DNA Hypermethylation at the Invasive Front of Oral Squamous Cell Carcinoma Confers Poorly Differentiated Characteristics and Promotes Migration of Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples and Patient Characteristics
2.2. Cell Lines and Culture
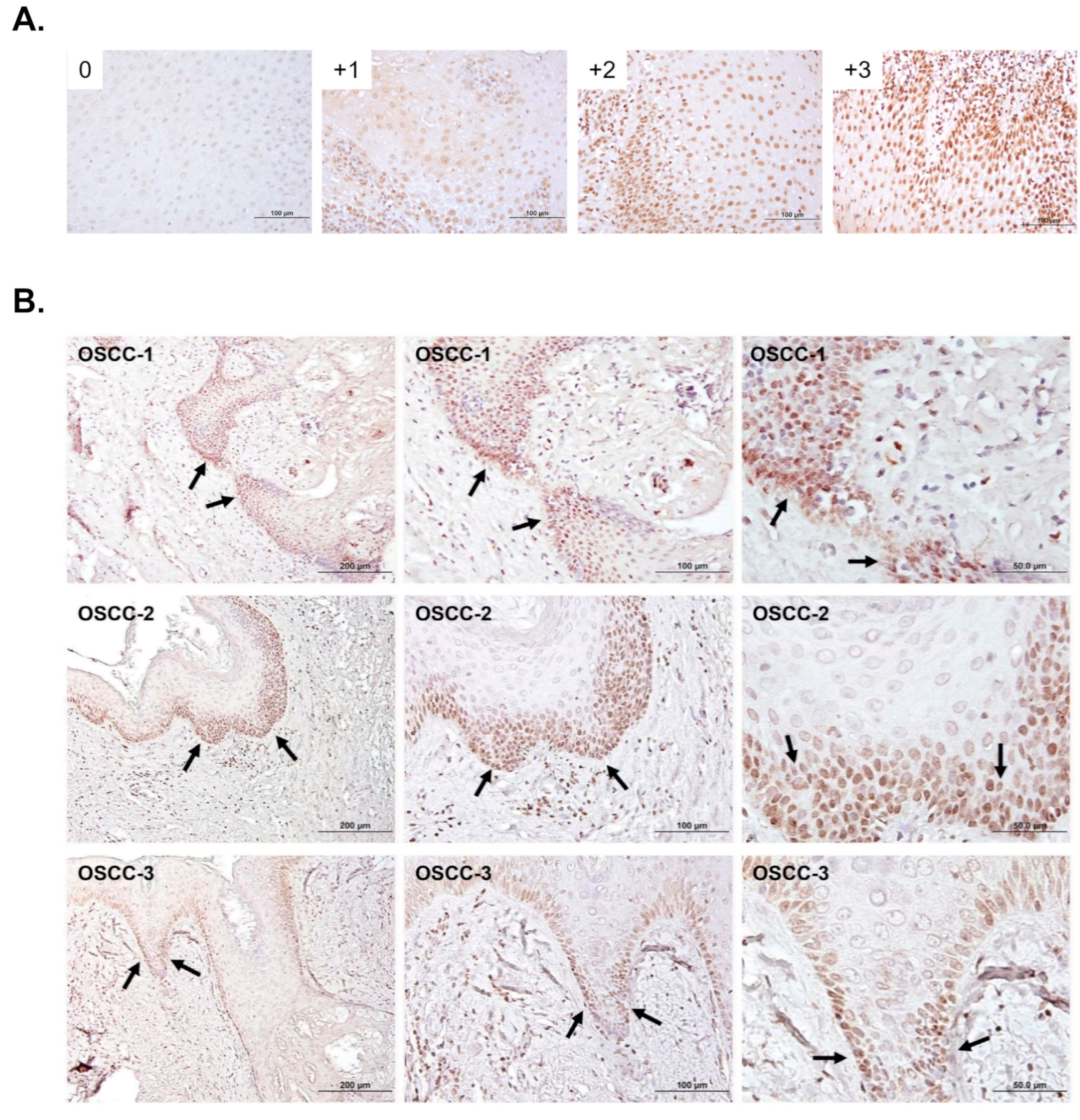
2.3. Immunohistochemical Analysis and Scoring
2.4. In Vitro Wound-Healing Assay
2.5. Genomic DNA Isolation and DNA Methylation Quantification
2.6. Drug Treatment and CAF-Conditioned Medium
2.7. RNA-Sequencing Analysis
2.8. Statistical Analysis
3. Results
3.1. The Association of 5-mC Levels with Clinicopathological Characteristics of OSCC Specimens
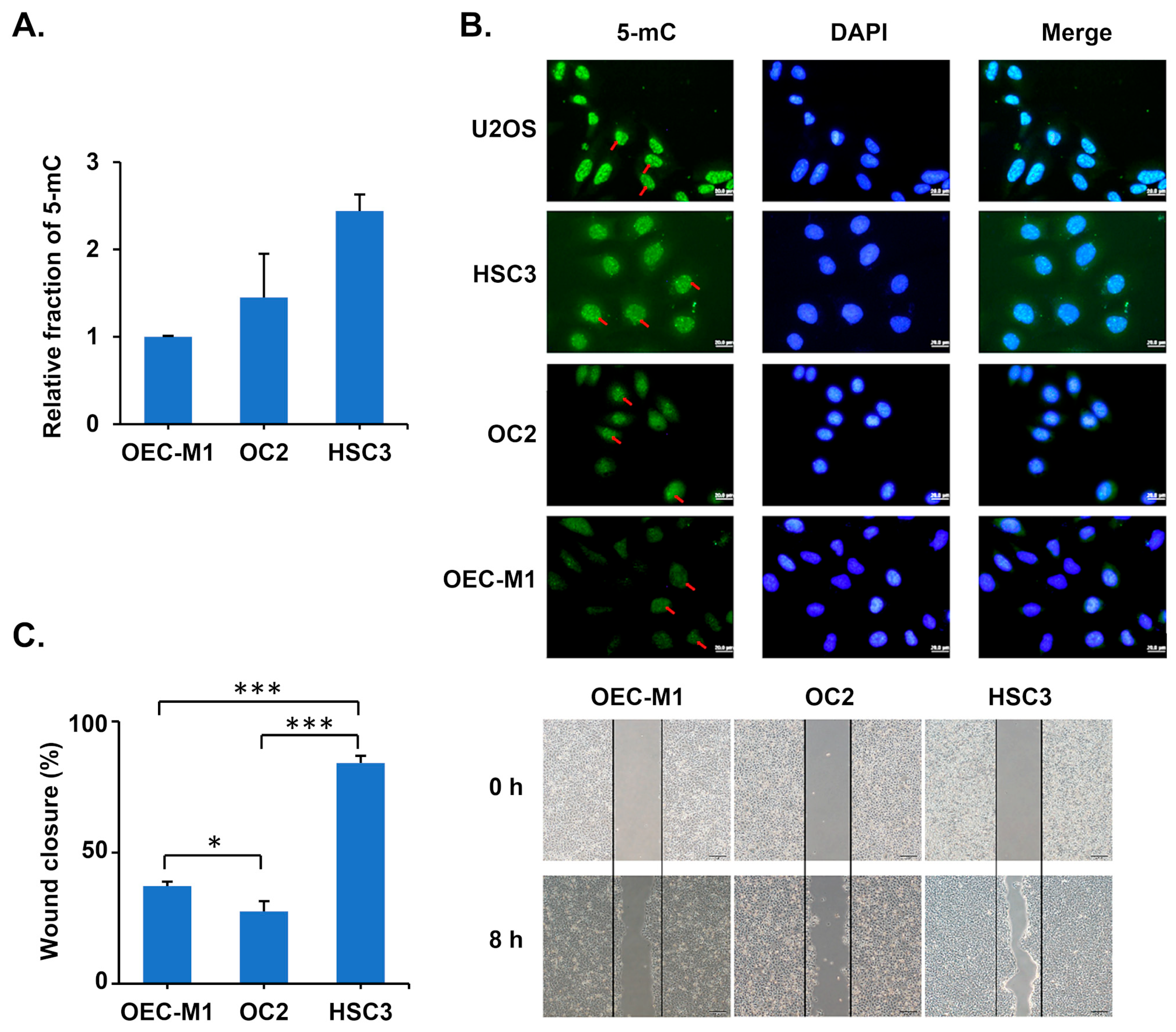
3.2. Oral Cancer Cells with High 5-mC Levels Exhibit High Migration Ability
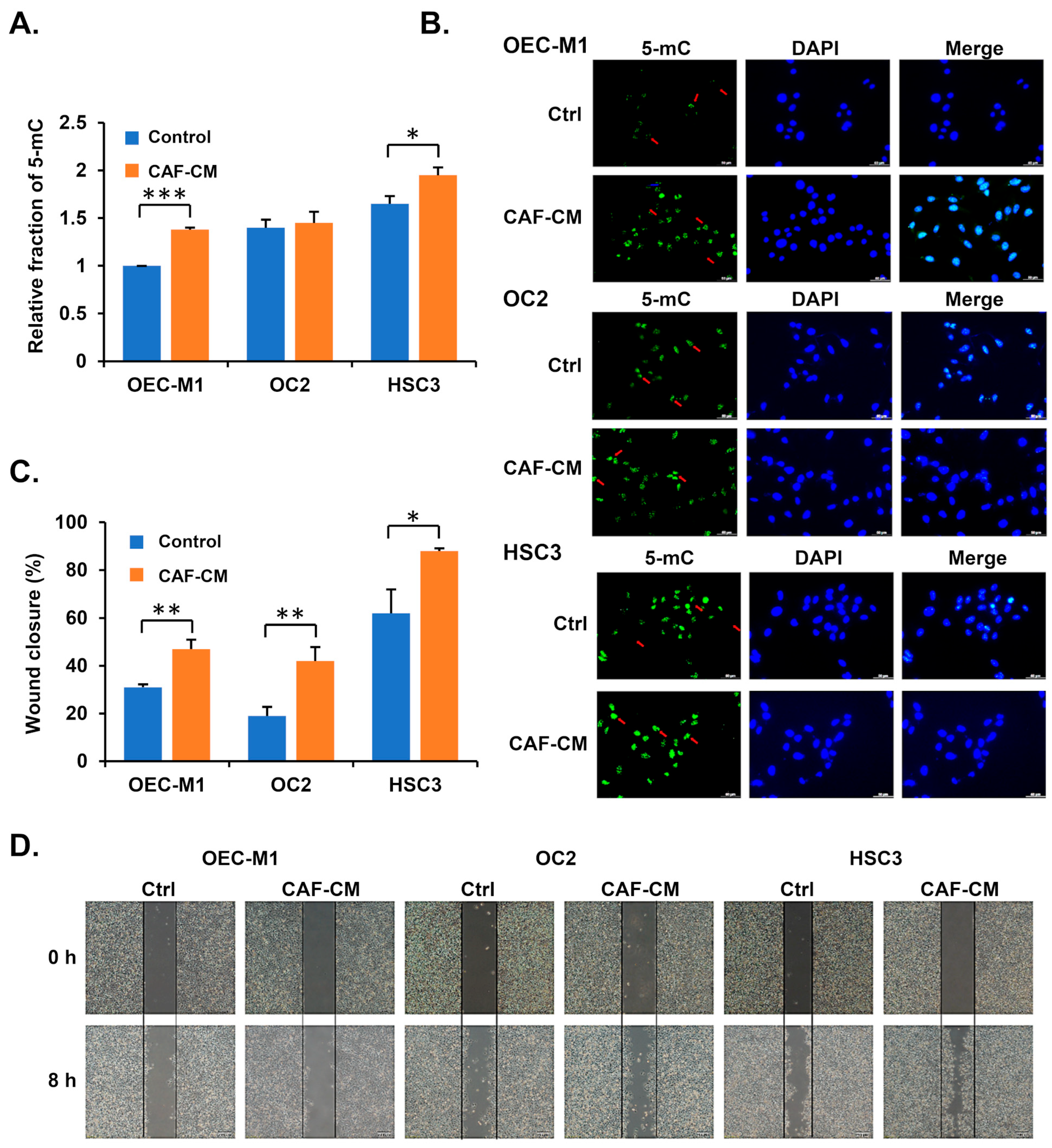
3.3. Cancer-Associated Fibroblasts Increase 5-mC Levels in Oral Cancer Cells
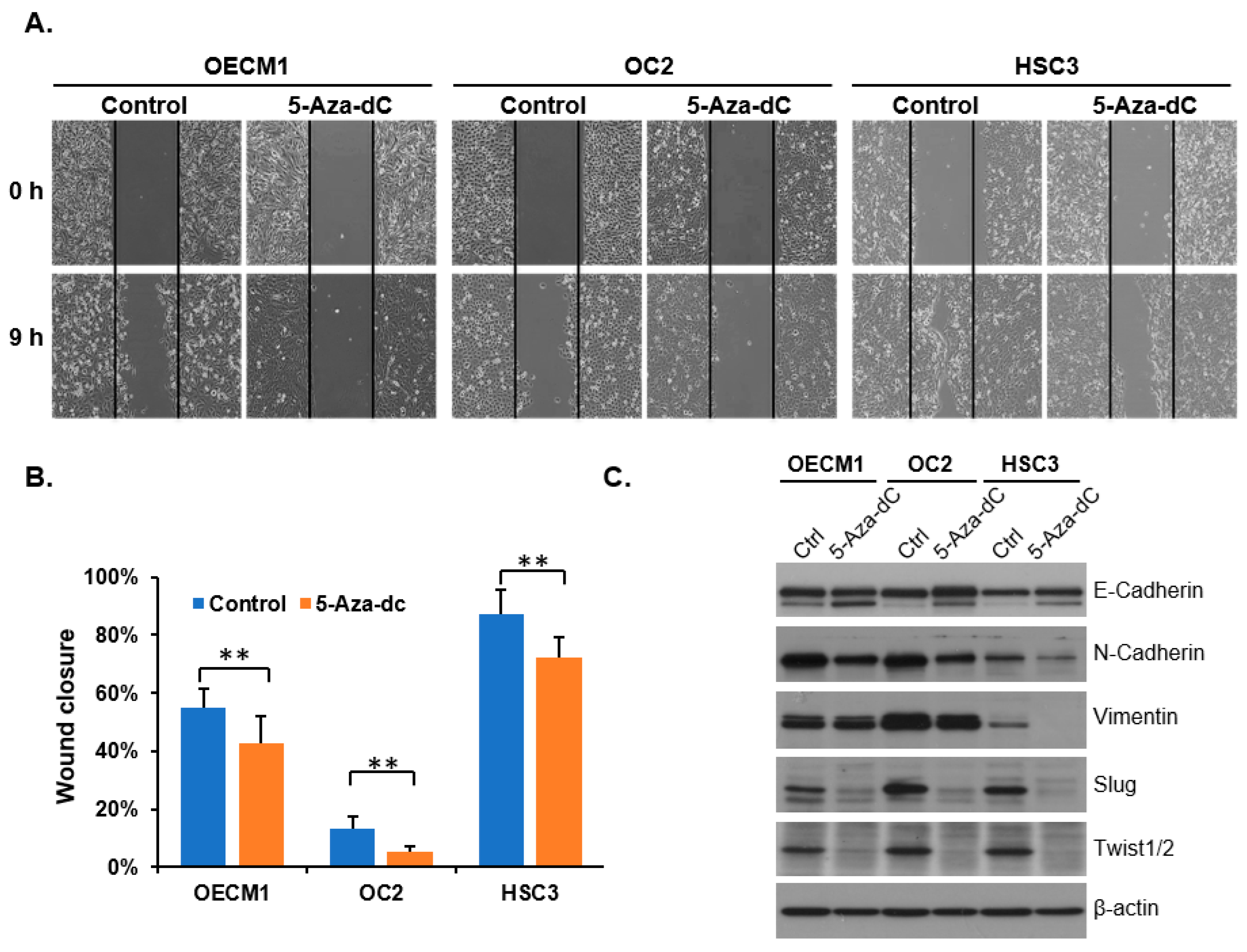
3.4. Decreasing 5-mC Reduces the Migration Ability of OSCC Cells
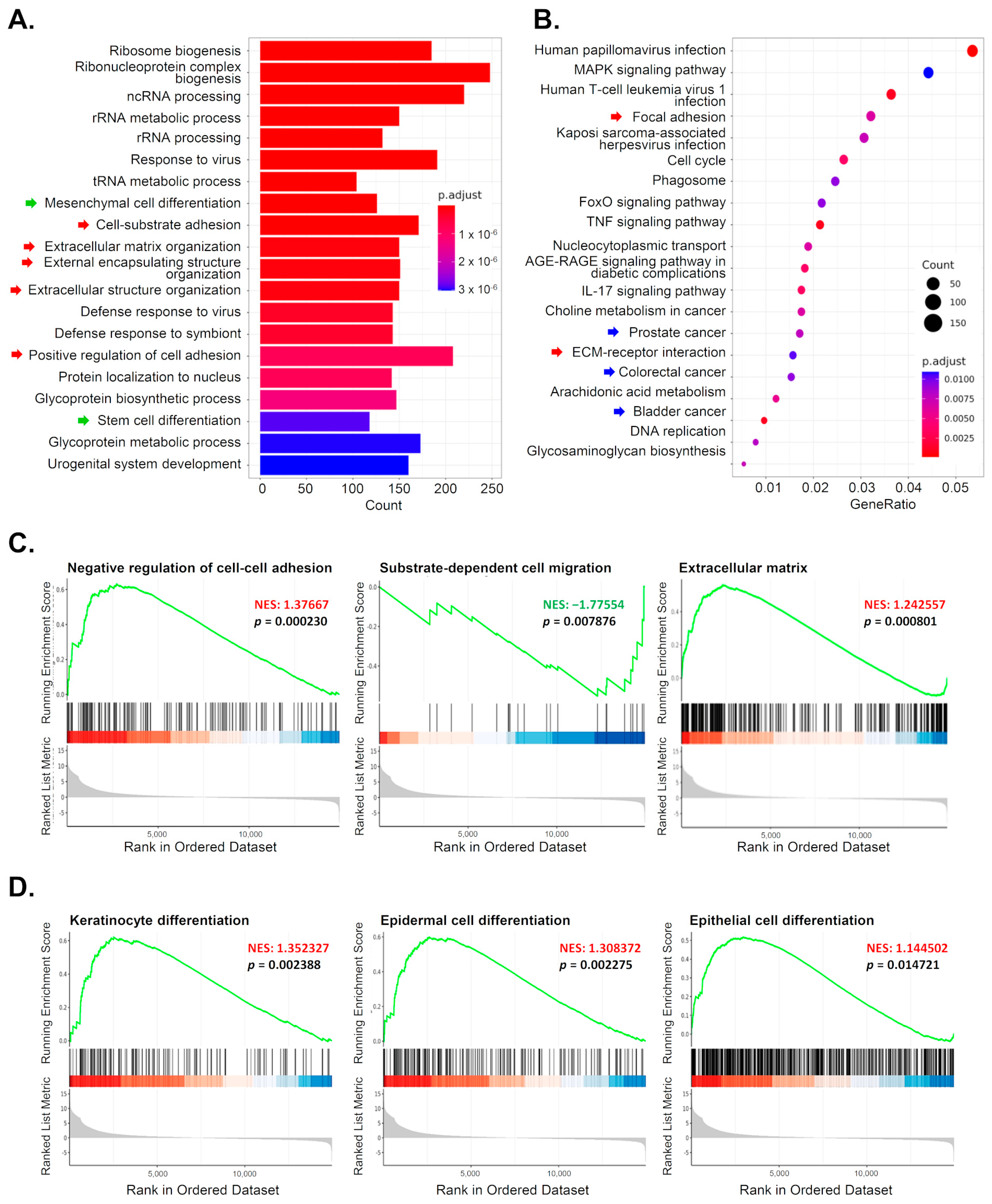
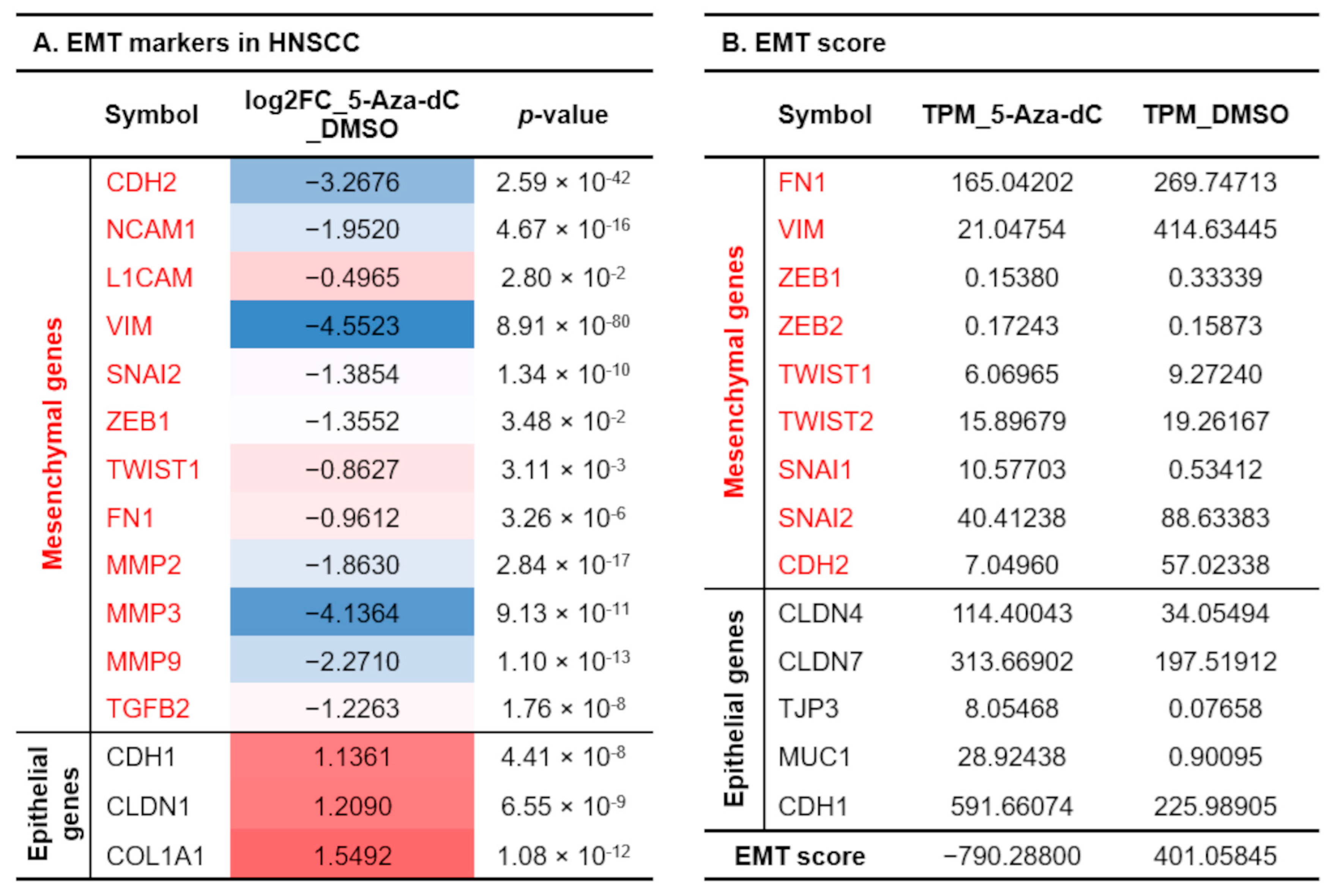
3.5. Reducing 5-mC Affects Gene Expression Related to Cell Differentiation and Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA A Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Bavle, R.M.; Venugopal, R.; Konda, P.; Muniswamappa, S.; Makarla, S. Molecular Classification of Oral Squamous Cell Carcinoma. J. Clin. Diagn. Res. 2016, 10, ZE18–ZE21. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis. 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes. Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef]
- Agarwal, N.; Jha, A.K. DNA hypermethylation of tumor suppressor genes among oral squamous cell carcinoma patients: A prominent diagnostic biomarker. Mol. Biol. Rep. 2024, 52, 44. [Google Scholar] [CrossRef]
