Laboratory Validation of a Fully Automated Point-of-Care Device for High-Order Multiplexing Real-Time PCR Detection of Respiratory Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. POCm Cartridge and Analyzer
2.2. POCm Testing Procedures
2.3. Assay Panel Design
2.4. PCR
2.5. Manually Performed RT-PCR Tests
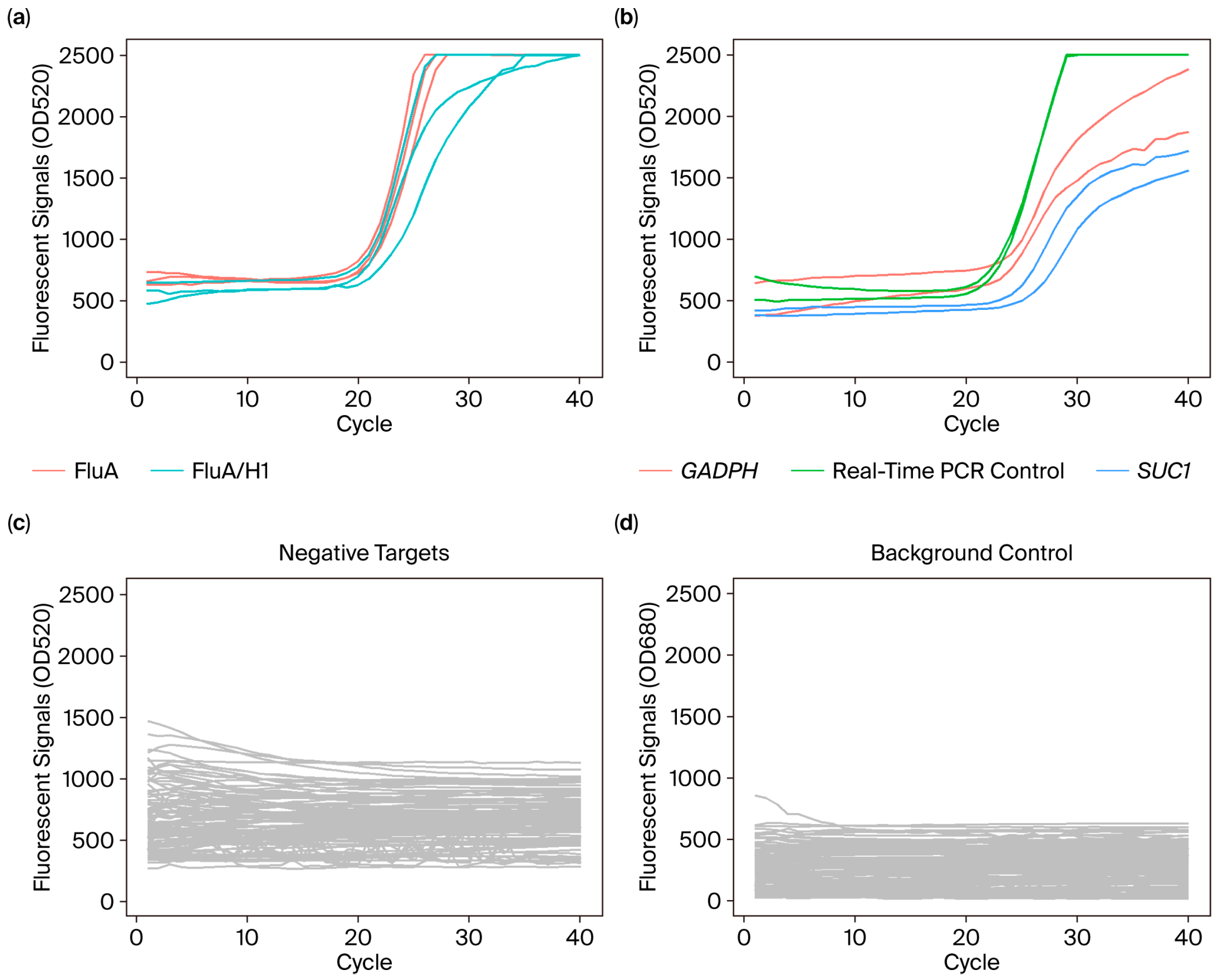
2.6. Preparation of Reference Controls
2.7. Evaluation with Clinical Samples
2.8. Statistical Analysis
3. Results and Discussion
3.1. Selection of Respiratory Pathogens for Multiplex Assay Panel
3.2. Analytical Detectability
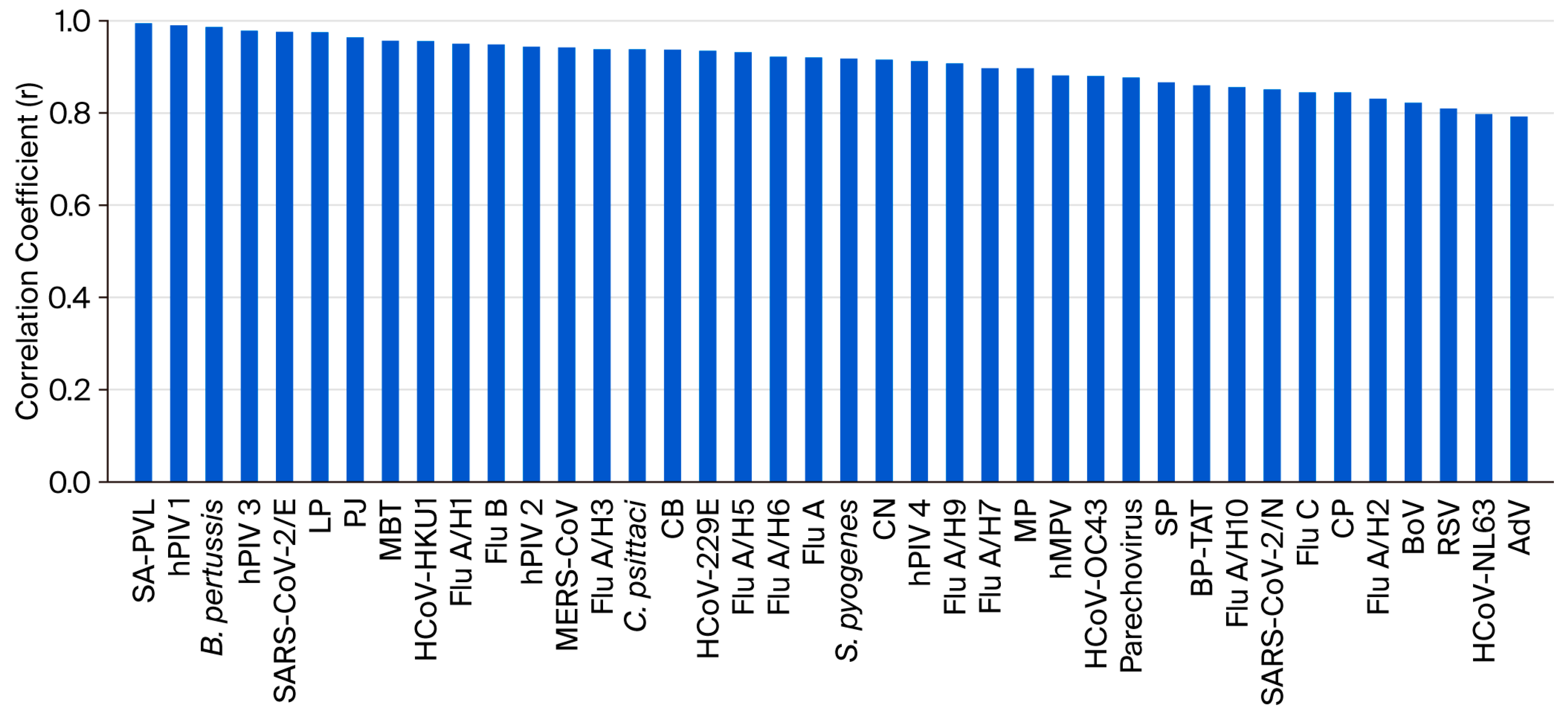
3.3. Linearity
3.4. Clinical Sample Evaluation
3.5. Pathogen Co-Infection Detection
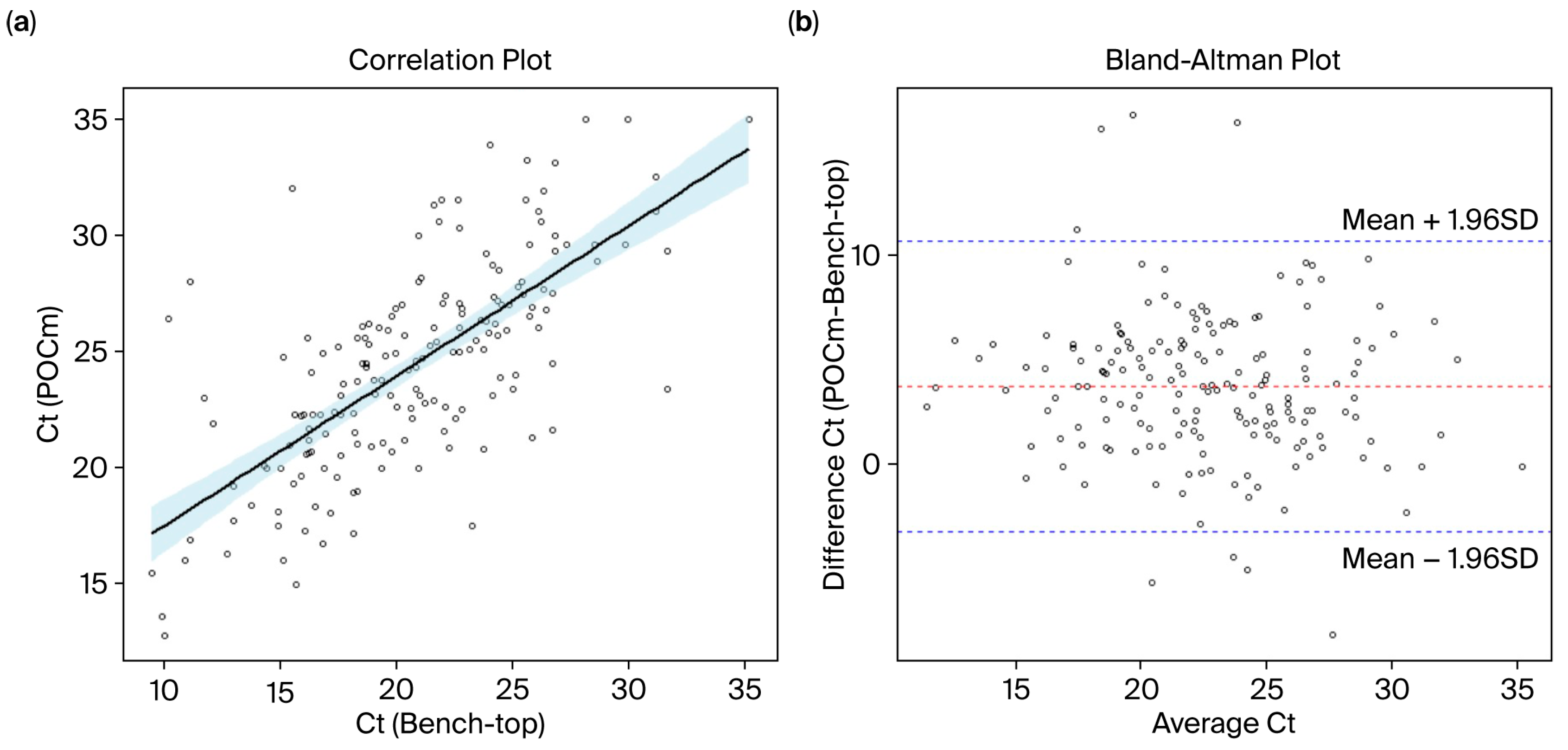
3.6. Measurement Agreement of Ct Values
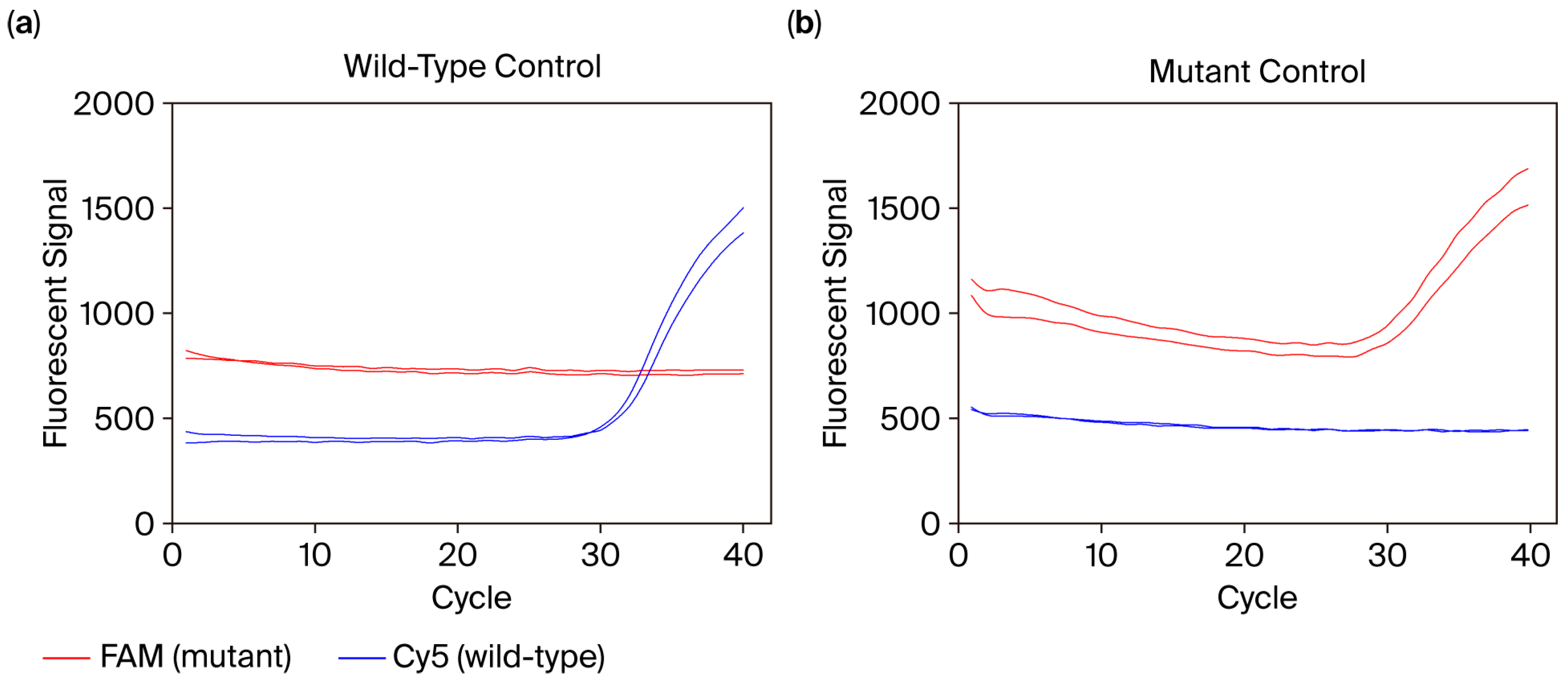
3.7. Mutation Genotyping
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-Care Testing for Infectious Diseases: Past, Present, and Future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef]
- Niemz, A.; Ferguson, T.M.; Boyle, D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011, 29, 240–250. [Google Scholar] [CrossRef]
- Nelson, P.P.; Rath, B.A.; Fragkou, P.C.; Antalis, E.; Tsiodras, S.; Skevaki, C. Current and Future Point-of-Care Tests for Emerging and New Respiratory Viruses and Future Perspectives. Front Cell Infect Microbiol. 2020, 10, 181. [Google Scholar] [CrossRef]
- Easley, C.J.; Karlinsey, J.M.; Bienvenue, J.M.; Legendre, L.A.; Roper, M.G.; Feldman, S.H.; Hughes, M.A.; Hewlett, E.L.; Merkel, T.J.; Ferrance, J.P.; et al. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc. Natl. Acad. Sci. USA 2006, 103, 19272–19277. [Google Scholar] [CrossRef]
- Charlton, C.L.; Babady, E.; Ginocchio, C.C.; Hatchette, T.F.; Jerris, R.C.; Li, Y.; Loeffelholz, M.; McCarter, Y.S.; Miller, M.B.; Novak-Weekley, S.; et al. Practical Guidance for Clinical Microbiology Laboratories: Viruses Causing Acute Respiratory Tract Infections. Clin. Microbiol. Rev. 2019, 32, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Tsang, P.Y.L.; Chu, S.L.H.; Li, L.C.W.; Tai, D.M.S.; Cheung, B.K.C.; Kebede, F.T.; Leung, P.Y.M.; Wong, W.; Chung, T.; Yip, C.C.Y.; et al. Automated System for Multiplexing Detection of COVID-19 and Other Respiratory Pathogens. IEEE J. Transl. Eng. Health Med. 2023, 11, 424–434. [Google Scholar] [CrossRef]
