Cytokine Profiles as Predictive Biomarkers of Disease Severity and Progression in Engineered Stone Silicosis: A Machine Learning Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects of the Study
2.2. Ethics
2.3. Clinical Data
2.3.1. Lung Function Measurements
2.3.2. Plasma Cytokine Analysis
2.4. Dataset
2.5. Statistical Methods
2.6. Machine Learning Models
2.6.1. Decision Tree
2.6.2. Random Forest
2.6.3. Gaussian Naïve Bayes
2.6.4. k-Nearest Neighbors
2.6.5. Linear Discriminant Analysis
2.6.6. Logistic Regression
2.6.7. Support Vector Machines
2.7. Data Preprocessing, Data Augmentation and Features Selection
2.8. Performance Metrics and Validation
2.9. Software
3. Results
3.1. Characteristics of the Study Population
3.2. Lung Function Tests
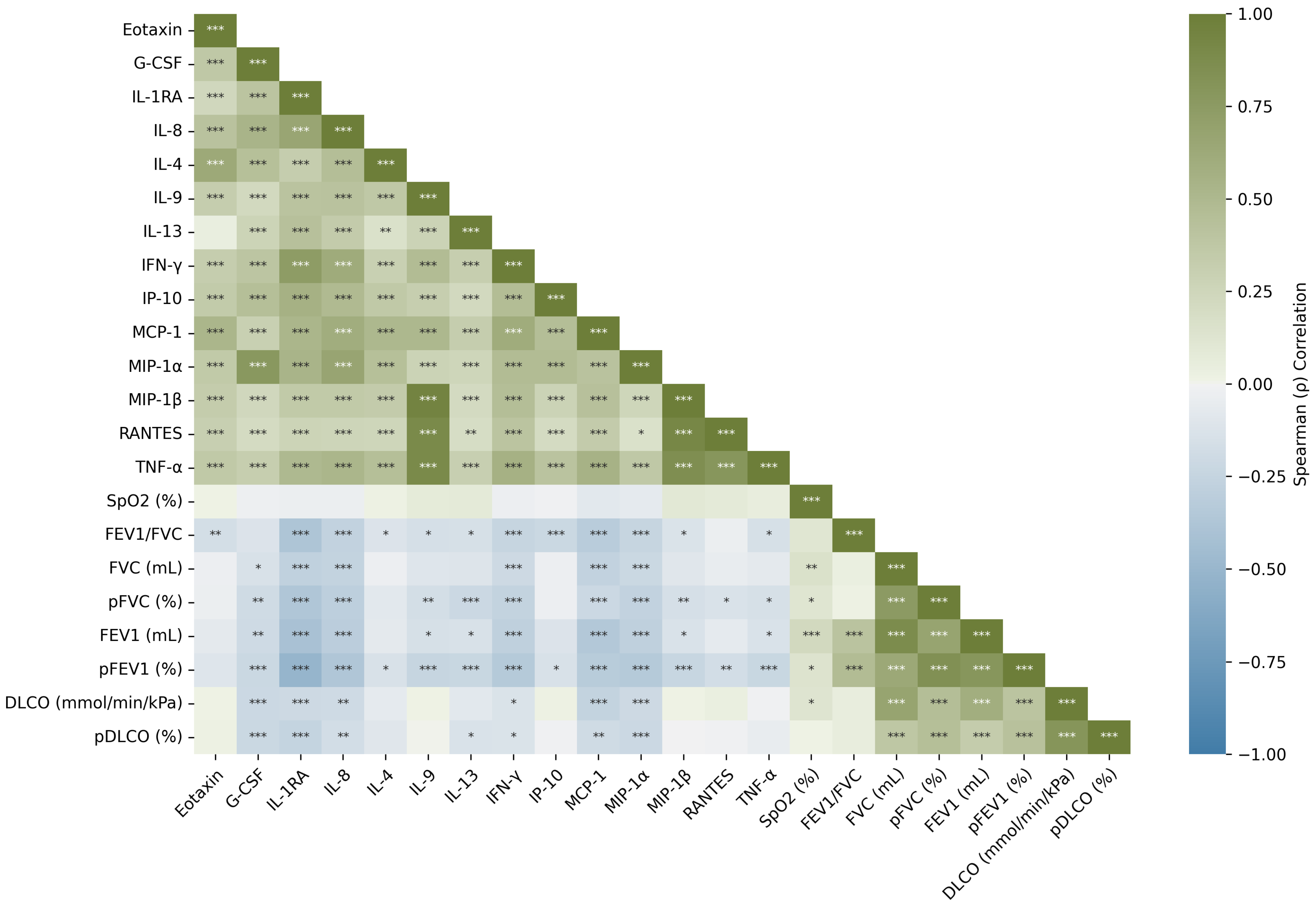
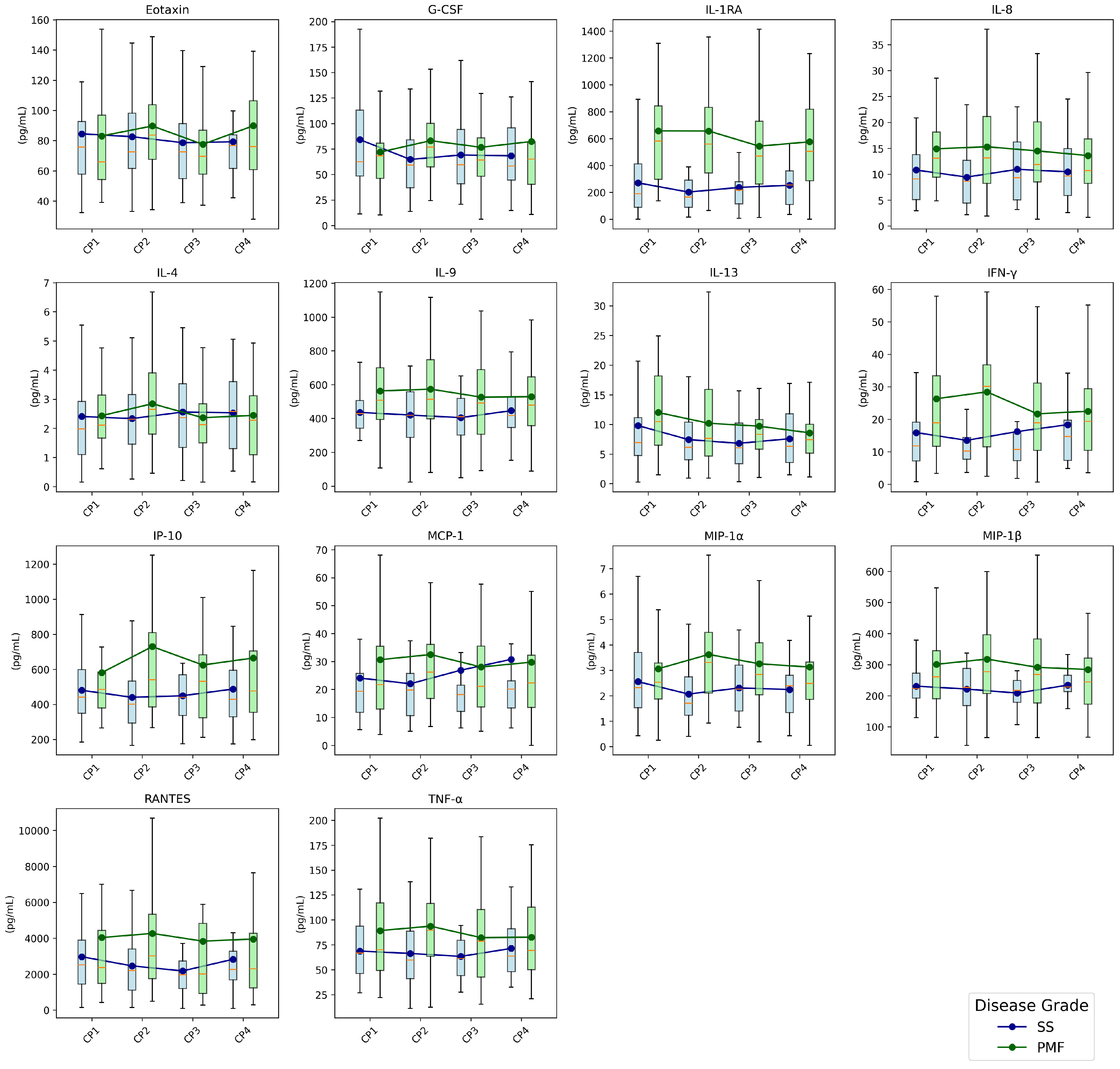
3.3. Cytokines
Cytokines Dynamics
3.4. Machine Learning Models
3.4.1. Disease Staging
3.4.2. Prognosis Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leung, C.C.; Yu, I.T.; Chen, W. Silicosis. Lancet 2012, 379, 2008–2018. [Google Scholar] [CrossRef]
- Hoy, R.F.; Chambers, D.C. Silica-related diseases in the modern world. Allergy 2020, 75, 2805–2817. [Google Scholar] [CrossRef]
- Kramer, M.R.; Blanc, P.D.; Fireman, E.; Amital, A.; Guber, A.; Rhahman, N.A.; Shitrit, D. Artificial stone silicosis [corrected]: Disease resurgence among artificial stone workers. Chest 2012, 142, 419–424. [Google Scholar] [CrossRef]
- Pérez-Alonso, A.; Córdoba-Doña, J.A.; Millares-Lorenzo, J.L.; Figueroa-Murillo, E.; García-Vadillo, C.; Romero-Morillos, J. Outbreak of silicosis in Spanish quartz conglomerate workers. Int. J. Occup. Environ. Health 2014, 20, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Heinzerling, A.; Cummings, K.J.; Flattery, J.; Weinberg, J.L.; Materna, B.; Harrison, R. Radiographic Screening Reveals High Burden of Silicosis among Workers at an Engineered Stone Countertop Fabrication Facility in California. Am. J. Respir. Crit. Care Med. 2021, 203, 764–766. [Google Scholar] [CrossRef]
- Kirby, T. Australia reports on audit of silicosis for stonecutters. Lancet 2019, 393, 861. [Google Scholar] [CrossRef]
- Leso, V.; Fontana, L.; Romano, R.; Gervetti, P.; Iavicoli, I. Artificial Stone Associated Silicosis: A Systematic Review. Int. J. Environ. Res. Public Health 2019, 16, 568. [Google Scholar] [CrossRef]
- Leon-Jimenez, A.; Hidalgo-Molina, A.; Conde-Sanchez, M.A.; Perez-Alonso, A.; Morales-Morales, J.M.; Garcia-Gamez, E.M.; Cordoba-Dona, J.A. Artificial Stone Silicosis: Rapid Progression Following Exposure Cessation. Chest 2020, 158, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Hoy, R.F.; Glass, D.C.; Dimitriadis, C.; Hansen, J.; Hore-Lacy, F.; Sim, M.R. Identification of early-stage silicosis through health screening of stone benchtop industry workers in Victoria, Australia. Occup. Environ. Med. 2021, 78, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Xue, C.; Yu, S.; Ye, Q. Artificial stone-associated silicosis in China: A prospective comparison with natural stone-associated silicosis. Respirology 2020, 25, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, X.; Xu, H.; Liu, H. Early identification, accurate diagnosis, and treatment of silicosis. Can. Respir. J. 2022, 2022, 3769134. [Google Scholar] [CrossRef]
- Lopes, A.J.; Mogami, R.; Capone, D.; Tessarollo, B.; de Melo, P.L.; Jansen, J.M. High-resolution computed tomography in silicosis: Correlation with chest radiography and pulmonary function tests. J. Bras. Pneumol. 2008, 34, 264–272. [Google Scholar] [CrossRef] [PubMed]
