Orlova et al., 2024 [12] | 456 | 107 | 23.46 | 23 | 5.04 |
| Sznajer et al., 2007 [33] | 272 | 104 | 38.24 | 19 | 18.2 |
| Tartaglia et al., 2002 [16] | 112 | 54 | 48.21 | 17 | 15.18 |
| Brasil et al., 2010 [34] | 95 | 42 | 44.21 | 11 | 11.58 |
| Zepeda-Olmos et al., 2024 [35] | 91 | 43 | 47.25 | 7 | 7.69 |
| Bertola et al., 2006 [32] | 50 | 21 | 42.00 | 0 | 0.00 |
| Yoshida et al., 2004 [36] | 45 | 18 | 40.00 | 2 | 4.44 |
| Ferreira et al., 2007 [37] | 33 | 16 | 48.48 | 5 | 15.15 |
| Kosaki et al., 2002 [38] | 21 | 7 | 33.33 | 1 | 4.76 |
| Athota et al., 2020 [21] | 363 | 107 | 29.47 | 12 | 3.30 |
| Ouboukss et al., 2024 [39] | 61 | 17 | 41.4% | 10 | 16.39 |
| Our study | 31 | Not known | Not known | 2 | 6.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazarie, F.V.; Miclea, D.; Șufană, C.; Botezatu, A.; Popp, R.A.; Pascanu, I.M.; Alkhzouz, C.; Bucerzan, S.; Lazăr, C.; Lazea, C.; et al. Clinical and Genetic Characterization of Noonan Syndrome in a Romanian Cohort from Transylvania: Details on PTPN11 c.922A>G Variant and Phenotypic Spectrum. Diagnostics 2025, 15, 2753. https://doi.org/10.3390/diagnostics15212753
Nazarie FV, Miclea D, Șufană C, Botezatu A, Popp RA, Pascanu IM, Alkhzouz C, Bucerzan S, Lazăr C, Lazea C, et al. Clinical and Genetic Characterization of Noonan Syndrome in a Romanian Cohort from Transylvania: Details on PTPN11 c.922A>G Variant and Phenotypic Spectrum. Diagnostics. 2025; 15(21):2753. https://doi.org/10.3390/diagnostics15212753
Chicago/Turabian StyleNazarie, Florina Victoria, Diana Miclea, Crina Șufană, Alina Botezatu, Radu Anghel Popp, Ionela Maria Pascanu, Camelia Alkhzouz, Simona Bucerzan, Călin Lazăr, Cecilia Lazea, and et al. 2025. "Clinical and Genetic Characterization of Noonan Syndrome in a Romanian Cohort from Transylvania: Details on PTPN11 c.922A>G Variant and Phenotypic Spectrum" Diagnostics 15, no. 21: 2753. https://doi.org/10.3390/diagnostics15212753
APA StyleNazarie, F. V., Miclea, D., Șufană, C., Botezatu, A., Popp, R. A., Pascanu, I. M., Alkhzouz, C., Bucerzan, S., Lazăr, C., Lazea, C., & Vulturar, R. (2025). Clinical and Genetic Characterization of Noonan Syndrome in a Romanian Cohort from Transylvania: Details on PTPN11 c.922A>G Variant and Phenotypic Spectrum. Diagnostics, 15(21), 2753. https://doi.org/10.3390/diagnostics15212753