- Khoury, M.J.; Iademarco, M.F.; Riley, W.T. Precision Public Health for the Era of Precision Medicine. Am. J. Prev. Med. 2016, 50, 398–401. [Google Scholar] [CrossRef]
- Gardy, J.L.; Loman, N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018, 19, 9–20. [Google Scholar] [CrossRef]
- Sozhamannan, S.; Holland, M.Y.; Hall, A.T.; Negron, D.A.; Ivancich, M.; Koehler, J.W.; Minogue, T.D.; Campbell, C.E.; Berger, W.J.; Christopher, G.W.; et al. Evaluation of Signature Erosion in Ebola Virus Due to Genomic Drift and Its Impact on the Performance of Diagnostic Assays. Viruses 2015, 7, 3130–3154. [Google Scholar] [CrossRef]
- Schmitz, J.E.; Stratton, C.W.; Persing, D.H.; Tang, Y.W. Forty Years of Molecular Diagnostics for Infectious Diseases. J. Clin. Microbiol. 2022, 60, e0244621. [Google Scholar] [CrossRef]
- Hayden, M.K.; Hanson, K.E.; Englund, J.A.; Lee, M.J.; Loeb, M.; Lee, F.; Morgan, D.J.; Patel, R.; El Mikati, I.K.; Iqneibi, S.; et al. The Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19: Molecular Diagnostic Testing (December 2023). Clin. Infect. Dis. 2024, 78, e385–e415. [Google Scholar] [CrossRef]
- Uyeki, T.M.; Bernstein, H.H.; Bradley, J.S.; Englund, J.A.; File, T.M.; Fry, A.M.; Gravenstein, S.; Hayden, F.G.; Harper, S.A.; Hirshon, J.M.; et al. Clinical Practice Guidelines by the Infectious Diseases Society of America: 2018 Update on Diagnosis, Treatment, Chemoprophylaxis, and Institutional Outbreak Management of Seasonal Influenzaa. Clin. Infect. Dis. 2019, 68, 895–902. [Google Scholar] [CrossRef]
- Force, U.S.P.S.T.; Davidson, K.W.; Barry, M.J.; Mangione, C.M.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; Krist, A.H.; et al. Screening for Chlamydia and Gonorrhea: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 326, 949–956. [Google Scholar] [CrossRef]
- Shane, A.L.; Mody, R.K.; Crump, J.A.; Tarr, P.I.; Steiner, T.S.; Kotloff, K.; Langley, J.M.; Wanke, C.; Warren, C.A.; Cheng, A.C.; et al. 2017 Infectious Diseases Society of America Clinical Practice Guidelines for the Diagnosis and Management of Infectious Diarrhea. Clin. Infect. Dis. 2017, 65, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.M.; Timbrook, T.T.; Hommel, B.; Prinzi, A.M. Breaking Boundaries in Pneumonia Diagnostics: Transitioning from Tradition to Molecular Frontiers with Multiplex PCR. Diagnostics 2024, 14, 752. [Google Scholar] [CrossRef]