- International Labour Organization (ILO). Guidelines for the Use of the ILO International Classification of Radiographs of Pneumoconioses, Rev version 2022; Technical Report; International Labour Organization: Geneve, Switzerland, 2022. [Google Scholar]
- SŞener, M.U.; Şimşek, C.; Özkara, Ş.; Evran, H.; Bursali, İ.; Gökçek, A. Comparison of the International Classification of High-resolution Computed Tomography for Occupational and Environmental Respiratory Diseases with the International Labor Organization International Classification of Radiographs of Pneumoconiosis. Ind. Health 2019, 57, 495–502. [Google Scholar] [CrossRef]
- Bodaghi, A.; Fattahi, N.; Ramazani, A. Biomarkers: Promising and valuable tools towards diagnosis, prognosis and treatment of Covid-19 and other diseases. Heliyon 2023, 9, e13323. [Google Scholar] [CrossRef]
- Zinellu, A.; Collu, C.; Nasser, M.; Paliogiannis, P.; Mellino, S.; Zinellu, E.; Traclet, J.; Ahmad, K.; Mangoni, A.A.; Carru, C.; et al. The Aggregate Index of Systemic Inflammation (AISI): A Novel Prognostic Biomarker in Idiopathic Pulmonary Fibrosis. J. Clin. Med. 2021, 10, 4134. [Google Scholar] [CrossRef] [PubMed]
- Kreuter, M.; Lee, J.S.; Tzouvelekis, A.; Oldham, J.M.; Molyneaux, P.L.; Weycker, D.; Atwood, M.; Kirchgaessler, K.U.; Maher, T.M. Monocyte Count as a Prognostic Biomarker in Patients with Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2021, 204, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Barratt, S.L.; Creamer, A.W.; Adamali, H.I.; Duckworth, A.; Fallon, J.; Fidan, S.; Nancarrow, T.; Wollerton, R.; Steward, M.; Gooptu, B.; et al. Use of peripheral neutrophil to lymphocyte ratio and peripheral monocyte levels to predict survival in fibrotic hypersensitivity pneumonitis (fHP): A multicentre retrospective cohort study. BMJ Open Respir. Res. 2021, 8, e001063. [Google Scholar] [CrossRef]
- Sanchez-Morillo, D.; León-Jiménez, A.; Guerrero-Chanivet, M.; Jiménez-Gómez, G.; Hidalgo-Molina, A.; Campos-Caro, A. Integrating routine blood biomarkers and artificial intelligence for supporting diagnosis of silicosis in engineered stone workers. Bioeng. Transl. Med. 2024, 9, e10694. [Google Scholar] [CrossRef]
- Jiménez-Gómez, G.; Campos-Caro, A.; García-Núñez, A.; Gallardo-García, A.; Molina-Hidalgo, A.; León-Jiménez, A. Analysis of Immune Cell Subsets in Peripheral Blood from Patients with Engineered Stone Silica-Induced Lung Inflammation. Int. J. Mol. Sci. 2024, 25, 5722. [Google Scholar] [CrossRef]
- García-Núñez, A.; Jiménez-Gómez, G.; Hidalgo-Molina, A.; Córdoba-Doña, J.A.; León-Jiménez, A.; Campos-Caro, A. Inflammatory indices obtained from routine blood tests show an inflammatory state associated with disease progression in engineered stone silicosis patients. Sci. Rep. 2022, 12, 8211. [Google Scholar] [CrossRef]
- Sun, G.k.; Xiang, Y.h.; Wang, L.; Xiang, P.p.; Wang, Z.x.; Zhang, J.; Wu, L. Development of a multi-laboratory integrated predictive model for silicosis utilizing machine learning: A retrospective case-control study. Front. Public Health 2025, 12, 1450439. [Google Scholar] [CrossRef]
- Hamilton, R.F., Jr.; Thakur, S.A.; Holian, A. Silica binding and toxicity in alveolar macrophages. Free Radic. Biol. Med. 2008, 44, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Peeters, P.; Perkins, T.; Wouters, E.; Mossman, B.; Reynaert, N. Silica induces NLRP3 inflammasome activation in human lung epithelial cells. Part. Fibre Toxicol. 2013, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Campos-Caro, A.; Jiménez-Gómez, G.; García-Núñez, A.; Hidalgo-Molina, A.; León-Jiménez, A. Plasma Cytokine Profiling Reveals Differences between Silicotic Patients with Simple Silicosis and Those with Progressive Massive Fibrosis Caused by Engineered Stone. Int. J. Mol. Sci. 2023, 24, 1541. [Google Scholar] [CrossRef]
