Machine Learning-Based Prediction of Three-Year Heart Failure and Mortality After Premature Ventricular Contraction Ablation
Abstract
1. Introduction
1.1. Predicting Mortality
1.2. Predicting Heart Failure
1.3. Contributions and Challenges of Machine Learning in Clinical Medicine
1.4. Features
1.5. Research Gap and Contribution
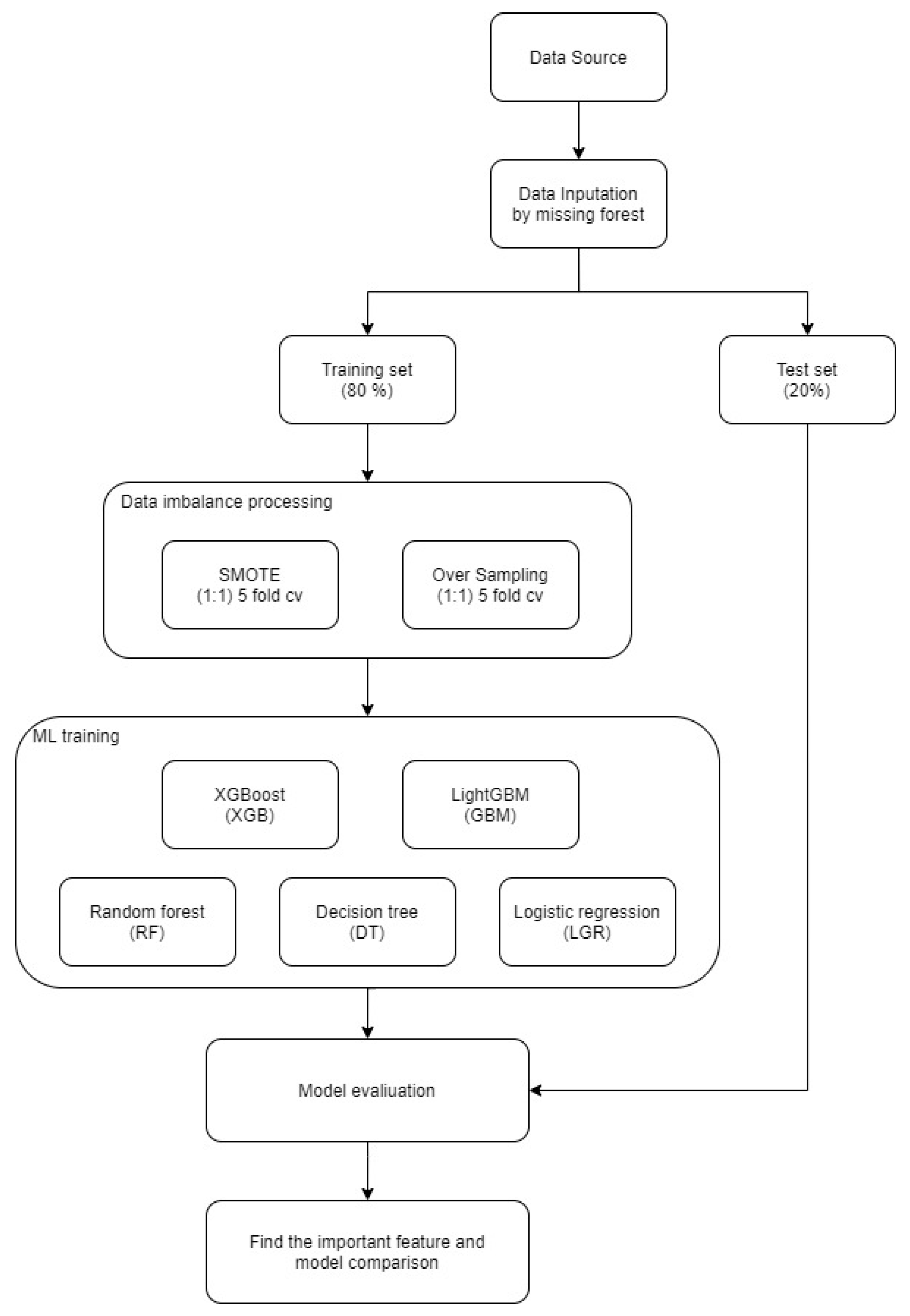
2. Materials and Methods
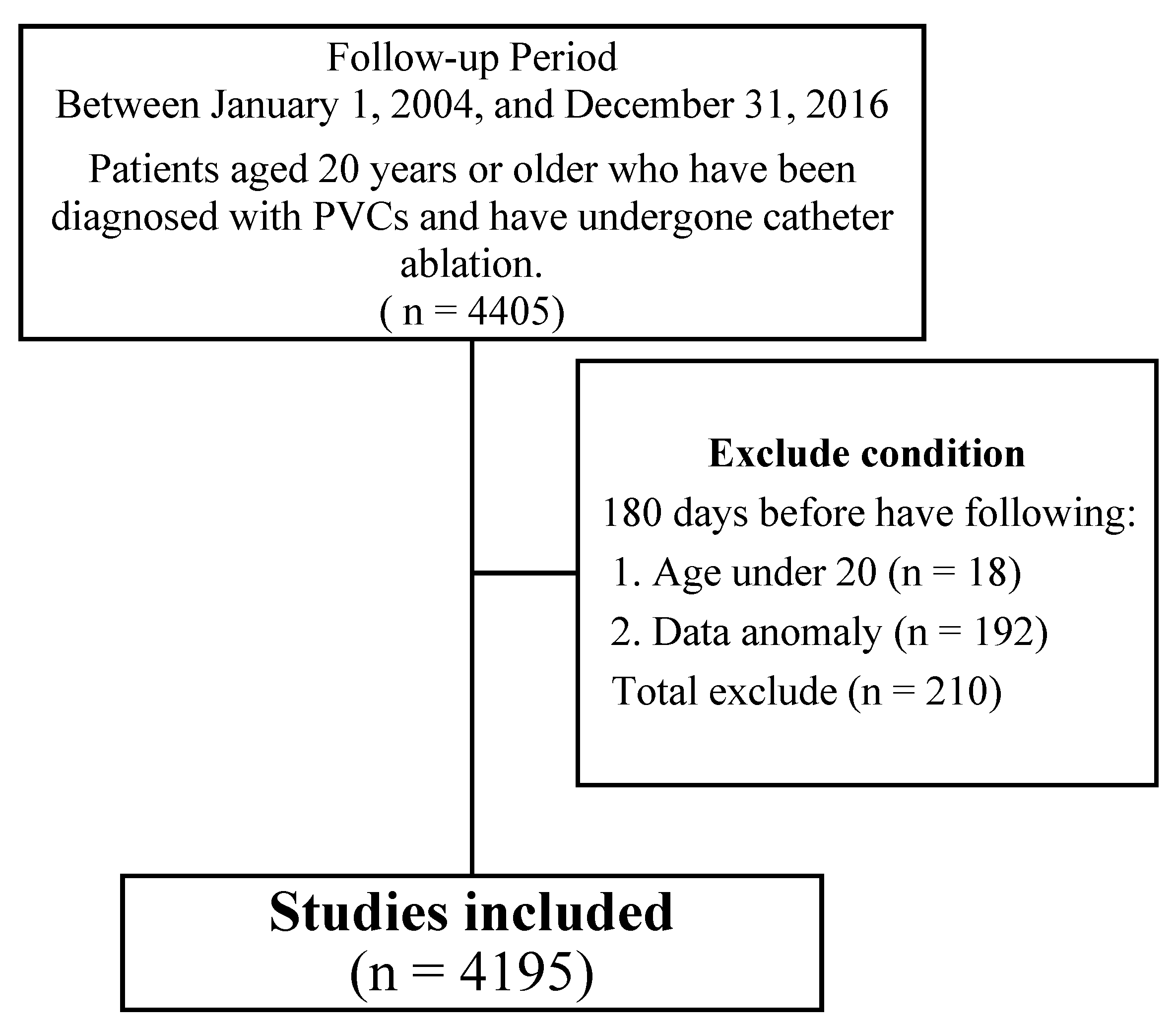
2.1. Collection of Demographics and Medical History
2.2. Study Design and Participants
2.3. Baseline Statistic Analysis
2.4. Method
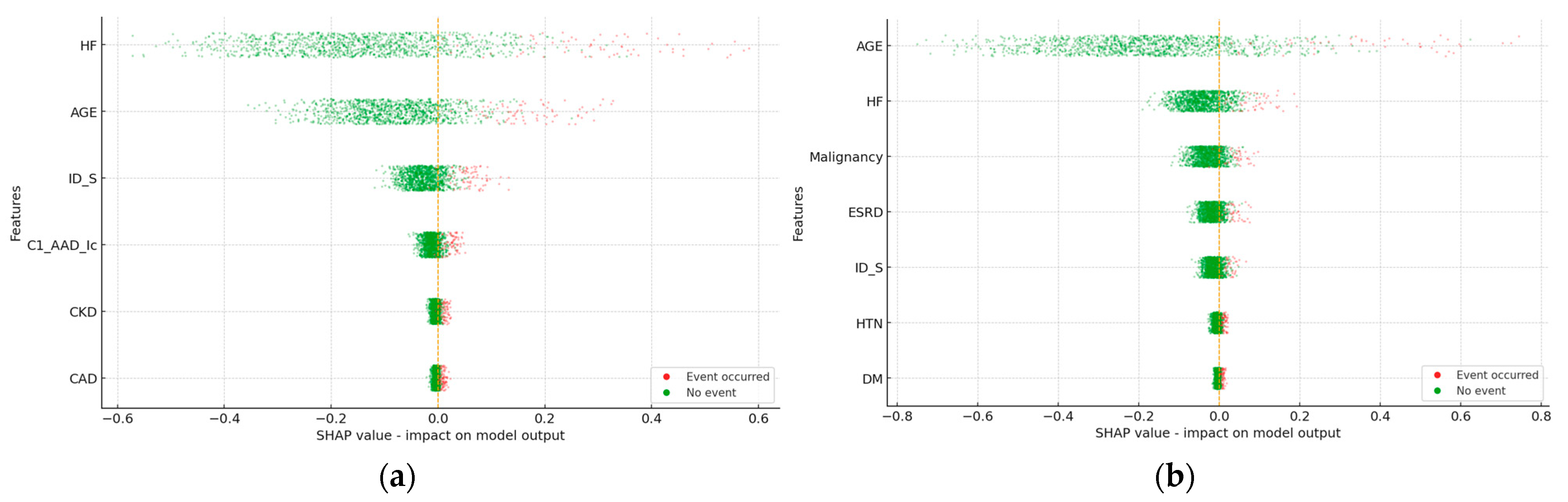
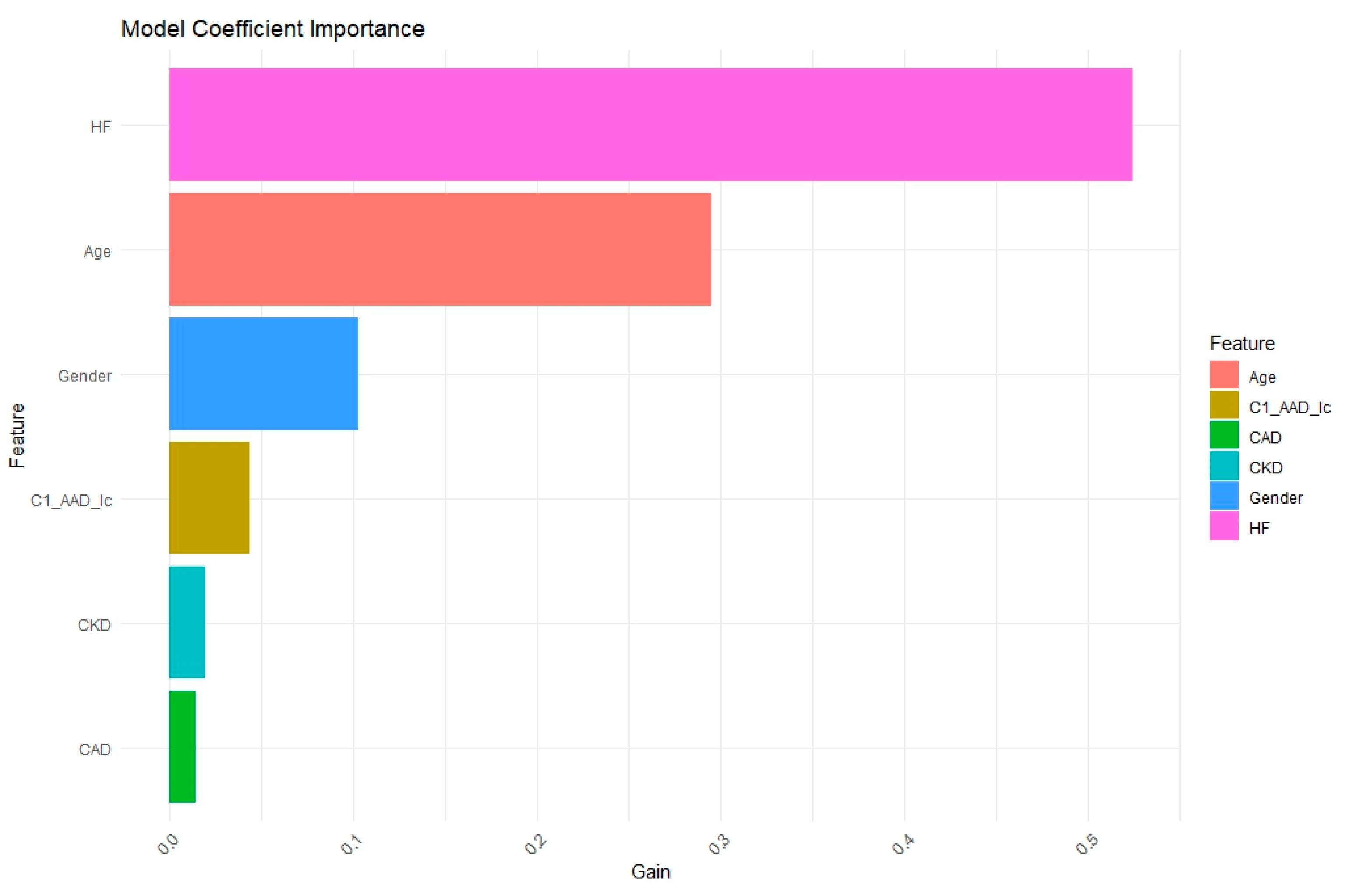
3. Results
3.1. Heart Failure in 3 Years
3.2. Mortality in 3 Years
4. Discussion
4.1. Data Imbalance and Discrimination Beyond ROC
4.2. Model Selection and Evaluation
4.3. Clinical Implications and Deployment
4.4. Limitations, Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PVC | Premature Ventricular Contraction |
| ML | Machine learning |
| HF | Heart failure |
| VT | ventricular tachycardia |
| ACS | Acute Coronary Syndrome |
| CAD | Coronary artery disease |
| PVD | Peripheral vascular disease |
| HTN | hypertension |
| DM | Diabetes mellitus |
| COPD | Chronic Obstruction Pulmonary Disease |
| CKD | Chronic kidney disease |
| ESRD | end stage renal disease |
| CVA | cerebrovascular accident |
| LD | moderate or severe liver disease |
| B_blocker | beta-blockers |
| C1_AAD_Ia & Ib | Class I antiarrhythmic drugs Ia, Ib |
| C1_AAD_Ic | Class I antiarrhythmic drugs Ic |
| C3_AAD | Class III antiarrhythmic drugs |
| CCB | Calcium channel blockers |
| F1 | F1 score |
| AUC | Area under curve |
| Logi | Logistic regression |
| Cart | Decision tree |
| RF | Random forest |
| ROSE | Random over-sampling |
| SMOTE | Synthetic Minority Oversampling Technique |
| XGB | Xgboost |
| SHAP | Shapley additive explanations |
Appendix A
| Hyperparameters | Setting |
|---|---|
| colsample_bytree | 0.8 |
| subsample | 0.8 |
| booster | gbtree |
| max_depth | 10 |
| eta | 0.01 |
| eval_metric | auc |
| eval_metric | error |
| objective | binary: logistic |
| gamma | 0.01 |
| lambda | 2 |
| min_child_weight | 1 |
| Feature | Racy |
|---|---|
| num_leaves | 3 |
| nthread | 1 |
| metric | auc |
| metric | binary_error |
| objective | binary |
| min_data | 1 |
| learning_rate | 0.1 |
Appendix B
| Logi _ROSE | Cart _ROSE | RF _ROSE | XGB _ROSE | LightGBM _ROSE | Logi_ SMOTE | Cart_ SMOTE | RF_ SMOTE | XGB_ SMOTE | LightGBM _SMOTE | |
|---|---|---|---|---|---|---|---|---|---|---|