- Samuel, L. Direct-from-Blood Detection of Pathogens: A Review of Technology and Challenges. J. Clin. Microbiol. 2023, 61, e0023121. [Google Scholar] [CrossRef]
- Ji, M.; Xia, Y.; Loo, J.F.; Li, L.; Ho, H.P.; He, J.; Gu, D. Automated multiplex nucleic acid tests for rapid detection of SARS-CoV-2, influenza A and B infection with direct reverse-transcription quantitative PCR (dirRT-qPCR) assay in a centrifugal microfluidic platform. RSC Adv. 2020, 10, 34088–34098. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, Y.; Su, X.; Liu, H.; Chen, H.; Chen, Z.; Jin, L.; He, N. A miniaturized and integrated dual-channel fluorescence module for multiplex real-time PCR in the portable nucleic acid detection system. Front Bioeng. Biotechnol. 2022, 10, 996456. [Google Scholar] [CrossRef]
- Shrestha, K.; Kim, S.; Han, J.; Florez, G.M.; Truong, H.; Hoang, T.; Parajuli, S.; Am, T.; Kim, B.; Jung, Y.; et al. Mobile Efficient Diagnostics of Infectious Diseases via On-Chip RT-qPCR: MEDIC-PCR. Adv. Sci. 2023, 10, e2302072. [Google Scholar] [CrossRef]
- Nguyen, P.Q.M.; Wang, M.; Ann Maria, N.; Li, A.Y.; Tan, H.Y.; Xiong, G.M.; Tan, M.M.; Bhagat, A.A.S.; Ong, C.W.M.; Lim, C.T. Modular micro-PCR system for the onsite rapid diagnosis of COVID-19. Microsyst. Nanoeng. 2022, 8, 82. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Alenazi, T.H.; AlOmaim, W.S.; Wazzan, A.; Alsufayan, A.; Hasanain, R.A.; Aldibasi, O.S.; Althawadi, S.; Altamimi, S.A.; Mutabagani, M.; et al. Portable RT-PCR System: A Rapid and Scalable Diagnostic Tool for COVID-19 Testing. J. Clin. Microbiol. 2021, 59, e03004-20. [Google Scholar] [CrossRef]
- Gradisteanu Pircalabioru, G.; Iliescu, F.S.; Mihaescu, G.; Cucu, A.I.; Ionescu, O.N.; Popescu, M.; Simion, M.; Burlibasa, L.; Tica, M.; Chifiriuc, M.C.; et al. Advances in the Rapid Diagnostic of Viral Respiratory Tract Infections. Front Cell Infect. Microbiol. 2022, 12, 807253. [Google Scholar] [CrossRef]
- Costales, C.; Crump, J.A.; Mremi, A.R.; Amsi, P.T.; Kalengo, N.H.; Kilonzo, K.G.; Kinabo, G.; Lwezaula, B.F.; Lyamuya, F.; Marandu, A.; et al. Performance of Xpert Ultra nasopharyngeal swab for identification of tuberculosis deaths in northern Tanzania. Clin. Microbiol. Infect. 2022, 28, 1150.E1–1150.E6. [Google Scholar] [CrossRef]
- To, K.K.; Wong, S.C.; Xu, T.; Poon, R.W.; Mok, K.Y.; Chan, J.F.; Cheng, V.C.; Chan, K.H.; Hung, I.F.; Yuen, K.Y. Use of nasopharyngeal aspirate for diagnosis of pneumocystis pneumonia. J. Clin. Microbiol. 2013, 51, 1570–1574. [Google Scholar] [CrossRef]