- Kim, S.Y.; Han, Y.K.; Song, J.M.; Lee, C.H.; Kang, K.; Yi, J.M.; Park, H.R. Aberrantly hypermethylated tumor suppressor genes were identified in oral squamous cell carcinoma (OSCC). Clin. Epigenetics 2019, 11, 116. [Google Scholar] [CrossRef]
- Flausino, C.S.; Daniel, F.I.; Modolo, F. DNA methylation in oral squamous cell carcinoma: From its role in carcinogenesis to potential inhibitor drugs. Crit. Rev. Oncol. Hematol. 2021, 164, 103399. [Google Scholar] [CrossRef]
- Chen, M.Y.; Chen, J.W.; Wu, L.W.; Huang, K.C.; Chen, J.Y.; Wu, W.S.; Chiang, W.F.; Shih, C.J.; Tsai, K.N.; Hsieh, W.T.; et al. Carcinogenesis of male oral submucous fibrosis alters salivary microbiomes. J. Dent. Res. 2021, 100, 397–405. [Google Scholar] [CrossRef]
- Mazloumi, Z.; Farahzadi, R.; Rafat, A.; Dizaji Asl, K.; Karimipour, M.; Montazer, M.; Movassaghpour, A.A.; Dehnad, A.; Nozad Charoudeh, H. Effect of aberrant DNA methylation on cancer stem cell properties. Exp. Mol. Pathol. 2022, 125, 104757. [Google Scholar] [CrossRef]
- Sato, N.; Fukushima, N.; Hruban, R.H.; Goggins, M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod. Pathol. 2008, 21, 238–244. [Google Scholar] [CrossRef]
- Inchanalkar, M.; Srivatsa, S.; Ambatipudi, S.; Bhosale, P.G.; Patil, A.; Schäffer, A.A.; Beerenwinkel, N.; Mahimkar, M.B. Genome-wide DNA methylation profiling of HPV-negative leukoplakia and gingivobuccal complex cancers. Clin. Epigenetics 2023, 15, 93. [Google Scholar] [CrossRef]
- Berdasco, M.; Esteller, M. Aberrant epigenetic landscape in cancer: How cellular identity goes awry. Dev. Cell 2010, 19, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Delpu, Y.; Cordelier, P.; Cho, W.C.; Torrisani, J. DNA methylation and cancer diagnosis. Int. J. Mol. Sci. 2013, 14, 15029–15058. [Google Scholar] [CrossRef] [PubMed]