| Logi_ ROSE | 1.000 | 0.001 | 0.021 | 0.036 | 0.796 | 0.615 | 0.000 | 0.045 | 0.000 | 0.316 |
| Cart_ ROSE | 0.001 | 1.000 | 0.003 | 0.781 | 0.001 | 0.001 | 0.193 | 0.002 | 0.256 | 0.001 |
| RF_ ROSE | 0.021 | 0.003 | 1.000 | 0.085 | 0.024 | 0.033 | 0.000 | 0.381 | 0.001 | 0.049 |
| XGB_ ROSE | 0.036 | 0.781 | 0.085 | 1.000 | 0.039 | 0.041 | 0.831 | 0.071 | 0.866 | 0.051 |
| LightGBM _ROSE | 0.796 | 0.001 | 0.024 | 0.039 | 1.000 | 0.793 | 0.000 | 0.053 | 0.000 | 0.415 |
| Logi_ SMOTE | 0.615 | 0.001 | 0.033 | 0.041 | 0.793 | 1.000 | 0.000 | 0.078 | 0.000 | 0.591 |
| Cart_ SMOTE | 0.000 | 0.193 | 0.000 | 0.831 | 0.000 | 0.000 | 1.000 | 0.000 | 0.850 | 0.000 |
| RF_ SMOTE | 0.045 | 0.002 | 0.381 | 0.071 | 0.053 | 0.078 | 0.000 | 1.000 | 0.001 | 0.122 |
| XGB_ SMOTE | 0.000 | 0.256 | 0.001 | 0.866 | 0.000 | 0.000 | 0.850 | 0.001 | 1.000 | 0.001 |
| LightGBM _SMOTE | 0.316 | 0.001 | 0.049 | 0.051 | 0.415 | 0.591 | 0.000 | 0.122 | 0.001 | 1.000 |
| Logi _ROSE | Cart _ROSE | RF _ROSE | XGB _ROSE | LightGBM _ROSE | Logi_ SMOTE | Cart_ SMOTE | RF_ SMOTE | XGB_ SMOTE | LightGBM _SMOTE | |
|---|---|---|---|---|---|---|---|---|---|---|
| Logi_ ROSE | 1 | 0.001 | 0.021 | 0.036 | 0.796 | 0.615 | 0.000 | 0.045 | 0.000 | 0.316 |
| Cart_ ROSE | 0.001 | 1 | 0.003 | 0.781 | 0.001 | 0.001 | 0.193 | 0.002 | 0.256 | 0.001 |
| RF_ ROSE | 0.021 | 0.003 | 1 | 0.085 | 0.024 | 0.033 | 0.000 | 0.381 | 0.001 | 0.049 |
| XGB_ ROSE | 0.036 | 0.781 | 0.085 | 1 | 0.039 | 0.041 | 0.831 | 0.071 | 0.866 | 0.051 |
| LightGBM _ROSE | 0.796 | 0.001 | 0.024 | 0.039 | 1 | 0.793 | 0.000 | 0.053 | 0.000 | 0.415 |
| Logi_ SMOTE | 0.615 | 0.001 | 0.033 | 0.041 | 0.793 | 1 | 0.000 | 0.078 | 0.000 | 0.591 |
| Cart_ SMOTE | 0.000 | 0.193 | 0.000 | 0.831 | 0.000 | 0.000 | 1 | 0.000 | 0.850 | 0.000 |
| RF_ SMOTE | 0.045 | 0.002 | 0.381 | 0.071 | 0.053 | 0.078 | 0.000 | 1 | 0.001 | 0.122 |
| XGB_ SMOTE | 0.000 | 0.256 | 0.001 | 0.866 | 0.000 | 0.000 | 0.850 | 0.001 | 1 | 0.001 |
| LightGBM _SMOTE | 0.316 | 0.001 | 0.049 | 0.051 | 0.415 | 0.591 | 0.000 | 0.122 | 0.001 | 1 |
Appendix C

References
- Li, R.; Shen, L.; Ma, W.; Li, L.; Yan, B.; Wei, Y.; Wang, Y.; Pan, C.; Yuan, J. Machine learning-based risk models for procedural complications of radiofrequency ablation for atrial fibrillation. BMC Med. Inform. Decis. Mak. 2023, 23, 257. [Google Scholar] [CrossRef] [PubMed]