- Bujang, M.A.; Adnan, T.H. Requirements for Minimum Sample Size for Sensitivity and Specificity Analysis. J. Clin. Diagn. Res. 2016, 10, YE01–YE06. [Google Scholar] [CrossRef]
- Chan, W.S.; Yau, S.K.; To, M.Y.; Leung, S.M.; Wong, K.P.; Lai, K.C.; Wong, C.Y.; Leung, C.P.; Au, C.H.; Wan, T.S.; et al. The Seasonality of Respiratory Viruses in a Hong Kong Hospital, 2014-2023. Viruses 2023, 15, 1820. [Google Scholar] [CrossRef]
- Poritz, M.A.; Blaschke, A.J.; Byington, C.L.; Meyers, L.; Nilsson, K.; Jones, D.E.; Thatcher, S.A.; Robbins, T.; Lingenfelter, B.; Amiott, E.; et al. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: Development and application to respiratory tract infection. PLoS ONE 2011, 6, e26047. [Google Scholar] [CrossRef]
- Kim, D.; Quinn, J.; Pinsky, B.; Shah, N.H.; Brown, I. Rates of Co-infection Between SARS-CoV-2 and Other Respiratory Pathogens. JAMA 2020, 323, 2085–2086. [Google Scholar] [CrossRef]
- Musuuza, J.S.; Watson, L.; Parmasad, V.; Putman-Buehler, N.; Christensen, L.; Safdar, N. Prevalence and outcomes of co-infection and superinfection with SARS-CoV-2 and other pathogens: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0251170. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ling, L.; Wong, S.H.; Wang, M.H.; Fitzgerald, J.R.; Zou, X.; Fang, S.; Liu, X.; Wang, X.; Hu, W.; et al. Outcomes of respiratory viral-bacterial co-infection in adult hospitalized patients. EClinicalMedicine 2021, 37, 100955. [Google Scholar] [CrossRef] [PubMed]
- Vasireddy, D.; Vanaparthy, R.; Mohan, G.; Malayala, S.V.; Atluri, P. Review of COVID-19 Variants and COVID-19 Vaccine Efficacy: What the Clinician Should Know? J. Clin. Med. Res. 2021, 13, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; de Oliveira, T.; Kosakovsky Pond, S.L.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Bender, A.T.; Boyle, D.S.; Drain, P.K.; Posner, J.D. Current state of commercial point-of-care nucleic acid tests for infectious diseases. Analyst 2021, 146, 2449–2462. [Google Scholar] [CrossRef] [PubMed]
- Lansbury, L.; Lim, B.; Baskaran, V.; Lim, W.S. Co-infections in people with COVID-19: A systematic review and meta-analysis. J. Infect. 2020, 81, 266–275. [Google Scholar] [CrossRef]
