Current Updates on Molecular Diagnostic Assays Used for Detection of Candida auris: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Search Strategy
2.3. Inclusion Criteria
- (1)
- Original research study involving molecular diagnostics of C. auris;
- (2)
- Studies involving DNA-based methodologies;
- (3)
- Either prospective or retrospective laboratory studies;
- (4)
- Studies published in the period between 1 January 2020 and 20 November 2024.
2.4. Exclusion Criteria
- (1)
- Non-human laboratory data;
- (2)
- Diagnostic methods other than molecular technologies (i.e., culture-based diagnostics);
- (3)
- Diagnostic and typing methods such as WGS, multilocus sequence typing (MLST), amplified fragment length polymorphism (AFLP), and pulsed-field gel electrophoresis (PFGE);
- (4)
- Studies other than original research (i.e., review articles);
- (5)
- Manuscripts written in languages other than English;
- (6)
- Studies published outside the defined study period.
2.5. Selection of Studies
2.6. Statistical Analysis
3. Results
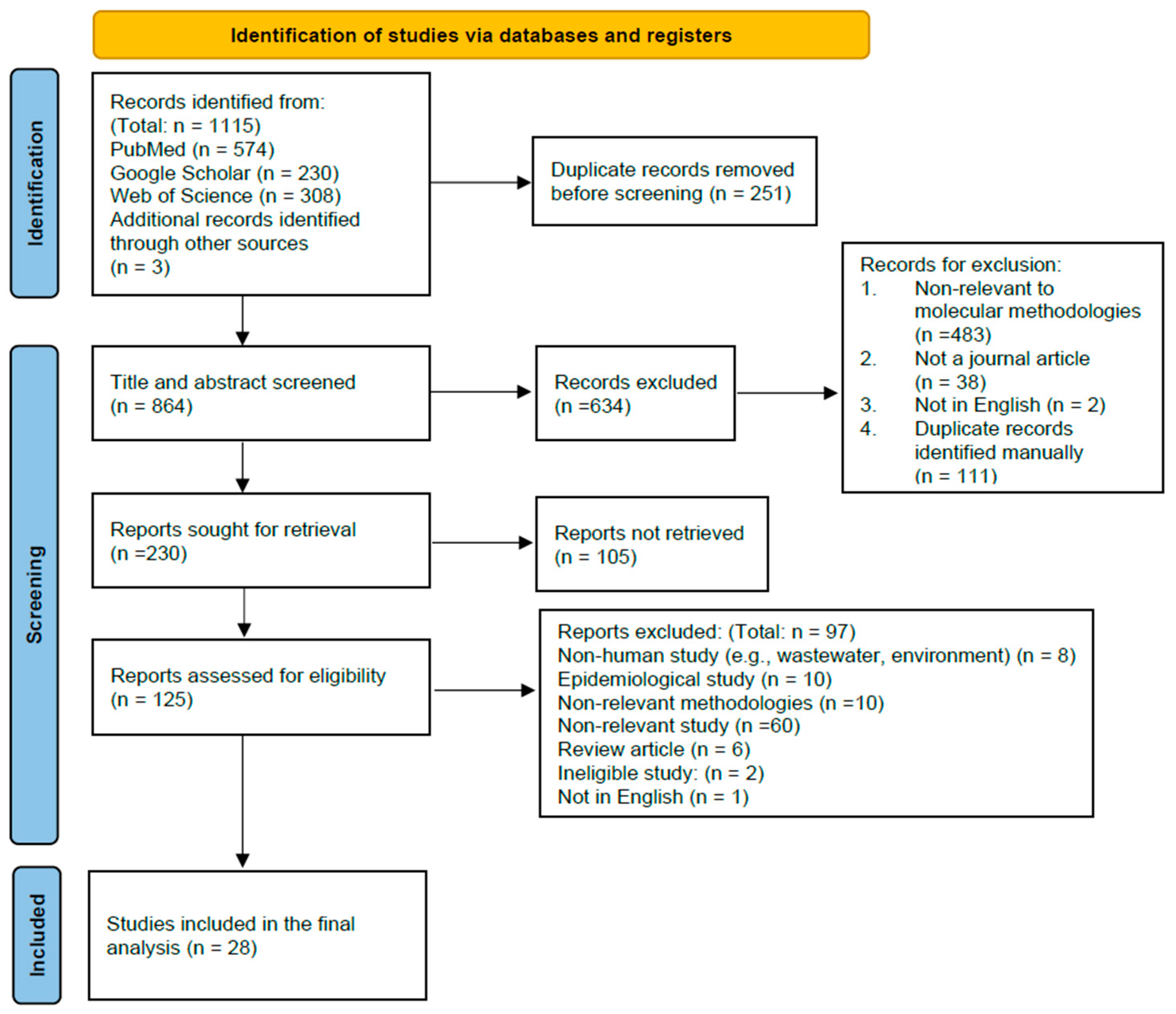
3.1. Study Selection and Identification of the Included Studies
3.2. Sample Pre-Extraction Procedures and Nucleic Acid Extraction
| Extraction Methods | Pre-Extraction Procedures | Extraction Kits Used | Equipment Used | Protocols | References |
|---|---|---|---|---|---|
| Column-based | / | Qiagen QIAamp kit, Qiagen QIAsymphony tissue kit (Qiagen, Hilden, Germany) | / | Manual extraction according to the manufacturer’s instructions. | [35] |
| Column-based | / | Qiagen Dneasy PowerLyzer microbial kit (Qiagen, Hilden, Germany) | / | Manual extraction according to the manufacturer’s instructions with the following modifications: (1) centrifuge the sample for 5 min; (2) incubate in elution buffer for 5 min; (3) elute the final sample in 50 μL. | [33] |
| Column-based | / | Qiagen kit (Qiagen, Hilden, Germany) | Qiagen BioRobot EZ1 (Qiagen, Tokyo, Japan) | Automated method according to the manufacturer’s instructions. | [13] |
| Column-based | Swabs were pooled in 1 mL water and vortex for 5 s | Qiagen virus mini kit (Qiagen, Hilden, Germany) | Qiagen BioRobot EZ1 | Automated method according to the manufacturer’s instructions. | [14] |
| Column-based | / | Qiagen DNeasy UltraClean Microbial kit (Qiagen, Hilden, Germany) | / | Manual extraction according to the manufacturer’s instructions. | [23] |
| Column-based and mechanical-based | Homogenize the sample prior to DNA extraction | FastDNA kit (MP Biomedicals, Irvine, CA, USA), QIAamp DNA mini kit (Qiagen, Hilden, Germany) | MP Biomedical FastPrep homogenizer (MP Biomedicals, Irvine, CA, USA) | After homogenization, lysed sample will proceed with nucleic acid extraction using the QIAamp DNA mini kit. | [31] |
| Extraction methods | Pre-extraction procedures | Extraction kits used | Equipment used | Protocols | References |
| Column-based and bead-beating | / | Zymo Quick-DNA Fungal/Bacterial Miniprep kit (Zymo Research, Irvine, CA, USA) | Vortex Genie 2 (Scientific Industries, Bohemia, NY, USA) | Manual extraction for isolates: (1) inoculate the colonies in a ZR BashingBead lysis tube with 750 μL of BashingBead buffer; (2) vortex at maximum speed for 10 min with a Vortex Genie 2; (3) spin it down and filter the supernatent through the Zymo-Spin filter through centrifugation; (4) mix the filtrate with genomic lysis buffer and then transfer to a column; (5) final elution volume is 100 μL. | [29] |
| Column-based and mechanical-based | Add swabs to 1 mL MP Biomedical Lysing Matrix A tube. Then, homogenize the sample for 60 s under the speed of 6 m/s prior to DNA extraction | Qiagen QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) | MP Biomedical FastPrep homogenizer | Semi-automated method: (1) quick spin the homogenized sample and pipette 200 μL into a 2 mL tube containing 20 μL of proteinase K; (2) mix 200 μL Buffer AL with 5 μL internal extraction control; (3) vortex the mixture for 15 s; (4) incubate at 56 °C for 10 min; (5) load the sample tube into the QIAcube and choose the “Body fluid spin protocol”; (6) extraction performed on the QIAcube; (7) final elution volume is 100 μL; (8) 6 μL DNA template for OLM AurisID real-time PCR assay. | This report |
| Chemical-based and thermal-based | / | MagCore Genomic DNA Whole Blood Kit (RBC Bioscience Corp., New Taipei City, Taiwan) | MagCore Plus II extractor (RBC Bioscience Corp., New Taipei City, Taiwan) | Manual extraction method includes cell lysis and protein degradation by heat-based lysis together with proteinase K and guanidine hydrochloride. | [9] |