- Nasab, E.M.; Sadeghian, S.; Farahani, A.V.; Sharif, A.Y.; Kabir, F.M.; Karvane, H.B.; Zahedi, A.; Bozorgi, A. Determining the recurrence rate of premature ventricular complexes and idiopathic ventricular tachycardia after radiofrequency catheter ablation with the help of designing a machine-learning model. Regen. Ther. 2024, 27, 32–38. [Google Scholar] [CrossRef]
- Mueller-Leisse, J.; Syrbius, G.; Hillmann, H.; Eiringhaus, J.; Hohmann, S.; Zormpas, C.; Karfoul, N.; Duncker, D.; Veltmann, C. Prediction of ablation success of idiopathic premature ventricular contractions with an inferior axis using the twelve-lead electrocardiogram. Europace 2023, 25, euad122.706. [Google Scholar] [CrossRef]
- Khalaji, A.; Behnoush, A.H.; Jameie, M.; Sharifi, A.; Sheikhy, A.; Fallahzadeh, A.; Sadeghian, S.; Pashang, M.; Bagheri, J.; Ahmadi Tafti, S.H. Machine learning algorithms for predicting mortality after coronary artery bypass grafting. Front. Cardiovasc. Med. 2022, 9, 977747. [Google Scholar] [CrossRef]
- Metsker, O.; Sikorsky, S.; Yakovlev, A.; Kovalchuk, S. Dynamic mortality prediction using machine learning techniques for acute cardiovascular cases. Procedia Comput. Sci. 2018, 136, 351–358. [Google Scholar] [CrossRef]
- Soffer, S.; Klang, E.; Barash, Y.; Grossman, E.; Zimlichman, E. Predicting in-hospital mortality at admission to the medical ward: A big-data machine learning model. Am. J. Med. 2021, 134, 227–234.e4. [Google Scholar] [CrossRef]
- Brajer, N.; Cozzi, B.; Gao, M.; Nichols, M.; Revoir, M.; Balu, S.; Futoma, J.; Bae, J.; Setji, N.; Hernandez, A. Prospective and External Evaluation of a Machine Learning Model to Predict In-Hospital Mortality. medRxiv 2019. medRxiv:19000133. [Google Scholar] [CrossRef]
- Weng, S.F.; Vaz, L.; Qureshi, N.; Kai, J. Prediction of premature all-cause mortality: A prospective general population cohort study comparing machine-learning and standard epidemiological approaches. PLoS ONE 2019, 14, e0214365. [Google Scholar] [CrossRef] [PubMed]
- Bjerre, D.S. Tree-based machine learning methods for modeling and forecasting mortality. ASTIN Bull. J. IAA 2022, 52, 765–787. [Google Scholar] [CrossRef]
- Cho, S.M.; Austin, P.C.; Ross, H.J.; Abdel-Qadir, H.; Chicco, D.; Tomlinson, G.; Taheri, C.; Foroutan, F.; Lawler, P.R.; Billia, F. Machine learning compared with conventional statistical models for predicting myocardial infarction readmission and mortality: A systematic review. Can. J. Cardiol. 2021, 37, 1207–1214. [Google Scholar] [CrossRef]