- Lai, C.C.; Wang, C.Y.; Hsueh, P.R. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infect. 2020, 53, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.W.; Ewings, S.; Medina, M.J.; Batham, S.; Curran, M.D.; Parmar, S.; Nicholson, K.G. Viral load is strongly associated with length of stay in adults hospitalised with viral acute respiratory illness. J. Infect. 2016, 73, 598–606. [Google Scholar] [CrossRef]
- Gu, L.; Qu, J.; Sun, B.; Yu, X.; Li, H.; Cao, B. Sustained Viremia and High Viral Load in Respiratory Tract Secretions Are Predictors for Death in Immunocompetent Adults with Adenovirus Pneumonia. PLoS ONE 2016, 11, e0160777. [Google Scholar] [CrossRef]
- Nickbakhsh, S.; Thorburn, F.; Mc, M.J.; Gunson, R.N.; Murcia, P.R. Extensive multiplex PCR diagnostics reveal new insights into the epidemiology of viral respiratory infections. Epidemiol. Infect. 2016, 144, 2064–2076. [Google Scholar] [CrossRef]
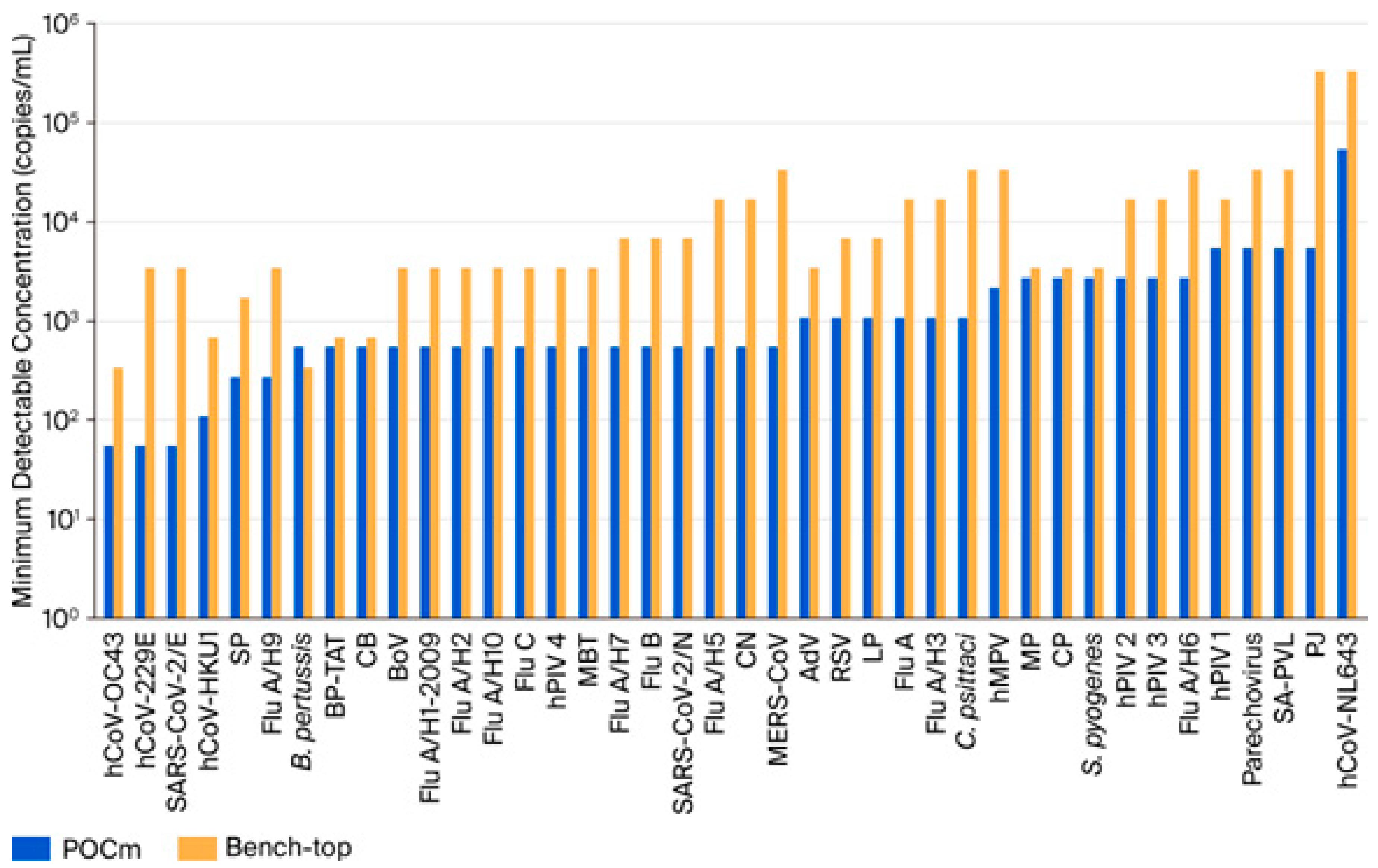
| Pathogens | Bench-Top | POCm 40-Plex | Fold Change | |||||
|---|---|---|---|---|---|---|---|---|
| Single-Plex a | 40-Plex a | Fold Change | ||||||
| Minimum Detectable Concentration b | Ct | Minimum Detectable Concentration a | Ct | 40-plex/ Single-Plex | Minimum Detectable Concentration a | Ct | (POCm 40-Plex/Bench-Top 40-Plex) | |
| (Copies/mL) | (Median) c | (Copies/mL) | (Median) b | (Copies/mL) | (Median) b | |||
| AdV | 333 | 33.28 | 3333 | 35.66 | 10.00 | 1067 | 29.73 | 3.20 |
| BoV | 3333 | 27.08 | 3333 | 28.17 | 1.00 | 533 | 25.99 | 0.16 |
| hCoV-229E | 333 | 24.97 | 3333 | 24.04 | 10.00 | 53 | 25.32 | 0.16 |
| hCoV-HKU1 | 667 | 24.05 | 667 | 25.52 | 1.00 | 107 | 25.8 | 0.16 |
| hCoV-NL63 | 33,333 | 28.11 | 333,333 | 28.06 | 10.00 | 53,333 | 25.02 | 1.60 |
| hCoV-OC43 | 333 | 27.53 | 333 | 24.83 | 1.00 | 53 | 26.98 | 0.16 |
| Flu A | 333 | 26.66 | 16,667 | 25.21 | 50.00 | 1067 | 26.55 | 3.20 |
| Flu A/H1-2009 | 333 | 23.97 | 3333 | 24.04 | 10.00 | 533 | 27.15 | 1.60 |
| Flu A/H2 | 333 | 28.94 | 3333 | 29.1 | 10.00 | 533 | 27.45 | 1.60 |
| Flu A/H3 | 333 | 26.1 | 16,667 | 26.96 | 50.00 | 1067 | 27.05 | 3.20 |
| Flu A/H5 | 3333 | 26.59 | 16,667 | 25.36 | 5.00 | 533 | 26.89 | 0.16 |
| Flu A/H6 | 3333 | 25.79 | 33,333 | 25.45 | 10.00 | 2667 | 26.78 | 0.80 |
| Flu A/H7 | 667 | 26.19 | 6667 | 28.2 | 10.00 | 533 | 27.77 | 0.80 |
| Flu A/H9 | 333 | 22.98 | 3333 | 27.16 | 10.00 | 267 | 28.03 | 0.80 |
| Flu A/H10 | 333 | 29.41 | 3333 | 30.29 | 10.00 | 533 | 29.07 | 1.60 |