| Magetic-based | / | Roche MagNA Pure 96 DNA and viral NA small volumn kit (Roche Diagnostics, North Ryde, NSW, Australia) | Roche MagNA Pure 96 (Roche Diagnostics, North Ryde, NSW, Australia) | (1) Add 4 mg/mL amplification control into 200 μL of sample; (2) DNA extracted by automated method and elute in final volume of 100 μL. | [47] |
| Magetic-based | / | / | easyMAG (bioMérieux, Durham, NC, USA) | Automated method according to the manufacturer’s instructions. | [25] |
| Extraction methods | Pre-extraction procedures | Extraction kits used | Equipment used | Protocols | References |
| Chemical-based | / | / | / | Manual extraction: (1) inoculate the swabs into 500 μL of Tris-EDTA (TE) pH 8.0 buffer; (2) for 50 μL of sample-containing TE buffer, add 5 μL of DiaSorin fungal lysis solution; (3) incubate the mixture at 60 °C for 30 min on a rocking incubator with 25 strokes per minute. | [26] |
| Chemical-based and thermal-based | Swab inoculate in wash solution | Kaneka DNA Extraction Kit 2 (Kaneka Co., Tokyo, Japan) | / | Manual extraction: (1) centrifuge 100 μL of sample at 13,000 r.p.m. for 10 min; (2) discard the supernatent and add 100 μL of Solution A to the pellets and vortex; (3) incubate at 98 °C for 8 min; (4) add 14 μL of Solution B and mix well; (5) 10 μL of DNA extract is used for LAMP reaction. | [15] |
| Thermal-based | / | Eazyplex LAMP kit (AmplexDiagnostics GmbH, Gars-Bahnhof, Germany) | / | (1) Addition of 25 μL of sample into the RALF buffer solution; (2) incubate at 99 °C for 5 min; (3) 25 μL of sample is used to test on the LAMP strip. | [27] |
| Mechanical-based | Cell suspension pre-wash by 500 μL of PBS | DM assay cartridge | OmniLyse device (Claremont BioSolutions, Upland, CA, USA) | Automated method based on the syringe-linked, bead-beating, and micro-motor-based principles; (1) 10 lysis cycles should be run for each of the samples; (2) mix 100 μL of lysate with 22 μL of magnetic bead buffer plus 1 μL of control DNA; (3) then, run it on a Point-of-Care handheld device with droplet magnetofluidics technology. | [29] |
| Mechanical-based | / | / | MP Biomedical FastPrep homogenizer | (1) Isolates were inoculated in 800 μL of solution (including 400 μL of TE buffer and 400 μL of phenol, chloroform, and isoamyl alcohol in a ratio of 25:24:1; (2) perform micro-bead shearing using the FastPrep homogenizer for 30 s under the speed of 6 m/s); (3) centrifuge the homogenate at 14,000 r.p.m. for 10 min; (4) 300 μL of the supernatent is then mix with 30 μL of 3 M sodium acetate and 900 μL of 100% ethanol; (5) cool the mixture at −20 °C for 30 min and centrifuge again at 14,000 r.p.m. for 10 min at 4 °C; (6) wash the pellet two times with 500 μL of 70% ethanol; (7) dry at 37 °C and dissolve the DNA in nuclease-free water. | [5] |
| Extraction methods | Pre-extraction procedures | Extraction kits used | Equipment used | Protocols | References |
| Mechanical-based | / | / | Precellys 24 homogenizer (Bertin Instruments, Montigny-le-Bretonneux, France) | Manual washing, freeze, thaw, and homogenization. (1) Centrifuge 200 μL of sample for 10 min at 15000 r.p.m. and remove the supernatant; (2) re-suspend the pellet in 100 μL of 1% PBS-BSA buffer; (3) add glass beads into the suspension; (4) freeze the suspension at −80 °C for 30 min and then incubate it on a heat block/water bath at 70 °C for 30 min; (5) homogenize the suspension for 20 s two times with 10 s intervals; (6) centrifuge the suspension for 10 min at 15,000 r.p.m; (7) then, proceed with PCR amplification for the supernatant. | [35] |
| Mechanical-based | Add swabs to 1 mL MP Biomedical Lysing Matrix A tube. Then, homogenize the sample for 60 s under the speed of 6 m/s before loading on the BD MAX | / | MP Biomedical FastPrep homogenizer | Automated method: (1) pipette 200 μL of the pre-treated specimen into a BD MAX ExK DNA-2 kit Sample Buffer Tube (SBT); (2) recap the inoculated SBT using a blue septum cap; (3) load the SBT on the BD MAX analyzer. | This report |
| Mechanical-based | Sample undergo ultrasonication before loading on the cobas Roche 6800 | / | TS5 tube ultrasonicator (Rinco Ultrasonics, Romanshorn, Switzerland) | Automated method: (1) load 5 sample tubes at one time into the ultrasonicator with the following settings: sonication time: 25 s, pause time: 5 s, 10 cycles; force: 600 N; sonication amplitude: 40%; cycle energy limits: min = 200 J; max = 6000 J; program energy limits: min = 4000 J; max = 30,000 J; (2) load the processed samples on the cobas Roche 6800. | This report |