- Leitheiser, M.; Capper, D.; Seegerer, P.; Lehmann, A.; Schüller, U.; Müller, K.R.; Klauschen, F.; Jurmeister, P.; Bockmayr, M. Machine learning models predict the primary sites of head and neck squamous cell carcinoma metastases based on DNA methylation. J. Pathol. 2022, 256, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Liouta, G.; Adamaki, M.; Tsintarakis, A.; Zoumpourlis, P.; Liouta, A.; Agelaki, S.; Zoumpourlis, V. DNA Methylation as a Diagnostic, Prognostic, and Predictive Biomarker in Head and Neck Cancer. Int. J. Mol. Sci. 2023, 24, 2996. [Google Scholar] [CrossRef]
- Sun, X.X.; Yu, Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol. Sin. 2015, 36, 1219–1227. [Google Scholar] [CrossRef]
- Tung, C.H.; Wu, J.E.; Huang, M.F.; Wang, W.L.; Wu, Y.Y.; Tsai, Y.T.; Hsu, X.R.; Lin, S.H.; Chen, Y.L.; Hong, T.M. Ubiquitin-specific peptidase 5 facilitates cancer stem cell-like properties in lung cancer by deubiquitinating β-catenin. Cancer Cell Int. 2023, 23, 207. [Google Scholar] [CrossRef]
- Wu, M.H.; Hong, H.C.; Hong, T.M.; Chiang, W.F.; Jin, Y.T.; Chen, Y.L. Targeting galectin-1 in carcinoma-associated fibroblasts inhibits oral squamous cell carcinoma metastasis by downregulating MCP-1/CCL2 expression. Clin. Cancer Res. 2011, 17, 1306–1316. [Google Scholar] [CrossRef]
- Hsu, X.R.; Wu, J.E.; Wu, Y.Y.; Hsiao, S.Y.; Liang, J.L.; Wu, Y.J.; Tung, C.H.; Huang, M.F.; Lin, M.S.; Yang, P.C.; et al. Exosomal long noncoding RNA MLETA1 promotes tumor progression and metastasis by regulating the miR-186-5p/EGFR and miR-497-5p/IGF1R axes in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2023, 42, 283. [Google Scholar] [CrossRef]