| Flu B | 333 | 27.49 | 6667 | 27.52 | 20.00 | 533 | 26.43 | 1.60 |
| Flu C | 333 | 27.16 | 3333 | 27.43 | 10.00 | 533 | 25.97 | 1.60 |
| MERS-CoV | 3333 | 26.99 | 33,333 | 25.24 | 10.00 | 533 | 26.4 | 0.16 |
| hMPV | 667 | 26.26 | 33,333 | 23.86 | 50.00 | 2133 | 27.73 | 3.20 |
| hPIV 1 | 3333 | 29.58 | 16,667 | 26.07 | 5.00 | 5333 | 25.37 | 1.60 |
| hPIV 2 | 3333 | 24.02 | 16,667 | 25.25 | 5.00 | 2667 | 26.66 | 0.80 |
| hPIV 3 | 3333 | 25.86 | 16,667 | 26.23 | 5.00 | 2667 | 27.23 | 0.80 |
| hPIV 4 | 667 | 26.02 | 3333 | 27.07 | 5.00 | 533 | 26.9 | 0.80 |
| Parechovirus | 3333 | 27.17 | 33,333 | 28.52 | 10.00 | 5333 | 27.87 | 1.60 |
| RSV | 333 | 27.37 | 6667 | 29.32 | 20.00 | 1067 | 25.4 | 3.20 |
| SARS-CoV-2/E | 333 | 20.98 | 3333 | 25.56 | 10.00 | 53 | 24.55 | 0.16 |
| SARS-CoV-2/N | 667 | 28.2 | 6667 | 27.28 | 10.00 | 533 | 24.7 | 0.80 |
| MTB | 333 | 25.97 | 3333 | 24.15 | 10.00 | 533 | 22.83 | 1.60 |
| MP | 3333 | 25.77 | 3333 | 24.32 | 1.00 | 2667 | 24.79 | 0.80 |
| LP | 6667 | 24.31 | 6667 | 23.1 | 1.00 | 1067 | 25.73 | 0.16 |
| B pertussis | 333 | 27.53 | 333 | 27.83 | 1.00 | 533 | 25.07 | 1.60 |
| C psittaci | 6667 | 30.35 | 33,333 | 26.24 | 5.00 | 1067 | 25.72 | 0.16 |
| BP-TAT | 667 | 26.94 | 667 | 26.45 | 1.00 | 533 | 24.13 | 0.80 |
| CB | 667 | 28.41 | 667 | 27.61 | 1.00 | 533 | 25.67 | 0.80 |
| CP | 3333 | 28.36 | 3333 | 26.21 | 1.00 | 2667 | 24.6 | 0.80 |
| SA-PVL | 3333 | 26.72 | 33,333 | 26.99 | 10.00 | 5333 | 25.92 | 1.60 |
| SP | 333 | 28.99 | 1667 | 25.56 | 5.00 | 267 | 23.73 | 0.80 |
| S pyogenes | 3333 | 27.18 | 3333 | 26.48 | 1.00 | 2667 | 23.53 | 0.80 |
| CN | 333 | 22.88 | 16,667 | 25.22 | 50.00 | 533 | 26.05 | 1.60 |
| PJ | 33,333 | 19.33 | 333,333 | 19.03 | 10.00 | 5333 | 22.17 | 0.16 |
| Target | Diagnosis a | POCm Result | Sensitivity | Specificity | Agreement b | Concordance c | PPV d | NPV d | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Result | n | Positive (n e) | Negative (n e) | % (95% CI) f | % (95% CI) | % (95% CI) | % (95% CI) | % (95% CI) | % (95% CI) | |
| AdV | Positive | 3 | 3 | 0 | Not analyzed g | 100 | 100 | 100 | 100 | 100 |
| Negative | 274 | 0 | 274 | (98.6–100) | (98.6–100) | (100–100) | - | (98.6–100) | ||
| Flu A | Positive | 49 | 44 | 5 | 89.8 | 100 | 98.2 | 93.6 | 100 | 97.9 |
| Negative | 234 | 0 | 234 | (78.2–95.6) | (98.4–100) | (95.9–99.2) | (88.0–99.1) | (92.0–100) | (95.2–99.1) | |