- Krittanawong, C.; Virk, H.U.H.; Kumar, A.; Aydar, M.; Wang, Z.; Stewart, M.P.; Halperin, J.L. Machine learning and deep learning to predict mortality in patients with spontaneous coronary artery dissection. Sci. Rep. 2021, 11, 8992. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Z.; Ren, Y.; Nie, H.; Lei, Y.; Qiu, H.; Xu, Z.; Pu, X. Machine learning based early mortality prediction in the emergency department. Int. J. Med. Inform. 2021, 155, 104570. [Google Scholar] [CrossRef]
- Mansur Huang, N.S.; Ibrahim, Z.; Mat Diah, N. Machine learning techniques for early heart failure prediction. Malays. J. Comput. (MJoC) 2021, 6, 872–884. [Google Scholar] [CrossRef]
- Saju, B.; Asha, V.; Prasad, A.; Kumar, H.; Rakesh, V.; Nirmala, A. Heart Disease Prediction Model using Machine Learning. In Proceedings of the 2022 International Conference on Automation, Computing and Renewable Systems (ICACRS), Pudukkottai, India, 13–15 December 2022; pp. 723–729. [Google Scholar]
- Zhuang, X.; Sun, X.; Zhong, X.; Zhou, H.; Zhang, S.; Liao, X. Deep phenotyping and prediction of long-term heart failure by machine learning. J. Am. Coll. Cardiol. 2019, 73, 690. [Google Scholar] [CrossRef]
- Garate Escamilla, A.K.; Hajjam El Hassani, A.; Andres, E. A comparison of machine learning techniques to predict the risk of heart failure. In Machine Learning Paradigms: Applications of Learning and Analytics in Intelligent Systems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 9–26. [Google Scholar]
- Nagavelli, U.; Samanta, D.; Chakraborty, P. Machine Learning Technology-Based Heart Disease Detection Models. J. Healthc. Eng. 2022, 2022, 7351061. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshinee, S.; Panda, M. Improving prediction of chronic heart failure using smote and machine learning. In Proceedings of the 2022 Second International Conference on Computer Science, Engineering and Applications (ICCSEA), Gunupur, India, 8 September 2022; pp. 1–6. [Google Scholar]
- Mohan, S.; Thirumalai, C.; Srivastava, G. Effective heart disease prediction using hybrid machine learning techniques. IEEE Access 2019, 7, 81542–81554. [Google Scholar] [CrossRef]
- Shinde, Y.; Kenchappago, A.; Patil, S.; Panchwatkar, Y.; Mishra, S. Hybrid Approach for Predicting Heart Failure using Machine Learning. In Proceedings of the 2021 2nd Global Conference for Advancement in Technology (GCAT), Bangalore, India, 1–3 October 2021; pp. 1–8. [Google Scholar]
- Koshiga, N.; Borugadda, P.; Shaprapawad, S. Prediction of Heart Disease Based on Machine Learning Algorithms. In Proceedings of the 2023 International Conference on Inventive Computation Technologies (ICICT), Lalitpur, Nepal, 26–28 April 2023; pp. 713–720. [Google Scholar]