- Spiegel, J.; Ehrlich, R.; Yassi, A.; Riera, F.; Wilkinson, J.; Lockhart, K.; Barker, S.; Kistnasamy, B. Using Artificial Intelligence for High-Volume Identification of Silicosis and Tuberculosis: A Bio-Ethics Approach. Ann. Glob. Health 2021, 87, 58. [Google Scholar] [CrossRef]
- Alowais, S.; Alghamdi, S.; Alsuhebany, N.; Algahtani, T.; Alshaya, A.; Almohareb, S.; Aldairem, A.; Alrashed, M.; Saleh, K.; Badreldin, H.; et al. Revolutionizing health care: The role of artificial intelligence in clinical practice. BMC Med. Educ. 2023, 23, 689. [Google Scholar] [CrossRef]
- Soerensen, P.; Christensen, H.; Gray Worsoe Laursen, S.; Hardahl, C.; Brandslund, I.; Madsen, J. Using artificial intelligence in a primary care setting to identify patients at risk for cancer: A risk prediction model based on routine laboratory tests. Clin. Chem. Lab. Med. 2022, 60, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, E.; Tabata, J.; Yanaihara, N.; Ishikawa, T.; Koseki, K.; Iida, Y.; Saito, M.; Komazaki, H.; Shapiro, J.; Goto, C.; et al. Application of artificial intelligence for preoperative diagnostic and prognostic prediction in epithelial ovarian cancer based on blood biomarkers. Clin. Cancer Res. 2019, 25, 3006–3015. [Google Scholar] [CrossRef]
- Luo, Y.; Xue, Y.; Song, H.; Tang, G.; Liu, W.; Bai, H.; Yuan, X.; Tong, S.; Wang, F.; Cai, Y.; et al. Machine learning based on routine laboratory indicators promoting the discrimination between active tuberculosis and latent tuberculosis infection. J. Infect. 2022, 84, 648–657. [Google Scholar] [CrossRef]
- Topol, E. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Abdar, M.; Ksiazek, W.; Acharya, U.; Tan, R.S.; Makarenkov, V.; Plawiak, P. A new machine learning technique for an accurate diagnosis of coronary artery disease. Comput. Methods Programs Biomed. 2019, 179, 104992. [Google Scholar] [CrossRef]
- Jinny, V.; Priya, R. Prediction Model for Respiratory Diseases Using Machine Learning Algorithms. Int. J. Adv. Sci. Technol. 2020, 29, 10083–10092. [Google Scholar]
- Asri, H.; Mousannif, H.; Al Moatassime, H.; Noel, T. Using Machine Learning Algorithms for Breast Cancer Risk Prediction and Diagnosis. Procedia Comput. Sci. 2016, 83, 1064–1069. [Google Scholar] [CrossRef]
- Sisodia, D.; Sisodia, D.S. Prediction of Diabetes using Classification Algorithms. Procedia Comput. Sci. 2018, 132, 1578–1585. [Google Scholar] [CrossRef]
- Bansal, D.; Chhikara, R.; Khanna, K.; Goopta, P. Comparative Analysis of Various Machine Learning Algorithms for Detecting Dementia. Procedia Comput. Sci. 2018, 132, 1497–1502. [Google Scholar] [CrossRef]
- Rahman, A.K.M.S.; Shamrat, F.M.J.M.; Tasnim, Z.; Roy, J.; Hossain, S.A. A Comparative Study on Liver Disease Prediction Using Supervised Machine Learning Algorithms. Int. J. Sci. Technol. Res. 2019, 8, 419–422. [Google Scholar]
- Tamura, T.; Suganuma, N.; Hering, K.; Vehmas, T.; Itoh, H.; Akira, M.; Takashima, Y.; Hirano, H.; Kusaka, Y. Relationships (I) of International Classification of High-resolution Computed Tomography for Occupational and Environmental Respiratory Diseases with the ILO International Classification of Radiographs of Pneumoconioses for parenchymal abnormalities. Ind. Health 2015, 53, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Suganuma, N.; Kusaka, Y.; Hering, K.; Vehmas, T.; Kraus, T.; Arakawa, H.; Parker, J.; Kivisaari, L.; Letourneux, M.; Gevenois, P.; et al. Reliability of the proposed international classification of high-resolution computed tomography for occupational and environmental respiratory diseases. J. Occup. Health 2009, 51, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Royston, P. Multiple imputation of missing values: Update of ice. Stata J. 2005, 5, 527–536. [Google Scholar] [CrossRef]