- Wu, M.H.; Hong, T.M.; Cheng, H.W.; Pan, S.H.; Liang, Y.R.; Hong, H.C.; Chiang, W.F.; Wong, T.Y.; Shieh, D.B.; Shiau, A.L.; et al. Galectin-1-mediated tumor invasion and metastasis, up-regulated matrix metalloproteinase expression, and reorganized actin cytoskeletons. Mol. Cancer Res. 2009, 7, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Pawlicka, M.; Gumbarewicz, E.; Błaszczak, E.; Stepulak, A. Transcription Factors and Markers Related to Epithelial-Mesenchymal Transition and Their Role in Resistance to Therapies in Head and Neck Cancers. Cancers 2024, 16, 1354. [Google Scholar] [CrossRef] [PubMed]
- Janiszewska, M.; Primi, M.C.; Izard, T. Cell adhesion in cancer: Beyond the migration of single cells. J. Biol. Chem. 2020, 295, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Drummond, M.L.; Guerrero-Juarez, C.F.; Tarapore, E.; MacLean, A.L.; Stabell, A.R.; Wu, S.C.; Gutierrez, G.; That, B.T.; Benavente, C.A.; et al. Single cell transcriptomics of human epidermis identifies basal stem cell transition states. Nat. Commun. 2020, 11, 4239. [Google Scholar] [CrossRef]
- Salt, M.B.; Bandyopadhyay, S.; McCormick, F. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 2014, 4, 86–99. [Google Scholar] [CrossRef]
- Towle, R.; Truong, D.; Hogg, K.; Robinson, W.P.; Poh, C.F.; Garnis, C. Global analysis of DNA methylation changes during progression of oral cancer. Oral. Oncol. 2013, 49, 1033–1042. [Google Scholar] [CrossRef]
- Rivera-Peña, B.; Folawiyo, O.; Turaga, N.; Rodríguez-Benítez, R.J.; Felici, M.E.; Aponte-Ortiz, J.A.; Pirini, F.; Rodríguez-Torres, S.; Vázquez, R.; López, R.; et al. Promoter DNA methylation patterns in oral, laryngeal and oropharyngeal anatomical regions are associated with tumor differentiation, nodal involvement and survival. Oncol. Lett. 2024, 27, 89. [Google Scholar] [CrossRef]