- Shamout, F.; Zhu, T.; Clifton, D.A. Machine learning for clinical outcome prediction. IEEE Rev. Biomed. Eng. 2020, 14, 116–126. [Google Scholar] [CrossRef]
- Gui, C.; Chan, V. Machine learning in medicine. Univ. West. Ont. Med. J. 2017, 86, 76–78. [Google Scholar] [CrossRef]
- Cohen, J.P.; Cao, T.; Viviano, J.D.; Huang, C.-W.; Fralick, M.; Ghassemi, M.; Mamdani, M.; Greiner, R.; Bengio, Y. Problems in the deployment of machine-learned models in health care. CMAJ 2021, 193, E1391–E1394. [Google Scholar] [CrossRef]
- Chen, P.-H.C.; Liu, Y.; Peng, L. How to develop machine learning models for healthcare. Nat. Mater. 2019, 18, 410–414. [Google Scholar] [CrossRef]
- Weng, W.-H. Machine learning for clinical predictive analytics. In Leveraging Data Science for Global Health; Springer: Berlin/Heidelberg, Germany, 2020; pp. 199–217. [Google Scholar]
- Crown, W.H. Potential application of machine learning in health outcomes research and some statistical cautions. Value Health 2015, 18, 137–140. [Google Scholar] [CrossRef]
- Orphanidou, C.; Wong, D. Machine learning models for multidimensional clinical data. In Handbook of Large-Scale Distributed Computing in Smart Healthcare; Springer: Berlin/Heidelberg, Germany, 2017; pp. 177–216. [Google Scholar]
- Sun, H.; Depraetere, K.; Meesseman, L.; Cabanillas Silva, P.; Szymanowsky, R.; Fliegenschmidt, J.; Hulde, N.; von Dossow, V.; Vanbiervliet, M.; De Baerdemaeker, J. Machine learning–based prediction models for different clinical risks in different hospitals: Evaluation of live performance. J. Med. Internet Res. 2022, 24, e34295. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Singh, H.; Verma, K. Using Machine Learning to Predict Patient Health Outcomes. In Proceedings of the 2023 14th International Conference on Computing Communication and Networking Technologies (ICCCNT), Delhi, India, 6–8 July 2023; pp. 1–7. [Google Scholar]
- Arora, A.; Basu, N. Machine Learning in Modern Healthcare. Int. J. Adv. Med. Sci. Technol. 2023, 3, 12–18. [Google Scholar] [CrossRef]
- Niu, X.N.; Wen, H.; Sun, N.; Zhao, R.; Wang, T.; Li, Y. Exploring risk factors of short-term readmission in heart failure patients: A cohort study. Front. Endocrinol. 2022, 13, 1024759. [Google Scholar] [CrossRef]
- Kamiya, K.; Sato, Y.; Takahashi, T.; Tsuchihashi-Makaya, M.; Kotooka, N.; Ikegame, T.; Takura, T.; Yamamoto, T.; Nagayama, M.; Goto, Y. Multidisciplinary cardiac rehabilitation and long-term prognosis in patients with heart failure. Circ. Heart Fail. 2020, 13, e006798. [Google Scholar] [CrossRef]