| Extraction methods | Pre-extraction procedures | Extraction kits used | Equipment used | Protocols | References |
| Mechanical-based and thermal-based | / | MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche Diagnostics, Indianapolis, IN, USA) | Disruptor Genie (Scientific Industries Inc., Bohemia, NY, USA), MagNA Pure LC 2.0 (Roche Diagnostics, Indianapolis, IN, USA) | Isolates: (1) inoculate 1 μL loopful of colonies in 500 μL of water with silica glass or zirconia beads; (2) heat the sample at 95 °C for 10 min; (3) mechanical lysis by the Disruptor Genie for 2 min; (4) 5 μL of processed sample is use for PCR. Surveillance swabs: (1) place the swabs or inoculate 60 μL of the liquid in 600 μL of Tris-EDTA neutralization buffer; (2) shake with a thermomixer at 14,000 r.p.m. for 6 min at 100 °C. Whole blood: (1) 200 μL of sample is used for extraction on the MagNA Pure, with a final elution volume of 100 μL. Urine: (1) centrifuge 5 mL of urine; (2) 250 μL of the sample is heated at 95 °C for 5 min; (3) 200 μL of the processed sample is used for extraction on the MagNA Pure, with a final elution volume of 100 μL. | [12] |
| Chemical-based and thermal-based | / | / | / | (1) Suspend the yeast colonies in 20 μL of 20 mM NaOH; (2) incubate the suspension at 95 °C for 15 min; (3) 3 μL of lysed suspension is used for each of the 25 μL PCR reactions. | [20] |
| Thermal-based, bead-based, and chemical-based | / | / | Precelleys homogenizer | Colonies: (1) Colonies suspended in 50 μL of water; (2) boil at 95 °C for 20 min; (3) centrifuge at 5000 r.p.m. for 5 min; (4) supernatent then used as the PCR template. Manual extraction of clinical specimens using the glass bead–phenol–chloroform DNA extraction method. (1) Sample transfer to a tube containing glass beads and 400 μL of lysis buffer, 300 μL of phenol–chloroform; (2) homogenize the sample at 6000 r.p.m. for 3 × 60 s; (3) centrifuge the lysed sample at 5000 r.p.m. for 5 min; (4) transfer the supernatant to 300 μL of phenol–chloroform, and centrifuge again at 5000 r.p.m. for 5 min; (5) transfer the supernatant to a new tube, add an equal volume of chloroform, and centrifuge again at 5000 r.p.m. for 5 min; (6) transfer the supernatant to a new tube and add 2.5 and 0.1 in volume of absolute ethanol and 3 M sodium acetate, respectively; (7) freeze at −20 °C for 1 h, and centrifuge at 12,000 r.p.m. for 10 min; (8) wash the pellet in 70% cold ethanol, and air dry; (9) dissolve the sample in 30 μL of water for downstream PCR. | [39,48] |
| Extraction methods | Pre-extraction procedures | Extraction kits used | Equipment used | Protocols | References |
| Thermal-based | / | / | / | Yeast colonies: (1) suspend the colonies in 100 μL of water; (2) boil at 100 °C for 10 min; (3) centrifuge at 2000 g for 3 min; (4) supernatent then used as PCR template; (5) check DNA purity at A260/A280 using NanoDrop with a ratio of 1.7 to 2.1; (6) 100 pg to 1 μg of genomic DNA is used for PCR amplification.. | [34] |
| Column-based and bead-based | / | Zymo Quick-DNA Fungal/Bacterial Miniprep kit | Vortex Genie 2 | Manual extraction for isolates: (1) colonies suspended in PBS to yield a concentration of 1 × 105 CFU/mL; (2) 200 μL of the sample is vortexed at maximum speed for 10 min with a Vortex Genie 2; (3) spin it down, and filter the supernatent through the Zymo-Spin filter through centrifugation; (4) mix the filtrate with a genomic lysis buffer and then transfer it to a column; (5) final elution volume is 35 μL. | [21] |
| Chemical-based | 250 μL of sample (1 × 105 CFU/mL) mix with 250 μL of binding buffer (4 M guanidine thiocyanate, 0.055 M Tris-HCL pH 7.5, 0.025 M pH 8 EDTA) and 10 μL of silica-coated magnetic beads | / | / | (1) Allow the samples to bind with the binding buffer for 10 min at room temperature; (2) pellet the silica-coated magnetic beads using a handheld magnet; (3) invert the tube to remove the solution; (4) resuspend the beads in 50 μL of wash solution with 0.05% Tween 20; (5) pellet the beads again, and remove the wash solution; (6) repeat for two times; (7) elute the beads in 10 μL of PCR buffer at 60 °C for 5 min. | [21] |
| Extraction methods | Pre-extraction procedures | Extraction kits used | Equipment used | Protocols | References |
| Thermal-based | Same as above | / | / | (1) Pre-heat 250 μL of the sample at 70 °C for 10 min before adding binding buffer and magnetic beads; (2) incubate for another 10 min; (3) same procedures as step 2 to 7 of the chemical-based protocol. | [21] |