- Brocks, D.; Assenov, Y.; Minner, S.; Bogatyrova, O.; Simon, R.; Koop, C.; Oakes, C.; Zucknick, M.; Lipka, D.B.; ICGC Early Onset Prostate Cancer Project; et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep. 2014, 8, 798–806. [Google Scholar] [CrossRef]
- Dong, N.; Shi, L.; Wang, D.C.; Chen, C.; Wang, X. Role of epigenetics in lung cancer heterogeneity and clinical implication. Semin. Cell Dev. Biol. 2017, 64, 18–25. [Google Scholar] [CrossRef]
- Kumar, R.; Liu, A.P.Y.; Orr, B.A.; Northcott, P.A.; Robinson, G.W. Advances in the classification of pediatric brain tumors through DNA methylation profiling: From research tool to frontline diagnostic. Cancer 2018, 124, 4168–4180. [Google Scholar] [CrossRef]
- Nowak, E.; Bednarek, I. Aspects of the Epigenetic Regulation of EMT Related to Cancer Metastasis. Cells 2021, 10, 3435. [Google Scholar] [CrossRef]
- Wen, G.; Wang, H.; Zhong, Z. Associations of RASSF1A, RARβ, and CDH1 promoter hypermethylation with oral cancer risk: A PRISMA-compliant meta-analysis. Medicine 2018, 97, e9971. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, K.; Leach, R. 5-Aza-dC treatment induces mesenchymal-to-epithelial transition in 1st trimester trophoblast cell line HTR8/SVneo. Biochem. Biophys. Res. Commun. 2013, 432, 116–122. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mesgari, H.; Esmaelian, S.; Nasiri, K.; Ghasemzadeh, S.; Doroudgar, P.; Payandeh, Z. Epigenetic Regulation in Oral Squamous Cell Carcinoma Microenvironment: A Comprehensive Review. Cancers 2023, 15, 5600. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.Y.; Chen, M.Y.; Chang, Y.H.; Huang, J.S.; Huang, T.T.; Wong, T.Y.; Hong, T.M.; Chen, Y.L. Growth-regulated oncogene-α from oral submucous fibrosis fibroblasts promotes malignant transformation of oral precancerous cells. J. Oral. Pathol. Med. 2018, 47, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, Y.; Weeraratna, A.T. Fibroblasts in cancer: Unity in heterogeneity. Cell 2023, 186, 1580–1609. [Google Scholar] [CrossRef]