- Sterne, J.; White, I.; Carlin, J.; Spratt, M.; Royston, P.; Kenward, M.; Wood, A.; Carpenter, J. Multiple imputation for missing data in epidemiological and clinical research: Potential and pitfalls. BMJ 2009, 338, b2393. [Google Scholar] [CrossRef]
- Royston, P. Multiple Imputation of Missing Values: Further Update of Ice, with an Emphasis on Interval Censoring. Stata J. 2007, 7, 445–464. [Google Scholar] [CrossRef]
- Xie, Y.; Meng, W.; Li, R.; Wang, Y.; Qian, X.; Chan, C.; Yu, Z.; Fan, X.; Pan, H.; Xie, C.; et al. Early lung cancer diagnostic biomarker discovery by machine learning methods. Transl. Oncol. 2021, 14, 100907. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, X.; Wei, M.; Wu, Y.; Zeng, J.; Liang, X.; Yi, J.; He, B.; Tu, Z. Screening of serum biomarkers and establishment of a decision tree in silica-exposed populations by surface-enhanced laser desorption ionization time-of-flight mass spectrometry. J. Occup. Environ. Med. 2007, 49, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2020, 45, 5–32. [Google Scholar] [CrossRef]
- Shah, C.; Jivani, A. Comparison of Data Mining Classification Algorithms for Breast Cancer Prediction. In Proceedings of the 2013 4th International Conference on Computing, Communications and Networking Technologies (ICCCNT), Tiruchengode, India, 4–6 July 2013; pp. 1–4. [Google Scholar] [CrossRef]
- Delizo, J.P.D.; Abisado, M.B.; Trinos, M.I.D. Philippine Twitter Sentiments during Covid-19 Pandemic using Multinomial Naïve-Bayes. Int. J. Adv. Trends Comput. Sci. Eng. 2020, 9, 408–412. [Google Scholar] [CrossRef]
- Villavicencio, C.; Macrohon, J.; Inbaraj, X.; Jeng, J.H.; Hsieh, J.G. Twitter Sentiment Analysis towards COVID-19 Vaccines in the Philippines Using Naïve Bayes. Information 2021, 12, 204. [Google Scholar] [CrossRef]
- Xuan, W.; Zheng, L.; Bunes, B.; Crane, N.; Zhou, F.; Zang, L. Engineering solutions to breath tests based on an e-nose system for silicosis screening and early detection in miners. J. Breath Res. 2022, 16, 036001. [Google Scholar] [CrossRef]
- Khorrami, M.; Prasanna, P.; Gupta, A.; Patil, P.; Velu, P.; Thawani, R.; Corredor, G.; Alilou, M.; Bera, K.; Fu, P.; et al. Changes in CT Radiomic Features Associated with Lymphocyte Distribution Predict Overall Survival and Response to Immunotherapy in Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 108–119. [Google Scholar] [CrossRef]
- Drera, G.; Freddi, S.; Emelianov, A.; Bobrinetskiy, I.; Chiesa, M.; Zanotti, M.; Pagliara, S.; Fedorov, F.; Nasibulin, A.; Montuschi, P.; et al. Exploring the performance of a functionalized CNT-based sensor array for breathomics through clustering and classification algorithms: From gas sensing of selective biomarkers to discrimination of chronic obstructive pulmonary disease. RSC Adv. 2021, 11, 30270–30282. [Google Scholar] [CrossRef]
- Khoshgoftaar, T.; Allen, E. Logistic regression modeling of software quality. Int. J. Reliab. Qual. Saf. Eng. 1999, 6, 303–317. [Google Scholar] [CrossRef]
- Richter, A.; Khoshgoftaar, T. A review of statistical and machine learning methods for modeling cancer risk using structured clinical data. Artif. Intell. Med. 2018, 90, 1–14. [Google Scholar] [CrossRef]
- El-Serag, H.; Kanwal, F.; Davila, J.; Kramer, J.; Richardson, P. A new laboratory-based algorithm to predict development of hepatocellular carcinoma in patients with hepatitis C and cirrhosis. Gastroenterology 2014, 146, 1249–1255. [Google Scholar] [CrossRef]