| Mechanical-based | Same as above | / | / | (1) Inoculate 250 μL of the sample into 250 μL of pre-filled zirconia/silica beads, (2) vortex at maximum speed for 10 min with a Vortex Genie 2 before adding binding buffer and magnetic beads; (3) incubate for another 10 min; (4) combine with 250 μL of binding buffer and 10 μL of silica-coated magnetic beads; (5) same procedures as step 2 to 7 of the chemical-based protocol. | [21] |
3.3. Commercial Molecular Assays
3.4. Laboratory-Developed Molecular Assays
| Name of Assays | Target/Method | Sample Types | No. of Samples | Approval | Real-Time PCR Machines/Analyzer | Performance | References |
|---|---|---|---|---|---|---|---|
| Cobas ePlex BCID-FP (GenMark Dx, (Roche Diagnostics, Indianapolis, IN, USA)) | Multiplex PCR | Positive blood culture (contrived) | 49 | FDA-cleared | cobas ePlex system (Roche Diagnostics, Indianapolis, IN, USA) | Sensitivity: 100%; Specificity: 100%; LoD:/; Reference method: culture | [49] |
| BioFire Filmarray BCID2 (bioMérieux, Durham, NC, USA) | Multiplex PCR | Positive blood culture | 152 | FDA-cleared | Filmarray system (bioMérieux, Durham, NC, USA) | Sensitivity: N.A.; Specificity: 100%; LoD:/; Reference method: culture | [50] |
| DiaSorin Molecular C. auris Simplexa (DiaSorin, Cypress, CA, USA) | ITS2/Real-time PCR | Axilla/groin | 282 | FDA De Novo granted in 7/2024 | DiaSorin LIAISON MDX (DiaSorin, Cypress, CA, USA) | Sensitivity: 100%; Specificity: 100%; LoD: 1–2 CFU/PCR reaction; Reference method: culture | [26] |
| DiaSorin Molecular C. auris Simplexa | ITS2/Real-time PCR | Axilla/groin | 60 | FDA De Novo granted in 7/2024 | DiaSorin LIAISON MDX | Sensitivity: 100%; Specificity: 100%; LoD: 26 CFU/PCR reaction; Reference method: culture | [10] |
| DiaSorin Molecular C. auris Simplexa | ITS2/Real-time PCR | Axilla/groin | 25 | FDA De Novo granted in 7/2024 | DiaSorin LIAISON MDX | Sensitivity: 95.6%; Specificity: 100%; LoD: 600 CFU/mL; Reference method: another real-time PCR assay | [37] |
| RealCycler Candida auris PCR (Progenie Molecular, Valencia, Spain) | Real-time PCR | Axilla/groin | 392 | CE-IVD | Bio-Rad CFX96 (Bio-Rad Laboratories, Hercules, CA, USA) | Sensitivity: N.A.; Specificity: N.A.; LoD: 1 copy/μL; Reference method: culture | [9] |
| Name of assays | Target/Method | Sample types | No. of samples | Approval | Real-time PCR machines/Analyzer | Performance | References |
| OLM AurisID (OLM, Newcastle Upon Tyne, UK) | 28S rRNA/Real-time PCR | Pharyngeal or axillary-rectal | 113 | CE-IVD | Bio-Rad CFX96 | Without prior DNA extraction; Sensitivity: 96.6%; Specificity: 100%; LoD: 1 CFU/PCR reaction (500 CFU/mL); Reference method: culture | [46] |
| OLM AurisID | 28S rRNA/Real-time PCR | Isolates | 29 | CE-IVD | ABI 7500 (Applied Biosystems, Foster City, CA, USA) | Sensitivity: N.A.; Specificity: 100%; LoD: 1 Copy/PCR reaction; Reference method: culture | [23] |
| OLM AurisID | 28S rRNA/Real-time PCR | Axilla/groin, Sample spiked with isolates | 95 | CE-IVD | Qiagen Rotor-Gene Q (Qiagen, Hilden, Germany) | Sensitivity: 94.9%; Specificity: 98.2%; LoD: 15 copies/PCR reaction; Reference method: culture | This report |
| Eazyplex Candida auris (AmplexDiagnostics GmbH, Gars-Bahnhof, Germany) | LAMP | Isolates/Pharyngeal, axillary-rectal | 51/152 | CE-IVD | Genie HT (OptiGene Limited, West Sussex, UK) | Initial results without resolving the discrepancy: Sensitivity: 76–76.9%; Specificity: 96.8–100%; LoD: 1 CFU/LAMP reaction; Reference method: culture | [27] |
| Fungiplex Candida Auris Real-Time PCR (Bruker, Bremen, Germany) | Mating locus alpha | Isolates | 29 | RUO | ABI 7500 | Sensitivity: N.A.; Specificity: 100%; LoD: 9 Copies/PCR reaction; Reference method: culture | [23] |
| Name of assays | Target/Method | Sample types | No. of samples | Approval | Real-time PCR machines/Analyzer | Performance | References |
| LAMPAuris | LAMP | Axilla/groin | 103 | / | Kaneka MyAbscope (Portable device) (Kaneka Co., Tokyo, Japan) | Sensitivity: 66–86%; Specificity: 97–100%; LoD: 20 copies/LAMP reaction; Reference method: culture and qPCR | [15] |
| Type of Methods | Target | Sample Types | No. of Samples | Internal Control(s) | Real-time PCR Machines | Performance | References |
|---|---|---|---|---|---|---|---|
| Real-time PCR | ITS2 | Nares, axilla/groin | 1414 | Human β-globin gene | QuantStudio 6 Flex (Thermo Fisher Scientific Inc., Singapore) | Sensitivity: 100%; Specificity: 100%; LoD: 1 CFU/PCR reaction; Reference method: culture | [35,51] |
| Conventional /Real-time PCR | ITS | Isolates/clinical samples * | 439/590 | / | LightCycler 96 (Roche Diagnostics, Mannheim, Germany) | Sensitivity: N.A.; Specificity: N.A.; Conventional PCR: 100 CFU/PCR reaction; Real-time PCR: LoD: 10 CFU/PCR raction; Reference method: N.A. | [39] |