- Ramón y Cajal, S.; Sesé, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellví, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef]
- Costa, P.M.D.S.; Sales, S.L.A.; Pinheiro, D.P.; Pontes, L.Q.; Maranhão, S.S.; Pessoa, C.D.Ó.; Furtado, G.P.; Furtado, C.L.M. Epigenetic reprogramming in cancer: From diagnosis to treatment. Front. Cell Dev. Biol. 2023, 11, 1116805. [Google Scholar] [CrossRef]
- Okada, N.; Steinberg, M.L.; Defendi, V. Re-expression of differentiated properties in SV40-infected human epidermal keratinocytes induced by 5-azacytidine. Exp. Cell Res. 1984, 153, 198–207. [Google Scholar] [CrossRef]
- Köhler, F.; Rodríguez-Paredes, M. DNA Methylation in Epidermal Differentiation, Aging, and Cancer. J. Investig. Dermatol. 2020, 140, 38–47. [Google Scholar] [CrossRef]
- Ehrlich, M.; Lacey, M. DNA methylation and differentiation: Silencing, upregulation and modulation of gene expression. Epigenomics 2013, 5, 553–568. [Google Scholar] [CrossRef]
- Valenzuela, M.A.; Canales, J.; Corvalán, A.H.; Quest, A.F. Helicobacter pylori-induced inflammation and epigenetic changes during gastric carcinogenesis. World J. Gastroenterol. 2015, 21, 12742–12756. [Google Scholar] [CrossRef]
- Tolg, C.; Sabha, N.; Cortese, R.; Panchal, T.; Ahsan, A.; Soliman, A.; Aitken, K.J.; Petronis, A.; Bägli, D.J. Uropathogenic E. coli infection provokes epigenetic downregulation of CDKN2A (p16INK4A) in uroepithelial cells. Lab. Investig. 2011, 91, 825–836. [Google Scholar] [CrossRef]