- Cirkovic, B.R.A.; Cvetkovic, A.M.; Ninkovic, S.M.; Filipovic, N.D. Prediction models for estimation of survival rate and relapse for breast cancer patients. In Proceedings of the 2015 IEEE 15th International Conference on Bioinformatics and Bioengineering (BIBE), Belgrade, Serbia, 2–4 November 2015; pp. 1–6. [Google Scholar] [CrossRef]
- Naraei, P.; Abhari, A.; Sadeghian, A. Application of Multilayer Perceptron Neural Networks and Support Vector Machines in Classification of Healthcare Data. In Proceedings of the 2016 Future Technologies Conference (FTC), San Francisco, CA, USA, 6–7 December 2016; pp. 848–852. [Google Scholar] [CrossRef]
- Syed, A.; Khan, T.; Alromema, N. A Hybrid Feature Selection Approach to Screen a Novel Set of Blood Biomarkers for Early COVID-19 Mortality Prediction. Diagnostics 2022, 12, 1604. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, R.; Lv, M.; Li, Z.; Jin, L.; Chen, X.; Han, Y.; Shi, C.; Jiang, Y.; Jin, S. Machine-Learning Algorithm-Based Prediction of Diagnostic Gene Biomarkers Related to Immune Infiltration in Patients With Chronic Obstructive Pulmonary Disease. Front. Immunol. 2022, 13, 740513. [Google Scholar] [CrossRef] [PubMed]
- Elreedy, D.; Atiya, A. A comprehensive analysis of synthetic minority oversampling technique (SMOTE) for handling class imbalance. Inf. Sci. 2019, 505, 32–64. [Google Scholar] [CrossRef]
- Blanco-Perez, J.; Blanco-Dorado, S.; Rodriguez-Garcia, J.; Gonzalez-Bello, M.; Salgado-Barreira, A.; Caldera-Diaz, A.; Pallares-Sanmartin, A.; Fernandez-Villar, A.; Gonzalez-Barcala, F. Serum levels of inflammatory mediators as prognostic biomarker in silica exposed workers. Sci. Rep. 2021, 11, 13348. [Google Scholar] [CrossRef]
- Jiang, P.R.; Cao, Z.; Qiu, Z.L.; Pan, J.W.; Zhang, N.; Wu, Y.F. Plasma levels of TNF-α and MMP-9 in patients with silicosis. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 1716–1720. [Google Scholar]
- Sato, T.; Saito, Y.; Inoue, S.; Shimosato, T.; Takagi, S.; Kaneko, T.; Ishigatsubo, Y. Serum heme oxygenase-1 as a marker of lung function decline in patients with chronic silicosis. J. Occup. Environ. Med. 2012, 54, 1461–1466. [Google Scholar] [CrossRef]
- Huang, H.B.; Huang, J.L.; Xu, X.T.; Huang, K.B.; Lin, Y.J.; Lin, J.B.; Zhuang, X.B. Serum neuron-specific enolase: A promising biomarker of silicosis. World J. Clin. Cases 2021, 9, 1016–1025. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Lu, Y.; Zhuang, H.; Gu, W.; Liu, B.; Liu, F.; Sun, J.; Yan, B.; Weng, D.; et al. IL-10-Producing CD1dhiCD5+ Regulatory B Cells May Play a Critical Role in Modulating Immune Homeostasis in Silicosis Patients. Front. Immunol. 2017, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Braz, N.F.; Carneiro, A.P.; Amorim, M.R.; de Oliveira Ferreira, F.; Lacerda, A.C.; Silva de Miranda, A.; Teixeira, M.M.; Teixeira, A.L.; Mendonça, V.A. Association between inflammatory biomarkers in plasma, radiological severity, and duration of exposure in patients with silicosis. J. Occup. Environ. Med. 2014, 56, 493–497. [Google Scholar] [CrossRef]
- Xue, C.; Wu, N.; Li, X.; Qiu, M.; Du, X.; Ye, Q. Serum concentrations of Krebs von den Lungen-6, surfactant protein D, and matrix metalloproteinase-2 as diagnostic biomarkers in patients with asbestosis and silicosis: A case-control study. BMC Pulm. Med. 2017, 17, 144. [Google Scholar] [CrossRef]
- Zhu, Y.; Duan, X.Y.; Cheng, Y.Q.; Yao, X.J.; Xu, H.; Zhang, K.S.; Li, F.S.; Yang, F.; Liu, L.H.; Yuan, X.J. Evaluation of differential serum expression of three factors and pulmonary function in patients with silicosis. Front. Immunol. 2021, 12, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.M.; Zhang, X.T.; Yan, Y.L.; He, E.Q.; Guo, P.; Zhang, Y.Y.; Zhao, D.K.; Yang, Z.G.; Chen, J.; Yao, M.Y.; et al. Change of serum TGF-β1 and TNF-α in silicosis patients. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 2011, 29, 606–607. [Google Scholar] [PubMed]