| Real-time PCR | ITS2 | Isolates | 104 | / | ABI 7500 | Sensitivity: N.A.; Specificity: 100%; LoD: 1 CFU/mL; Reference method: N.A. | [34] |
| Real-time PCR | ITS2 | Sample spiked with isolates/groin, axilla | 15/106 | Amplificaton control | Roche LightCycler 480 (Roche Diagnostics, Mannheim, Germany) | Sensitivity: N.A.; Specificity: 100%; LoD: 1 CFU/PCR reaction; Reference method: culture | [47] |
| Real-time PCR | ITS2 | Isolates/whole blood/Urine/Swabs | 32/30/30/90 | PhHV1 glycoprotein B gene | Roche LightCycler 480 | Sensitivity: 93.3–100%; Specificity: 100%; LoD: 4–37 CFU/PCR reaction; Reference method: contrived samples spiked with colonies | [12] |
| Real-time PCR (Point-of-care) | ITS2 | Isolates | 16 | Bicoid gene | POC.auris Droplet magnetofluidic device | POC.auris assay: Sensitivity: N.A.; Specificity: 100%; LoD: 300 CFU/mL; Reference method: manual real-time PCR assay | [29] |
| Type of methods | Target | Sample types | No. of samples | Internal control(s) | Real-time PCR machines | Performance | References |
| Real-time PCR | ITS1/ITS2 | Nares, throat, axilla/groin, Urine | 10,818 (pooled swabs) 4792 (ruine) | Human beta-actin | QuantStudio 7 (Thermo Fisher Scientific Inc., Singapore) | Sensitivity: 100%; Specificity: 99.4%; LoD: 100 CFU/mL; Reference method: culture | [14] |
| Conventional PCR | ITS | Isolates | 23 | / | Labcycler Basic thermocycler (Bioké, Leiden, The Netherlands). | Clade-specific PCR for rapid identification of C. auris and differentiation of 5 different clades. Results were confirmed by Sanger sequencing. | [5] |
| Real-time PCR | ITS2 | Skin swabs, Sample spiked with isolates | 145 | Proprietary internal control | GeneXpert (LDT-Automated) (Cepheid, Sunnyvale, CA, USA) | CaurisSurV cartridge; Sensitivity: 92 to 97.5%; Specificity: 100%; LoD: 10.5 to 14.8 CFU/mL; Reference method: BD MAX PCR assay | [4] |
| Real-time PCR | ITS2 | Axilla/groin | 56 | Proprietary internal control | Hologic Panther Fusion (LDT-Automated) (Hologic, Marlborough, MA, USA) | Sensitivity: 100%; Specificity: 97.96%; LoD: 0.47 CFU/mL; Reference method: culture | [6] |
| Real-time PCR | ITS2 | Nares, axilla/groin | 110 | Bicoid gene | BD Max open system (LDT-Automated) (BD Diagnostics, Sparks, MD, USA) | Optimum extraction temperature on BD MAX is 75 °C for 20 min; Sensitivity: 96%; Specificity: 92%; LoD: 1 CFU/PCR reaction; Reference method: manual real-time PCR assay | [36,51] |
| Real-time PCR | ITS | Axilla/groin, Sample spiked with isolates | 2566 | Proprietary internal control | BD Max open system (LDT-Automated) | Sensitivity: 94.6%; Specificity: 100%; LoD: 31.25 copies/PCR reaction; Reference method: culture | This report |
| Type of methods | Target | Sample types | No. of samples | Internal control(s) | Real-time PCR machines | Performance | References |
| Real-time PCR | 5.8s rRNA, ITS2 | Axilla/groin, Sample spiked with isolates | 104 | Proprietary internal control | Roche 6800 cobas omni utility channel (LDT-Automated) (Roche Diagnostics, Indianapolis, IN, USA) | Sensitivity: 97.4%; Specificity: 100%; LoD: 270 copies/mL; Reference method: another real-time PCR assay | This report |
| Real-time PCR | 5.8S rRNA, ITS2, 28S rRNA | Anterior nares | 70 | Lambda phage | Bio-Rad CFX 96 | Sensitivity: 100%; Specificity: 100%; LoD: 10 CFU/PCR reaction; Reference method: culture | [24,33,52] |
| Real-time PCR | GPI | Isolates, Stool | 128 1053 | N.A. | Bio-Rad CFX 96 | Sensitivity: N.A.; Specificity: 100%; LoD: 13 CFU/PCR reaction; Reference method: N.A. | [13] |
| Real-time PCR | GPI | Isolates | 155 | / | ABI 7500 | Sensitivity: N.A.; Specificity: N.A.; LoD: 5 CFU/PCR reaction; Reference method: N.A. | [20] |
| Real-time PCR | / | Axilla/groin | 195 | / | / | Sensitivity: 44%; Specificity: 99%; LoD: N.A.; Reference method: culture | [38] |
| Real-time PCR | CDC protocol | Sample spiked with isolates | 222 | Bicoid gene | Bio-Rad CFX 96 | Isolates (100 CFU/mL): Sensitivity: 100%; Specificity: 100%; LoD: 27.11 CFU/mL; Reference method: sample spiked with colonies. Isolates (10 CFU/mL): Sensitivity: 82.1%; Specificity: 100%; LoD: 27.11 CFU/mL; Reference method: sample specimens spiked with colonies. | [25] |
4. Discussion
4.1. Sample Pre-Extraction Procedures and Nucleic Acid Extraction
4.2. Commercial Molecular Assays and Laboratory-Developed Molecular Assays
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action. Available online: https://www.who.int/publications/i/item/9789240060241 (accessed on 18 November 2024).