| Flu A/H1 | Positive | 42 | 40 | 2 | 95.2 | 100 | 99.3 | 97.1 | 100 | 99.2 |
| Negative | 239 | 0 | 239 | (84.2–98.7) | (98.4–100) | (97.4–99.8) | (93.2–100.0) | (91.2–100) | (97.0–99.8) | |
| Flu A/H3 | Positive | 7 | 7 | 0 | Not analyzed | 100 | 100 | 100 | 100 | 100 |
| Negative | 276 | 0 | 276 | (98.6–100) | (98.7–100) | (100–100) | (64.6–100) | (98.6–100) | ||
| Flu B | Positive | 10 | 9 | 1 | Not analyzed | 100 | 99.6 | 94.6 | 100 | 99.6 |
| Negative | 273 | 0 | 273 | (98.6–100) | (98.0–99.9) | (83.9–100) | (70.1–100) | (97.9–99.9) | ||
| hMPV | Positive | 5 | 5 | 0 | Not analyzed | 100 | 100 | 100 | 100 | 100 |
| Negative | 272 | 0 | 272 | (98.6–100) | (98.6–100) | (100–100) | (56.6–100) | (98.6–100) | ||
| hPIV1 | Positive | 8 | 8 | 0 | Not analyzed | 100 | 100 | 100 | 100 | 100 |
| Negative | 269 | 0 | 269 | (98.6–100) | (98.6–100) | (100–100) | (67.6–100) | (98.6–100) | ||
| hPIV3 | Positive | 11 | 11 | 0 | Not analyzed | 100 | 100 | 100 | 100.0 | 100 |
| Negative | 266 | 0 | 266 | (98.6–100) | (98.6–100) | (100–100) | (74.1–100) | (98.6–100) | ||
| RSV | Positive | 10 | 9 | 1 | Not analyzed | 100 | 99.6 | 94.6 | 100 | 99.6 |
| Negative | 267 | 0 | 267 | (98.6–100) | (98.0–99.9) | (83.9–100) | (70.1–100) | (97.9–99.9) | ||
| SARS-CoV-2 | Positive | 24 | 22 | 2 | 91.7 | 100 | 99.3 | 95.3 | 100 | 99.2 |
| Negative | 253 | 0 | 253 | (74.2–97.7) | (98.5–100) | (97.4–99.8) | (88.7–100) | (85.1–100) | (97.2–99.8) | |
| Mycoplasma pneumoniae (MP) | Positive | 2 | 1 | 1 | Not analyzed | 100 | 99.6 | 66.5 | 100 | 99.6 |
| Negative | 275 | 0 | 275 | (98.6–100) | (98.0–99.9) | (0.97–100) | - | (98.0–99.9) | ||
| Sample | Specimen Type a | Diagnosed Pathogen | POCm | FilmArray | Agreement | |
|---|---|---|---|---|---|---|
| Target | Ct Value | Target | ||||
| 20QM0214 | NPA | Flu A/H1 | Flu A (matrix) | 18.03 | Flu A/H1 | Concordant |
| Flu A/H1 | 17.28 | |||||
| 20QM0055 | NPS | Flu A/H1 | Flu A (matrix) | 21.16 | Flu A/H1 | Concordant |
| Flu A/H1 | 20.99 | |||||
| 20QM0176 | NPA | Flu A/H3 | Flu A (matrix) | 23.76 | Flu A/H3 | Concordant |
| Flu A/H3 | 23.75 | |||||
| 21QM0002 | NPA | Flu B | Flu B | 25.90 | Flu B | Concordant |
| 20QM0219 | NPS | hPIV1 | hPIV1 | 26.87 | hPIV1 | Concordant |
| 20QM0175 | NPA | hPIV3 | hPIV3 | 22.23 | hPIV3 | Concordant |
| 20QM0062 | NPA | hMPV | hMPV | 18.94 | hMPV | Concordant |