- Thongtip, S.; Siviroj, P.; Prapamontol, T.; Deesomchok, A.; Wisetborisut, A.; Nangola, S.; Khacha-Ananda, S. Club cell protein 16 as biomarker from crystalline silica exposure among Thai stone-carving workers. Toxicol. Ind. Health 2020, 36, 287–296. [Google Scholar] [CrossRef]
- Ferreira, V.L.; Borba, H.H.; de F. Bonetti, A.; Leonart, L.P.; Pontarolo, R. Cytokines and Interferons: Types and Functions. In Autoantibodies and Cytokines; Khan, W.A., Ed.; IntechOpen: Rijeka, Croatia, 2018; Chapter 4. [Google Scholar] [CrossRef]
- Yucesoy, B.; Vallyathan, V.; Landsittel, D.P.; Simeonova, P.; Luster, M.I. Cytokine polymorphisms in silicosis and other pneumoconioses. Mol. Cell. Biochem. 2002, 234/235, 219–224. [Google Scholar] [CrossRef]
- Zhou, Y.; Kang, Y.; Zhang, Z.; Liu, J. IL-1RA +2018 polymorphism and susceptibility to pneumoconiosis: A meta-analysis. Int. J. Clin. Exp. Med. 2014, 7, 2204–2208. [Google Scholar]
- Fouad, M.M.; Ramadan, M.A. Serum intracellular adhesion molecule-1 and interleukin-8 as predictors of pulmonary impairment among copper smelter workers. Int. Arch. Occup. Environ. Health 2021, 95, 365–375. [Google Scholar] [CrossRef]
- Lee, J.S.; Shin, J.H.; Choi, B.S. Serum levels of IL-8 and ICAM-1 as biomarkers for progressive massive fibrosis in coal workers’ pneumoconiosis. J. Korean Med. Sci. 2015, 30, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-γ: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef]
- Green, D.S.; Young, H.A.; Valencia, J.C. Current prospects of type II interferon γ signaling and autoimmunity. J. Biol. Chem. 2017, 292, 13925–13933. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Duan, M.; Tang, Y.; Wu, J.; Zhao, K.; Zhong, Y.; He, Z.; Meng, J.; Chen, F.; Xiao, X.; et al. Impaired interferon-γ signaling promotes the development of silicosis. iScience 2022, 25, 104647. [Google Scholar] [CrossRef]
- Pryhuber, G.S.; Huyck, H.L.; Baggs, R.; Oberdorster, G.; Finkelstein, J.N. Induction of chemokines by low-dose intratracheal silica is reduced in TNFR I (p55) null mice. Toxicol. Sci. 2003, 72, 150–157. [Google Scholar] [CrossRef]
- Liu, S.; Liu, C.; Wang, Q.; Liu, S.; Min, J. CC chemokines in idiopathic pulmonary fibrosis: Pathogenic role and therapeutic potential. Biomolecules 2023, 13, 333. [Google Scholar] [CrossRef]
- Yonker, L.M.; Cigana, C.; Hurley, B.P.; Bragonzi, A. Host-pathogen interplay in the respiratory environment of cystic fibrosis. J. Cyst. Fibros. 2015, 14, 431–439. [Google Scholar] [CrossRef]
- Cockx, M.; Gouwy, M.; Van Damme, J.; Struyf, S. Chemoattractants and cytokines in primary ciliary dyskinesia and cystic fibrosis. Cell. Mol. Immunol. 2018, 15, 312–323. [Google Scholar] [CrossRef]
- Tahtinen, S.; Tong, A.J.; Himmels, P.; Oh, J.; Paler-Martinez, A.; Kim, L.; Wichner, S.; Oei, Y.; McCarron, M.J.; Freund, E.C.; et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat. Immunol. 2023, 23, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Sato, S.; Takehara, K. Augmented production of MCP-1, MIP-1α and MIP-1β in systemic sclerosis patients: Involvement in pulmonary fibrosis. Clin. Exp. Immunol. 1999, 117, 159–165. [Google Scholar] [CrossRef]
- Pérez-Alonso, A.; González-Domínguez, M.E.; Novalbos-Ruiz, J.P.; León-Jiménez, A.; Córdoba-Doña, J.A. Artificial Stone Silicosis: Accumulation of errors in the resurgence of an occupational disease: A qualitative study. Work 2021, 70, 433–442. [Google Scholar] [CrossRef] [PubMed]