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Geremia, N.; Brugnaro, P.; Solinas, M.; Scarparo, C.; Panese, S. Candida auris as an Emergent Public Health Problem: A Current Update on European Outbreaks and Cases. Healthcare 2023, 11, 425. [Google Scholar] [CrossRef] [PubMed]
- Banik, S.; Ozay, B.; Trejo, M.; Zhu, Y.; Kanna, C.; Santellan, C.; Shaw, B.; Chandrasekaran, S.; Chaturvedi, S.; Vejar, L.; et al. A simple and sensitive test for Candida auris colonization, surveillance, and infection control suitable for near patient use. J. Clin. Microbiol. 2024, 62, e0052524. [Google Scholar] [CrossRef] [PubMed]
- Carolus, H.; Jacobs, S.; Lobo Romero, C.; Deparis, Q.; Cuomo, C.A.; Meis, J.F.; Van Dijck, P. Diagnostic Allele-Specific PCR for the Identification of Candida auris Clades. J. Fungi 2021, 7, 754. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, F.M.; Bertsch, J.; DeMaet, M.A.; York, T.; McDougal, A.; Patel, J.A.; Ren, P. Enhancing Candida auris Surveillance in High-Risk Settings by Implementing a High-Throughput Molecular Assay on the Hologic Fusion Open Access Platform. J. Fungi 2024, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.P.; Nguyen, T.A.M.; Kidd, S.P.; Chambers, J.M.; Alastruey-Izquierdo, A.P.; Shin, J.M.; Dao, A.P.; Forastiero, A.M.; Wahyuningsih, R.M.; Chakrabarti, A.M.; et al. Candida auris—A systematic review to inform the world health organization fungal priority pathogens list. Med. Mycol. 2024, 62, myae042. [Google Scholar] [CrossRef] [PubMed]
- Keighley, C.; Garnham, K.; Harch, S.A.J.; Robertson, M.; Chaw, K.; Teng, J.C.; Chen, S.C. Candida auris: Diagnostic Challenges and Emerging Opportunities for the Clinical Microbiology Laboratory. Curr. Fungal Infect. Rep. 2021, 15, 116–126. [Google Scholar] [CrossRef]
- Magnasco, L.; Mikulska, M.; Sepulcri, C.; Ullah, N.; Giacobbe, D.R.; Vena, A.; Di Pilato, V.; Willison, E.; Orsi, A.; Icardi, G.; et al. Frequency of Detection of Candida auris Colonization Outside a Highly Endemic Setting: What Is the Optimal Strategy for Screening of Carriage? J. Fungi 2023, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, J.D.; Wang, C.Y.; Bolton, D.; Liggayu, B.; Schaefer, S.; Patel, G.; Javaid, W.; Cordon-Cardo, C.; Firpo-Betancourt, A.; Sordillo, E.M.; et al. Molecular Detection of Candida auris Using DiaSorin Molecular Simplexa(®) Detection Kit: A Diagnostic Performance Evaluation. J. Fungi 2023, 9, 849. [Google Scholar] [CrossRef] [PubMed]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Abbasi, A.F.; Prakash, S.; Mangat, J.; Hosein, Z.; Haider, N.; Chan, J. Candida auris: An Overview of the Emerging Drug-Resistant Fungal Infection. Infect. Chemother. 2022, 54, 236–246. [Google Scholar] [CrossRef]
- Walchak, R.C.; Buckwalter, S.P.; Zinsmaster, N.M.; Henn, K.M.; Johnson, K.M.; Koelsch, J.M.; Herring, S.A.; Steinmetz, L.K.; Reed, K.A.; Barth, J.E.; et al. Candida auris Direct Detection from Surveillance Swabs, Blood, and Urine Using a Laboratory-Developed PCR Method. J. Fungi 2020, 6, 224. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Baron, S.A.; Yousfi, H.; Hadjadj, L.; Lalaoui, R.; Morand, S.; Rolain, J.M.; Bittar, F. Development and standardization of a specific real-time PCR assay for the rapid detection of Candida auris. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1547–1551. [Google Scholar] [CrossRef]
- Taori, S.K.; Rhodes, J.; Khonyongwa, K.; Szendroi, A.; Smith, M.; Borman, A.M.; Kumarage, J.; Brown, C.S.; Moore, G.; Desai, N. First experience of implementing Candida auris real-time PCR for surveillance in the UK: Detection of multiple introductions with two international clades and improved patient outcomes. J. Hosp. Infect. 2022, 127, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Alshahni, M.M.; Komori, A.; Mimaki, M.; Makimura, K. Assessment of LAMPAuris for Rapid Detection of Candida auris in Clinical Specimens. Mycopathologia 2024, 189, 87. [Google Scholar] [CrossRef]
- Jeffery-Smith, A.; Taori, S.K.; Schelenz, S.; Jeffery, K.; Johnson, E.M.; Borman, A.; Manuel, R.; Brown, C.S. Candida auris: A Review of the Literature. Clin. Microbiol. Rev. 2018, 31, e00029. [Google Scholar] [CrossRef] [PubMed]
- Candida auris: A Review of Recent Literature. Available online: https://www.gov.uk/government/consultations/candida-auris-update-to-management-guidance/candida-auris-a-review-of-recent-literature (accessed on 1 December 2024).