- Roger, V.L. Epidemiology of heart failure: A contemporary perspective. Circ. Res. 2021, 128, 1421–1434. [Google Scholar] [CrossRef]
- Mirkin, K.A.; Enomoto, L.M.; Caputo, G.M.; Hollenbeak, C.S. Risk factors for 30-day readmission in patients with congestive heart failure. Heart Lung 2017, 46, 357–362. [Google Scholar] [CrossRef]
- Bansal, N.; Zelnick, L.; Bhat, Z.; Dobre, M.; He, J.; Lash, J.; Jaar, B.; Mehta, R.; Raj, D.; Rincon-Choles, H. Burden and outcomes of heart failure hospitalizations in adults with chronic kidney disease. J. Am. Coll. Cardiol. 2019, 73, 2691–2700. [Google Scholar] [CrossRef]
- Dhuny, S.; Wu, H.H.; David, M.; Chinnadurai, R. Hypertrophic Cardiomyopathy and Chronic Kidney Disease: An Updated Review. Cardiogenetics 2024, 14, 26–37. [Google Scholar] [CrossRef]
- Wang, X.; Shapiro, J.I. Evolving concepts in the pathogenesis of uraemic cardiomyopathy. Nat. Rev. Nephrol. 2019, 15, 159–175. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Menardi, G.; Torelli, N. Training and assessing classification rules with imbalanced data. Data Min. Knowl. Discov. 2014, 28, 92–122. [Google Scholar] [CrossRef]
- Lunardon, N.; Menardi, G.; Torelli, N. ROSE: A package for binary imbalanced learning. R J. 2014, 6, 79–89. [Google Scholar] [CrossRef]
- Branco, P.; Ribeiro, R.P.; Torgo, L. UBL: An R package for utility-based learning. arXiv 2016, arXiv:1604.08079. [Google Scholar] [CrossRef]
- Weng, S.F.; Reps, J.; Kai, J.; Garibaldi, J.M.; Qureshi, N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS ONE 2017, 12, e0174944. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L.; Friedman, J.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Chapman and Hall/CRC: New York, NY, USA, 2017. [Google Scholar]
- Ghiasi, M.M.; Zendehboudi, S.; Mohsenipour, A.A. Decision tree-based diagnosis of coronary artery disease: CART model. Comput. Methods Programs Biomed. 2020, 192, 105400. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chuang, C.-W.; Wu, C.-K.; Wu, C.-H.; Shia, B.-C.; Chen, M. Machine Learning in Predicting Cardiac Events for ESRD Patients: A Framework for Clinical Decision Support. Diagnostics 2025, 15, 1063. [Google Scholar] [CrossRef]
- Benedetto, U.; Dimagli, A.; Sinha, S.; Cocomello, L.; Gibbison, B.; Caputo, M.; Gaunt, T.; Lyon, M.; Holmes, C.; Angelini, G.D. Machine learning improves mortality risk prediction after cardiac surgery: Systematic review and meta-analysis. J. Thorac. Cardiovasc. Surg. 2022, 163, 2075–2087.e9. [Google Scholar] [CrossRef]