| Data | SS (n = 36) | PMF (n = 36) | p-Value |
|---|---|---|---|
| Age | 41.58 ± 7.75 | 41.86 ± 6.45 | 0.186 |
| Years of exposure | 13.75 ± 6.94 | 13.25 ± 6.22 | 0.157 |
| Years since first exposure | 21.06 ± 5.80 | 21.03 ± 5.80 | 0.171 |
| Years since last exposure | 7.25 ± 2.79 | 7.81 ± 2.83 | 0.086 |
| Years from first exposure to diagnosis | 15.75 ± 6.69 | 14.78 ± 5.81 | 0.143 |
| Years from last exposure to diagnosis | 1.86 ± 3.08 | 1.53 ± 3.51 | 0.200 |
| Years since diagnosis | 5.22 ± 2.51 | 6.17 ± 2.30 | 0.082 |
| SpO2 | 97.4 ± 1.1 | 97.7 ± 1.0 | 0.441 |
| FEV1/FVC | 0.77 ± 0.06 | 0.73 ± 0.08 | 0.045 * |
| FEV1 (mL) | 3388.06 ± 638.69 | 2958.88 ± 650.67 | 0.045 * |
| FEV1 (%) | 90.10 ± 12.92 | 77.34 ± 16.24 | 0.005 * |
| FVC (mL) | 4391.39 ± 736.80 | 4044.40 ± 824.71 | 0.156 |
| FVC (%) | 94.56 ± 12.34 | 85.52 ± 17.01 | 0.045 * |
| DLCO (mmol/min/kPa) | 9.21 ± 1.97 | 8.54 ± 1.51 | 0.375 |
| DLCO (%) | 87.46 ± 17.90 | 80.41 ± 16.00 | 0.181 |
| Cytokine | SS (n = 36) | PMF (n = 36) | p-Value |
|---|---|---|---|
| Eotaxin | 84.48 ± 41.81 | 83.09 ± 43.94 | 0.654 |
| GCSF | 84.17 ± 54.48 | 71.91 ± 43.26 | 0.474 |
| IL-1RA | 270.49 ± 244.48 | 657.37 ± 472.12 | p < 0.001 *** |
| IL-4 | 2.40 ± 1.77 | 2.43 ± 1.22 | 0.474 |
| IL-8 | 10.82 ± 7.50 | 14.92 ± 8.57 | 0.045 * |
| IL-9 | 435.70 ± 151.14 | 562.66 ± 261.17 | 0.045 * |
| IL-13 | 9.80 ± 7.53 | 12.00 ± 7.28 | 0.217 |
| IFN- | 15.90 ± 12.62 | 26.31 ± 20.20 | 0.045 * |
| IP-10 | 480.17 ± 184.34 | 580.93 ± 334.26 | 0.474 |
| MCP-1 | 24.08 ± 22.35 | 30.64 ± 31.06 | 0.441 |
| MIP-1 | 2.56 ± 1.41 | 3.06 ± 2.03 | 0.441 |
| MIP-1 | 230.61 ± 65.29 | 301.07 ± 179.85 | 0.234 |
| RANTES | 2972.50 ± 1999.45 | 4040.01 ± 4389.79 | 0.942 |
| TNF- | 68.77 ± 27.14 | 89.28 ± 53.67 | 0.376 |
| Cytokine | Variable | F-Statistic | df | p-Value |
|---|---|---|---|---|
| IL-1RA | Checkpoint | 2.9746 | 3 | 0.072 |
| Disease Grade | 4.1158 | 1 | 0.072 | |
| Years with Disease | 7.6291 | 1 | 0.030 * | |
| Years with Disease × Disease Grade | 1.6627 | 1 | 0.248 | |
| Checkpoint x Disease Grade | 0.9918 | 3 | 0.397 | |
| MIP-1 | Checkpoint | 2.0350 | 3 | 0.171 |
| Disease Grade | 2.2290 | 1 | 0.171 | |
| Years with Disease | 6.7780 | 1 | 0.040 * | |
| Years with Disease x Disease Grade | 1.1840 | 1 | 0.277 | |
| Checkpoint x Disease Grade | 3.5050 | 3 | 0.040 * |
| Data | SS (n = 22) | SS That Progress (n = 14) | p-Value |
|---|---|---|---|
| Eotaxin | 75.10 ± 35.62 | 99.21 ± 47.69 | 0.661 |
| G-CSF | 69.96 ± 36.49 | 106.50 ± 70.44 | 0.661 |
| IL-1RA | 252.72 ± 211.39 | 298.42 ± 295.55 | 1.000 |
| IL4 | 2.27 ± 1.76 | 2.61 ± 1.83 | 1.000 |
| IL-8 | 9.98 ± 5.83 | 12.13 ± 9.67 | 1.000 |
| IL-9 | 451.82 ± 175.83 | 410.37 ± 102.14 | 1.000 |
| IL-13 | 10.69 ± 6.12 | 8.39 ± 9.42 | 0.661 |
| INF- | 15.69 ± 12.03 | 16.21 ± 13.96 | 1.000 |
| IP-10 | 483.50 ± 192.86 | 474.94 ± 177.08 | 1.000 |
| MCP-1 | 25.75 ± 26.97 | 21.46 ± 12.59 | 1.000 |
| MIP-1 | 2.30 ± 1.25 | 2.95 ± 1.60 | 1.000 |
| MIP-1 | 237.17 ± 72.20 | 220.31 ± 53.55 | 1.000 |
| RANTES | 3169.25 ± 2162.24 | 2663.34 ± 1744.25 | 1.000 |
| TNF- | 69.28 ± 28.61 | 67.96 ± 25.68 | 1.000 |
| SpO2 (%) | 97.41 ± 1.05 | 97.39 ± 1.21 | 1.000 |
| FEV1/FVC | 0.78 ± 0.05 | 0.76 ± 0.07 | 1.000 |
| FEV1 (mL) | 3455.00 ± 631.81 | 3282.86 ± 658.73 | 1.000 |
| FEV1 (%) | 90.88 ± 12.48 | 88.88 ± 13.97 | 1.000 |
| FVC1 (mL) | 4397.73 ± 655.78 | 4381.43 ± 875.63 | 1.000 |
| FVC1 (%) | 94.44 ± 9.90 | 94.75± 15.87 | 1.000 |
| DLCO (mmol/min/kPa) | 9.23 ± 1.96 | 9.17 ± 2.05 | 1.000 |
| DLCO (%) | 86.39 ± 16.52 | 89.14 ± 20.42 | 1.000 |
| Model | Selected Cytokines | Se | Sp | AUC | F1-Score | Pr | Acc |
|---|---|---|---|---|---|---|---|