- Suphavilai, C.; Ko, K.K.K.; Lim, K.M.; Tan, M.G.; Boonsimma, P.; Chu, J.J.K.; Goh, S.S.; Rajandran, P.; Lee, L.C.; Tan, K.Y.; et al. Detection and characterisation of a sixth Candida auris clade in Singapore: A genomic and phenotypic study. Lancet Microbe 2024, 5, 100878. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.; Fisher, M.C. Global epidemiology of emerging Candida auris. Curr. Opin. Microbiol. 2019, 52, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, M.; Bartolomé Álvarez, J.; Lockhart, S.R.; Valentín, E.; Ruiz-Gaitán, A.C.; Eraso, E.; de Groot, P.W.J. Identification of Candida auris and related species by multiplex PCR based on unique GPI protein-encoding genes. Mycoses 2021, 64, 194–202. [Google Scholar] [CrossRef]
- Jin, M.; Trick, A.Y.; Totten, M.; Lee, P.W.; Zhang, S.X.; Wang, T.H. Streamlined instrument-free lysis for the detection of Candida auris. Sci. Rep. 2023, 13, 21848. [Google Scholar] [CrossRef]
- Vallabhaneni, S.; Jackson, B.R.; Chiller, T.M. Candida auris: An Emerging Antimicrobial Resistance Threat. Ann. Intern. Med. 2019, 171, 432–433. [Google Scholar] [CrossRef]
- Sattler, J.; Noster, J.; Brunke, A.; Plum, G.; Wiegel, P.; Kurzai, O.; Meis, J.F.; Hamprecht, A. Comparison of Two Commercially Available qPCR Kits for the Detection of Candida auris. J. Fungi 2021, 7, 154. [Google Scholar] [CrossRef]
- Kordalewska, M.; Perlin, D.S. Molecular Diagnostics in the Times of Surveillance for Candida auris. J. Fungi 2019, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Komorowski, A.S.; Aftanas, P.; Porter, V.; Katz, K.; Kozak, R.A.; Li, X.X. Verification, Analytical Sensitivity, Cost-effectiveness, and Comparison of 4 Candida auris Screening Methods. Open Forum Infect. Dis. 2024, 11, ofae017. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.C.; Ahmed, M.; Kendra, C.G.; Sperling, R.M.; Van Benten, K.; Lavik, J.P.; Emery, C.L.; Relich, R.F.; Gavina, K. Validation of a qualitative real-time PCR assay for the detection of Candida auris in hospital inpatient screening. J. Clin. Microbiol. 2024, 62, e0015824. [Google Scholar] [CrossRef]
- Hernández Felices, F.J.; Tormo Palop, N.; Salvador García, C.; Mulet Bayona, J.V.; Guna Serrano, M.R.; Gimeno Cardona, C. Evaluation of Eazyplex® LAMP test for fast Candida auris direct detection of colonized patients. Mycoses 2024, 67, e13665. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, S.; Luchian, I.; Damian, C.; Goriuc, A.; Porumb-Andrese, E.; Popa, C.G.; Cobzaru, R.G.; Ripa, C.; Ursu, R.G. Candida auris Updates: Outbreak Evaluation through Molecular Assays and Antifungal Stewardship-A Narrative Review. Curr. Issues Mol. Biol. 2024, 46, 6069–6084. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.W.; Totten, M.; Chen, L.; Chen, F.E.; Trick, A.Y.; Shah, K.; Ngo, H.T.; Jin, M.; Hsieh, K.; Zhang, S.X.; et al. A Portable Droplet Magnetofluidic Device for Point-of-Care Detection of Multidrug-Resistant Candida auris. Front. Bioeng. Biotechnol. 2022, 10, 826694. [Google Scholar] [CrossRef] [PubMed]
- Osei Sekyere, J. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen 2018, 7, e00578. [Google Scholar] [CrossRef] [PubMed]
- González-Durán, E.; Contreras-Pérez, C.U.; Caceres, D.H.; Ríos-Rosas, C.; Piñón-Ortega, J.J.; Téllez-Saucedo, M.D.; Marín-Suro, E.S.; Wong-Arámbula, C.E.; Moreno-Escobar, E.A.; Ramírez-González, J.E.; et al. The use of readily available laboratory tests for the identification of the emerging yeast Candida auris in Mexico. Arch. Microbiol. 2022, 204, 592. [Google Scholar] [CrossRef]
- Ruiz-Gaitán, A.; Moret, A.M.; Tasias-Pitarch, M.; Aleixandre-López, A.I.; Martínez-Morel, H.; Calabuig, E.; Salavert-Lletí, M.; Ramírez, P.; López-Hontangas, J.L.; Hagen, F.; et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses 2018, 61, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Georgacopoulos, O.; Nunnally, N.S.; Le, N.; Lysen, C.; Welsh, R.M.; Kordalewska, M.; Perlin, D.S.; Berkow, E.L.; Sexton, D.J. Performance Evaluation of Culture-Independent SYBR Green Candida auris Quantitative PCR Diagnostics on Anterior Nares Surveillance Swabs. J. Clin. Microbiol. 2020, 58, e00690-20. [Google Scholar] [CrossRef] [PubMed]
- Jafarian, H.; Khodadadi, H.; Badiee, P. Development a hydrolysis probe-based quantitative PCR assay for the specific detection and quantification of Candida auris. Curr. Med. Mycol. 2020, 6, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Malczynski, M.; Dowllow, N.; Rezaeian, S.; Rios, J.; Dirnberger, L.; Zembower, J.A.; Zhu, A.; Qi, C. Optimizing a real-time PCR assay for rapid detection of Candida auris in nasal and axillary/groin samples. J. Med. Microbiol. 2020, 69, 824–829. [Google Scholar] [CrossRef]
- Leach, L.; Russell, A.; Zhu, Y.; Chaturvedi, S.; Chaturvedi, V. A Rapid and Automated Sample-to-Result Candida auris Real-Time PCR Assay for High-Throughput Testing of Surveillance Samples with the BD Max Open System. J. Clin. Microbiol. 2019, 57, e00630. [Google Scholar] [CrossRef] [PubMed]
- Rosa, R.; Jimenez, A.; Andrews, D.; Dinh, H.; Parra, K.; Martinez, O.; Abbo, L.M. Impact of In-house Candida auris Polymerase Chain Reaction Screening on Admission on the Incidence Rates of Surveillance and Blood Cultures with, C. auris and Associated Cost Savings. Open Forum Infect. Dis. 2023, 10, ofad567. [Google Scholar] [CrossRef] [PubMed]