- Farzipour, A.; Elmi, R.; Nasiri, H. Detection of Monkeypox cases based on symptoms using XGBoost and Shapley additive explanations methods. Diagnostics 2023, 13, 2391. [Google Scholar] [CrossRef]
- Zheng, C.; Tian, J.; Wang, K.; Han, L.; Yang, H.; Ren, J.; Li, C.; Zhang, Q.; Han, Q.; Zhang, Y. Time-to-event prediction analysis of patients with chronic heart failure comorbid with atrial fibrillation: A LightGBM model. BMC Cardiovasc. Disord. 2021, 21, 379. [Google Scholar] [CrossRef]
- Ghaheri, P.; Nasiri, H.; Shateri, A.; Homafar, A. Diagnosis of Parkinson’s disease based on voice signals using SHAP and hard voting ensemble method. Comput. Methods Biomech. Biomed. Eng. 2024, 27, 1858–1874. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D. Ten quick tips for machine learning in computational biology. BioData Min. 2017, 10, 35. [Google Scholar] [CrossRef]
- Ozenne, B.; Subtil, F.; Maucort-Boulch, D. The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J. Clin. Epidemiol. 2015, 68, 855–859. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]

| Features | Total Number (n = 4195) |
|---|---|
| Gender (Female) | 2124 (50.6%) |
| Age | 52.38 ± 14.67 |
| Comorbidities (%) | |
| HF | 308 (7.3%) |
| VT | 234 (5.6%) |
| ACS | 103 (2.5%) |
| CAD | 382 (9.1%) |
| PVD | 53 (1.3%) |
| HTN | 1418 (33.8%) |
| DM | 484 (11.5%) |
| Hyperlipidemia | 986 (23.5%) |
| COPD | 422 (10.1%) |
| CKD | 195 (4.6%) |
| ESRD | 141 (3.4%) |
| Malignancy | 185 (4.4%) |
| CVA | 251 (6.0%) |
| Rheumatic | 67 (1.6%) |
| LD | 303 (7.2%) |
| Medications (%) | |
| B_blocker | 2226 (53.1%) |
| C1_AAD_Ia & Ib | 724 (17.3%) |
| C1_AAD_Ic | 987 (23.5%) |
| C3_AAD | 335 (8.0%) |
| CCB | 1922 (45.8%) |
| Model & Data Processing Method | Accuracy | Sensitivity | Specificity | F1 | AUC |
|---|---|---|---|---|---|
| Logi_ROSE | 0.809 (0.085) | 0.707 (0.081) | 0.813 (0.093) | 0.316 (0.041) | 0.817 (0.012) |
| Cart_ROSE | 0.816 (0.013) | 0.493 (0.060) | 0.836 (0.018) | 0.236 (0.053) | 0.665 (0.026) |
| RF_ROSE | 0.756 (0.072) | 0.663 (0.077) | 0.760 (0.080) | 0.241 (0.027) | 0.752 (0.021) |
| XGB_ROSE | 0.753 (0.086) | 0.504 (0.043) | 0.768 (0.091) | 0.197 (0.043) | 0.584 (0.026) |
| LightGBM_ROSE | 0.782 (0.051) | 0.735 (0.043) | 0.784 (0.055) | 0.281 (0.036) | 0.822 (0.018) |
| Logi_SMOTE | 0.824 (0.066) | 0.688 (0.072) | 0.831 (0.072) | 0.321 (0.032) | 0.805 (0.026) |
| Cart_SMOTE | 0.873 (0.005) | 0.44 (0.069) | 0.9 (0.011) | 0.281 (0.037) | 0.67 (0.03) |