| DT | IL-1RA, IL-13, IFN-, IP-10, MIP-1, RANTES, IL-1RA, MIP-1, RANTES, IL-4, IFN-, MIP-1, RANTES | 0.664 | 0.793 | 0.729 | 0.663 | 0.673 | 0.749 |
| RF | IL-1RA, IL-8, RANTES | 0.774 | 0.794 | 0.812 | 0.727 | 0.705 | 0.781 |
| GNB | IL-1RA, IP-10, MIP-1, MIP-1, TNF, IL-8 | 0.903 | 0.572 | 0.781 | 0.707 | 0.608 | 0.704 |
| KNN | G-CSF, IL-1RA, IL-9, IL-13, IFN-, RANTES, TNF, Eotaxin, G-CSF, IL-1RA, IL-8, IL-4, IP-10, MIP-1, MIP-1, RANTES, Eotaxin, IL-9, IL-13, IP-10, MIP-1, MIP-1, Years with the Disease | 0.874 | 0.716 | 0.807 | 0.750 | 0.683 | 0.775 |
| LDA | IL-1RA, IL-8, IFN-, MCP-1, MIP-1, RANTES, Eotaxin, G-CSF, IL-1RA, IFN-, MCP-1, MIP-1, MIP-1, RANTES, TNF-, IL-1RA, IL-4, IL-13, TNF, Years with the Disease | 0.879 | 0.778 | 0.847 | 0.784 | 0.725 | 0.824 |
| LR | IL-1RA, IL-8, IL-4, IFN-, MCP-1, MIP-1, Eotaxin, G-CSF, IL-9, TNF-, IL-1RA, IL-4, IL-13, Years with the Disease | 0.852 | 0.788 | 0.839 | 0.768 | 0.720 | 0.813 |
| SVM | IL-1RA, IL-8, IFN-, IP-10, MCP-1, MIP-1, MIP-1, RANTES, IFN-, MIP-1, RANTES, IL-1RA, IL-4, IL-13, Years with the Disease | 0.840 | 0.821 | 0.851 | 0.778 | 0.741 | 0.827 |
| Model | Selected Cytokines | Se | Sp | AUC | F1-Score | Pr | Acc |
|---|---|---|---|---|---|---|---|
| DT | G-CSF, IL-8, IL-13, IFN-, RANTES, Eotaxin, G-CSF, IL-1RA, MIP-1, TNF-, Eotaxin, G-CSF, IL-8, IL-4, IL-13, IFN-, RANTES | 0.562 | 0.836 | 0.699 | 0.549 | 0.544 | 0.765 |
| RF | G-CSF, IL-9, IL-13, IL-1RA, IL-13, RANTES, G-CSF, IP-10, RANTES | 0.535 | 0.916 | 0.674 | 0.572 | 0.643 | 0.800 |
| GNB | IL-9, MCP-1, RANTES, IL-9, IL-13, MCP-1, RANTES, IL-1RA, IFN-, IP-10, RANTES | 0.815 | 0.527 | 0.638 | 0.573 | 0.465 | 0.616 |
| KNN | G-CSF, IL-9, RANTES | 0.718 | 0.690 | 0.689 | 0.618 | 0.552 | 0.706 |
| LDA | G-CSF, IL-1RA, IL-13, IFN-, IP-10, MIP-1, RANTES, IL-1RA, IL-13, RANTES, G-CSF, IL-1RA, IL-8, IL-13, IFN-, MCP-1, MIP-1, TNF-, Years with the Disease | 0.642 | 0.749 | 0.746 | 0.571 | 0.550 | 0.704 |
| LR | G-CSF, IL-9, IL-13, IFN-, RANTES, G-CSF, RANTES | 0.652 | 0.783 | 0.732 | 0.640 | 0.685 | 0.752 |
| SVM | G-CSF, IL-1RA, IL-9, IL-13, IFN-, TNF-, Eotaxin, IL-1RA, IL-4, IL-13, Years with the Disease | 0.740 | 0.807 | 0.817 | 0.667 | 0.647 | 0.772 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez-Morillo, D.; Martín-Carrillo, A.; Priego-Torres, B.; Sopo-Lambea, I.; Jiménez-Gómez, G.; León-Jiménez, A.; Campos-Caro, A. Cytokine Profiles as Predictive Biomarkers of Disease Severity and Progression in Engineered Stone Silicosis: A Machine Learning Approach. Diagnostics 2025, 15, 2413. https://doi.org/10.3390/diagnostics15182413
Sanchez-Morillo D, Martín-Carrillo A, Priego-Torres B, Sopo-Lambea I, Jiménez-Gómez G, León-Jiménez A, Campos-Caro A. Cytokine Profiles as Predictive Biomarkers of Disease Severity and Progression in Engineered Stone Silicosis: A Machine Learning Approach. Diagnostics. 2025; 15(18):2413. https://doi.org/10.3390/diagnostics15182413
Chicago/Turabian StyleSanchez-Morillo, Daniel, Ana Martín-Carrillo, Blanca Priego-Torres, Iris Sopo-Lambea, Gema Jiménez-Gómez, Antonio León-Jiménez, and Antonio Campos-Caro. 2025. "Cytokine Profiles as Predictive Biomarkers of Disease Severity and Progression in Engineered Stone Silicosis: A Machine Learning Approach" Diagnostics 15, no. 18: 2413. https://doi.org/10.3390/diagnostics15182413
APA StyleSanchez-Morillo, D., Martín-Carrillo, A., Priego-Torres, B., Sopo-Lambea, I., Jiménez-Gómez, G., León-Jiménez, A., & Campos-Caro, A. (2025). Cytokine Profiles as Predictive Biomarkers of Disease Severity and Progression in Engineered Stone Silicosis: A Machine Learning Approach. Diagnostics, 15(18), 2413. https://doi.org/10.3390/diagnostics15182413