- Qin, W.; Scicluna, B.P.; van der Poll, T. The Role of Host Cell DNA Methylation in the Immune Response to Bacterial Infection. Front. Immunol. 2021, 12, 696280. [Google Scholar] [CrossRef]
- Yin, L.; Chung, W.O. Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol. 2011, 4, 409–419. [Google Scholar] [CrossRef]
- Fernández-Ramos, D.; Lopitz-Otsoa, F.; Lu, S.C.; Mato, J.M. S-Adenosylmethionine: A Multifaceted Regulator in Cancer Pathogenesis and Therapy. Cancers 2025, 17, 535. [Google Scholar] [CrossRef]
| Clinicopathological Parameters | Number of Patients | 5-Methylcytosine Level | p-Value | |
|---|---|---|---|---|
| Low No = 52 | High No = 58 | Fisher’s Exact Test | ||
| Age (years) | ||||
| >50 | 61 | 31 | 30 | 0.4462 |
| ≤50 | 49 | 21 | 28 | |
| Tumor stage | ||||
| T1–T2 | 85 | 38 | 47 | 0.367 |
| T3–T4 | 25 | 14 | 11 | |
| Differentiation # | ||||
| Gx-G1 | 71 | 41 | 30 | 0.0081 ** |
| ≥G2 | 37 | 11 | 26 | |
| N stage | ||||
| N (−) | 81 | 40 | 41 | 0.8234 |
| N (+) | 26 | 12 | 14 | |
| Oral submucous fibrosis | ||||
| Yes | 52 | 23 | 29 | 0.5713 |
| No | 58 | 29 | 29 | |
| Site of tumor | ||||
| Buccal mucosa | 44 | 24 | 20 | |
| Gingiva/Mouth floor/Lip | 28 | 11 | 17 | 0.6223 |
| Tongue/Others | 37 | 17 | 20 | |
| Gender | ||||
| Male | 106 | 50 | 56 | 1 |
| Female | 4 | 2 | 2 | |
| Basal keratinocyte markers | Spinous cell markers | Granular keratinocyte markers | ||||||
|---|---|---|---|---|---|---|---|---|
| Symbol | log2FC_5-Aza-dC _DMSO | p-Value | Symbol | log2FC_5-Aza-dC _DMSO | p-Value | Symbol | log2FC_5-Aza-dC _DMSO | p-Value |
| KRT5 | −0.6006 | 3.42 × 10−3 | KRT1 | 4.5040 | 1.52 × 10−17 | IVL | 1.7620 | 2.12 × 10−5 |
| CDC20 | −0.6351 | 2.17 × 10−3 | CDH1 | 1.1361 | 4.41 × 10−8 | ZNF750 | 3.3005 | 7.56 × 10−33 |
| RRM2 | −0.4995 | 1.56 × 10−2 | DEFB1 | 7.8417 | 6.17 × 10−43 | SPINK5 | 0.9962 | 1.08 × 10−4 |