| 20QM0153 | NPS | RSV | RSV | 23.72 | RSV | Concordant |
| 20QM0042 | NPA | AdV | AdV | 24.76 | AdV | Concordant |
| 20QM0212 | NPA | MP | MP | 22.11 | MP | Concordant |
| 20M048 | NPS | Normal control | Negative | - | Negative | Concordant |
| 20M049 | NPS | Normal control | Negative | - | Negative | Concordant |
| 20M057 | NPS | Normal control | Negative | - | Negative | Concordant |
| Sample | Specimen Type a | Diagnosed Pathogens | POCm | |
|---|---|---|---|---|
| Positive Target | Ct Value | |||
| 20QM0123 b | NPS | SARS-CoV-2, hMPV | SARS-CoV-2/E | 25.48 |
| SARS-CoV-2/N | 27.69 | |||
| hMPV | 29.30 | |||
| 20QM0113 | NPA | Flu A/H1, hPIV4 | Flu A | 19.04 |
| Flu A/H1 | 18.34 | |||
| hPIV4 | 23.12 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.C.W.; Tai, D.M.S.; Yee, A.; Tsui, N.B.Y.; Tsang, P.Y.L.; Chu, S.L.H.; Leung, C.T.; Leung, B.K.W.; Wong, W.; Kebede, F.T.; et al. Laboratory Validation of a Fully Automated Point-of-Care Device for High-Order Multiplexing Real-Time PCR Detection of Respiratory Pathogens. Diagnostics 2025, 15, 2445. https://doi.org/10.3390/diagnostics15192445
Li LCW, Tai DMS, Yee A, Tsui NBY, Tsang PYL, Chu SLH, Leung CT, Leung BKW, Wong W, Kebede FT, et al. Laboratory Validation of a Fully Automated Point-of-Care Device for High-Order Multiplexing Real-Time PCR Detection of Respiratory Pathogens. Diagnostics. 2025; 15(19):2445. https://doi.org/10.3390/diagnostics15192445
Chicago/Turabian StyleLi, Libby C. W., Deborah M. S. Tai, Anita Yee, Nancy B. Y. Tsui, Parker Y. L. Tsang, Sunny L. H. Chu, Chui Ting Leung, Bernice K. W. Leung, Winston Wong, Firaol Tamiru Kebede, and et al. 2025. "Laboratory Validation of a Fully Automated Point-of-Care Device for High-Order Multiplexing Real-Time PCR Detection of Respiratory Pathogens" Diagnostics 15, no. 19: 2445. https://doi.org/10.3390/diagnostics15192445
APA StyleLi, L. C. W., Tai, D. M. S., Yee, A., Tsui, N. B. Y., Tsang, P. Y. L., Chu, S. L. H., Leung, C. T., Leung, B. K. W., Wong, W., Kebede, F. T., Leung, P. Y. M., Chung, T., Yip, C. C. Y., Chen, J. H. K., Poon, R. W. S., To, K. K. W., Yuen, K.-Y., Fok, M., Lau, J. Y. N., & Lau, L. T. (2025). Laboratory Validation of a Fully Automated Point-of-Care Device for High-Order Multiplexing Real-Time PCR Detection of Respiratory Pathogens. Diagnostics, 15(19), 2445. https://doi.org/10.3390/diagnostics15192445