- Shuping, L.; Maphanga, T.G.; Naicker, S.D.; Mpembe, R.; Ngoma, N.; Velaphi, S.; Nakwa, F.; Wadula, J.; Jaglal, P.; Govender, N.P. High Prevalence of Candida auris Colonization during Protracted Neonatal Unit Outbreak, South Africa. Emerg. Infect. Dis. 2023, 29, 1913–1916. [Google Scholar] [CrossRef] [PubMed]
- Safari, F.; Madani, M.; Kheirollahi, M.; Mirhendi, H. Molecular investigation of the incidence of Candida auris infections at selected hospitals in Iran. Mycoses 2022, 65, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar] [CrossRef] [PubMed]
- Spivak, E.S.; Hanson, K.E. Candida auris: An Emerging Fungal Pathogen. J. Clin. Microbiol. 2018, 56, e01588-17. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Bmj 2021, 372, n71. [Google Scholar] [CrossRef]
- Leeflang, M.M.G.; Kraaijpoel, N. Systematic reviews of diagnostic test accuracy in CMI. Clin. Microbiol. Infect. 2018, 24, 1115–1116. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2019, 10, 2993. [Google Scholar] [CrossRef] [PubMed]
- Calderone, R.A.; Cihlar, R.L. Methods in Molecular Biology Candida Species Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Mulet Bayona, J.V.; Salvador García, C.; Tormo Palop, N.; Gimeno Cardona, C. Validation and implementation of a commercial real-time PCR assay for direct detection of Candida auris from surveillance samples. Mycoses 2021, 64, 612–615. [Google Scholar] [CrossRef]
- Crawford, L.C.; Kidd, S.E.; Anninos, T.M.; Turra, M.; Weldhagen, G.F. Candida auris PCR for high-throughput infection control screening. Med. Mycol. 2022, 60, myac057. [Google Scholar] [CrossRef]
- Aboutalebian, S.; Ahmadikia, K.; Fakhim, H.; Chabavizadeh, J.; Okhovat, A.; Nikaeen, M.; Mirhendi, H. Direct Detection and Identification of the Most Common Bacteria and Fungi Causing Otitis Externa by a Stepwise Multiplex PCR. Front. Cell Infect. Microbiol. 2021, 11, 644060. [Google Scholar] [CrossRef]
- Zhang, S.X.; Carroll, K.C.; Lewis, S.; Totten, M.; Mead, P.; Samuel, L.; Steed, L.L.; Nolte, F.S.; Thornberg, A.; Reid, J.L.; et al. Multicenter Evaluation of a PCR-Based Digital Microfluidics and Electrochemical Detection System for the Rapid Identification of 15 Fungal Pathogens Directly from Positive Blood Cultures. J. Clin. Microbiol. 2020, 58, e02096-19. [Google Scholar] [CrossRef]
- Caméléna, F.; Péan de Ponfilly, G.; Pailhoriès, H.; Bonzon, L.; Alanio, A.; Poncin, T.; Lafaurie, M.; Dépret, F.; Cambau, E.; Godreuil, S.; et al. Multicenter Evaluation of the FilmArray Blood Culture Identification 2 Panel for Pathogen Detection in Bloodstream Infections. Microbiol. Spectr. 2023, 11, e0254722. [Google Scholar] [CrossRef] [PubMed]
- Leach, L.; Zhu, Y.; Chaturvedi, S. Development and Validation of a Real-Time PCR Assay for Rapid Detection of Candida auris from Surveillance Samples. J. Clin. Microbiol. 2018, 56, e01223-17. [Google Scholar] [CrossRef]
- Kordalewska, M.; Zhao, Y.; Lockhart, S.R.; Chowdhary, A.; Berrio, I.; Perlin, D.S. Rapid and Accurate Molecular Identification of the Emerging Multidrug-Resistant Pathogen Candida auris. J. Clin. Microbiol. 2017, 55, 2445–2452. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.; Widen, R.; Vestal, G.; Uy, D.; Silbert, S. A TaqMan Probe-Based Real-Time PCR Assay for the Rapid Identification of the Emerging Multidrug-Resistant Pathogen Candida auris on the BD Max System. J. Clin. Microbiol. 2019, 57, e01604-18. [Google Scholar] [CrossRef]
- Adams, E.; Quinn, M.; Tsay, S.; Poirot, E.; Chaturvedi, S.; Southwick, K.; Greenko, J.; Fernandez, R.; Kallen, A.; Vallabhaneni, S.; et al. Candida auris in Healthcare Facilities, New York, USA, 2013–2017. Emerg Infect Dis 2018, 24, 1816–1824. [Google Scholar] [CrossRef] [PubMed]
- Berlau, A.; Stoll, S.; Edel, B.; Löffler, B.; Rödel, J. Evaluation of the Eazyplex(®)Candida ID LAMP Assay for the Rapid Diagnosis of Positive Blood Cultures. Diagnostics 2024, 14, 2125. [Google Scholar] [CrossRef] [PubMed]
- Sexton, D.J.; Bentz, M.L.; Welsh, R.M.; Litvintseva, A.P. Evaluation of a new T2 Magnetic Resonance assay for rapid detection of emergent fungal pathogen Candida auris on clinical skin swab samples. Mycoses 2018, 61, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Guidance for Detection of C. auris Colonization. Available online: https://www.cdc.gov/candida-auris/hcp/laboratories/detection-colonization.html (accessed on 30 November 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, R.C.-W.; Lee, A.L.-H.; Cheung, I.Y.-Y.; Chow, V.C.-Y.; Ip, M.; Lai, C.K.-C. Current Updates on Molecular Diagnostic Assays Used for Detection of Candida auris: A Systematic Review. Diagnostics 2025, 15, 140. https://doi.org/10.3390/diagnostics15020140
Wong RC-W, Lee AL-H, Cheung IY-Y, Chow VC-Y, Ip M, Lai CK-C. Current Updates on Molecular Diagnostic Assays Used for Detection of Candida auris: A Systematic Review. Diagnostics. 2025; 15(2):140. https://doi.org/10.3390/diagnostics15020140
Chicago/Turabian StyleWong, River Chun-Wai, Alfred Lok-Hang Lee, Ingrid Yu-Ying Cheung, Viola Chi-Ying Chow, Margaret Ip, and Christopher Koon-Chi Lai. 2025. "Current Updates on Molecular Diagnostic Assays Used for Detection of Candida auris: A Systematic Review" Diagnostics 15, no. 2: 140. https://doi.org/10.3390/diagnostics15020140
APA StyleWong, R. C.-W., Lee, A. L.-H., Cheung, I. Y.-Y., Chow, V. C.-Y., Ip, M., & Lai, C. K.-C. (2025). Current Updates on Molecular Diagnostic Assays Used for Detection of Candida auris: A Systematic Review. Diagnostics, 15(2), 140. https://doi.org/10.3390/diagnostics15020140