| RF_SMOTE | 0.724 (0.118) | 0.707 (0.129) | 0.723 (0.132) | 0.243 (0.047) | 0.758 (0.025) |
| XGB_SMOTE | 0.757 (0.065) | 0.61 (0.048) | 0.765 (0.069) | 0.227 (0.03) | 0.675 (0.043) |
| LightGBM_SMOTE | 0.748 (0.093) | 0.726 (0.077) | 0.748 (0.104) | 0.257 (0.038) | 0.801 (0.018) |
| Model & Data Processing Method | Accuracy | Sensitivity | Specificity | F1 | AUC |
|---|---|---|---|---|---|
| Logi_ROSE | 0.826 (0.046) | 0.847 (0.084) | 0.826 (0.048) | 0.253 (0.076) | 0.886 (0.047) |
| Cart_ROSE | 0.897 (0.015) | 0.483 (0.134) | 0.912 (0.008) | 0.231 (0.036) | 0.698 (0.071) |
| RF_ROSE | 0.807 (0.069) | 0.773 (0.081) | 0.807 (0.072) | 0.219 (0.048) | 0.831 (0.047) |
| XGB_ROSE | 0.735 (0.303) | 0.61 (0.171) | 0.74 (0.311) | 0.205 (0.114) | 0.68 (0.18) |
| LightGBM_ROSE | 0.797 (0.032) | 0.879 (0.06) | 0.793 (0.033) | 0.224 (0.059) | 0.882 (0.044) |
| Logi_SMOTE | 0.78 (0.084) | 0.877 (0.095) | 0.776 (0.091) | 0.221 (0.064) | 0.878 (0.046) |
| Cart_SMOTE | 0.908 (0.018) | 0.403 (0.098) | 0.927 (0.022) | 0.227 (0.075) | 0.665 (0.048) |
| RF_SMOTE | 0.769 (0.071) | 0.847 (0.038) | 0.767 (0.074) | 0.204 (0.069) | 0.845 (0.042) |
| XGB_SMOTE | 0.826 (0.062) | 0.625 (0.11) | 0.832 (0.065) | 0.2 (0.062) | 0.669 (0.067) |
| LightGBM_SMOTE | 0.764 (0.086) | 0.889 (0.057) | 0.759 (0.091) | 0.208 (0.047) | 0.87 (0.038) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-Y.; Lai, Y.-T.; Chuang, C.-W.; Yu, C.-H.; Lo, C.-Y.; Chen, M.; Shia, B.-C. Machine Learning-Based Prediction of Three-Year Heart Failure and Mortality After Premature Ventricular Contraction Ablation. Diagnostics 2025, 15, 2693. https://doi.org/10.3390/diagnostics15212693
Lin C-Y, Lai Y-T, Chuang C-W, Yu C-H, Lo C-Y, Chen M, Shia B-C. Machine Learning-Based Prediction of Three-Year Heart Failure and Mortality After Premature Ventricular Contraction Ablation. Diagnostics. 2025; 15(21):2693. https://doi.org/10.3390/diagnostics15212693
Chicago/Turabian StyleLin, Chung-Yu, Yu-Te Lai, Chien-Wei Chuang, Chih-Hsien Yu, Chiung-Yun Lo, Mingchih Chen, and Ben-Chang Shia. 2025. "Machine Learning-Based Prediction of Three-Year Heart Failure and Mortality After Premature Ventricular Contraction Ablation" Diagnostics 15, no. 21: 2693. https://doi.org/10.3390/diagnostics15212693
APA StyleLin, C.-Y., Lai, Y.-T., Chuang, C.-W., Yu, C.-H., Lo, C.-Y., Chen, M., & Shia, B.-C. (2025). Machine Learning-Based Prediction of Three-Year Heart Failure and Mortality After Premature Ventricular Contraction Ablation. Diagnostics, 15(21), 2693. https://doi.org/10.3390/diagnostics15212693