| HELLS | −0.4273 | 4.45 × 10−2 | FXYD3 | 2.1689 | 2.56 × 10−17 | CALML5 | 4.8699 | 1.28 × 10−55 |
| UHRF1 | −0.7751 | 2.41 × 10−4 | CCND1 | −0.8113 | 8.82 × 10−5 | |||
| COL17A1 | −0.5664 | 6.05 × 10−3 | ||||||
| GJB2 | 0.9503 | 4.27 × 10−6 | ||||||
| KRT16 | 2.4534 | 1.82 × 10−28 | ||||||
| ASS1 | 3.0584 | 5.59 × 10−24 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.-P.; Li, C.-Y.; Wu, Y.-H.; Chen, M.-Y.; Hsieh, Y.-P.; Huang, T.-T.; Hong, T.-M.; Chen, Y.-L. DNA Hypermethylation at the Invasive Front of Oral Squamous Cell Carcinoma Confers Poorly Differentiated Characteristics and Promotes Migration of Cancer Cells. Diagnostics 2025, 15, 2477. https://doi.org/10.3390/diagnostics15192477
Wang L-P, Li C-Y, Wu Y-H, Chen M-Y, Hsieh Y-P, Huang T-T, Hong T-M, Chen Y-L. DNA Hypermethylation at the Invasive Front of Oral Squamous Cell Carcinoma Confers Poorly Differentiated Characteristics and Promotes Migration of Cancer Cells. Diagnostics. 2025; 15(19):2477. https://doi.org/10.3390/diagnostics15192477
Chicago/Turabian StyleWang, Li-Po, Chien-Ya Li, Yu-Hsueh Wu, Meng-Yen Chen, Yi-Ping Hsieh, Tze-Ta Huang, Tse-Ming Hong, and Yuh-Ling Chen. 2025. "DNA Hypermethylation at the Invasive Front of Oral Squamous Cell Carcinoma Confers Poorly Differentiated Characteristics and Promotes Migration of Cancer Cells" Diagnostics 15, no. 19: 2477. https://doi.org/10.3390/diagnostics15192477
APA StyleWang, L.-P., Li, C.-Y., Wu, Y.-H., Chen, M.-Y., Hsieh, Y.-P., Huang, T.-T., Hong, T.-M., & Chen, Y.-L. (2025). DNA Hypermethylation at the Invasive Front of Oral Squamous Cell Carcinoma Confers Poorly Differentiated Characteristics and Promotes Migration of Cancer Cells. Diagnostics, 15(19), 2477. https://doi.org/10.3390/diagnostics15192477






