Duplex Probe-Based Fluorescence Melting Curve Analysis for Simultaneous Genotyping of rs1126728 and rs11208257 in the Phosphoglucomutase-1 Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. DNA Samples
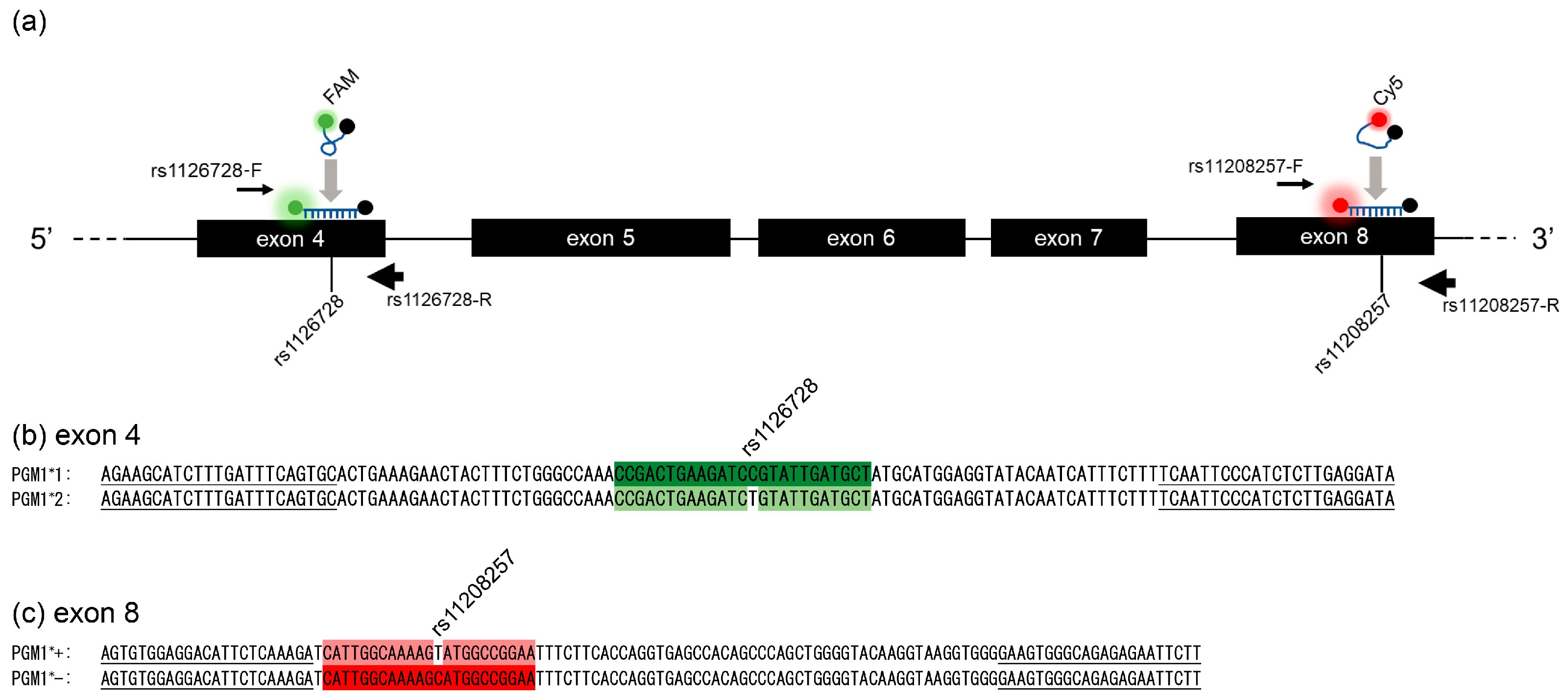
2.3. Probes and Primers for a Symmetric Real-Time PCR for Amplification of rs1126728 and rs11208257 in PGM1
2.4. Genotyping of rs1126728 and rs11208257 in PGM1 by Sanger Sequencing
2.5. Genotyping of rs1126728 and rs11208257 in PGM1 by FMCA
3. Results
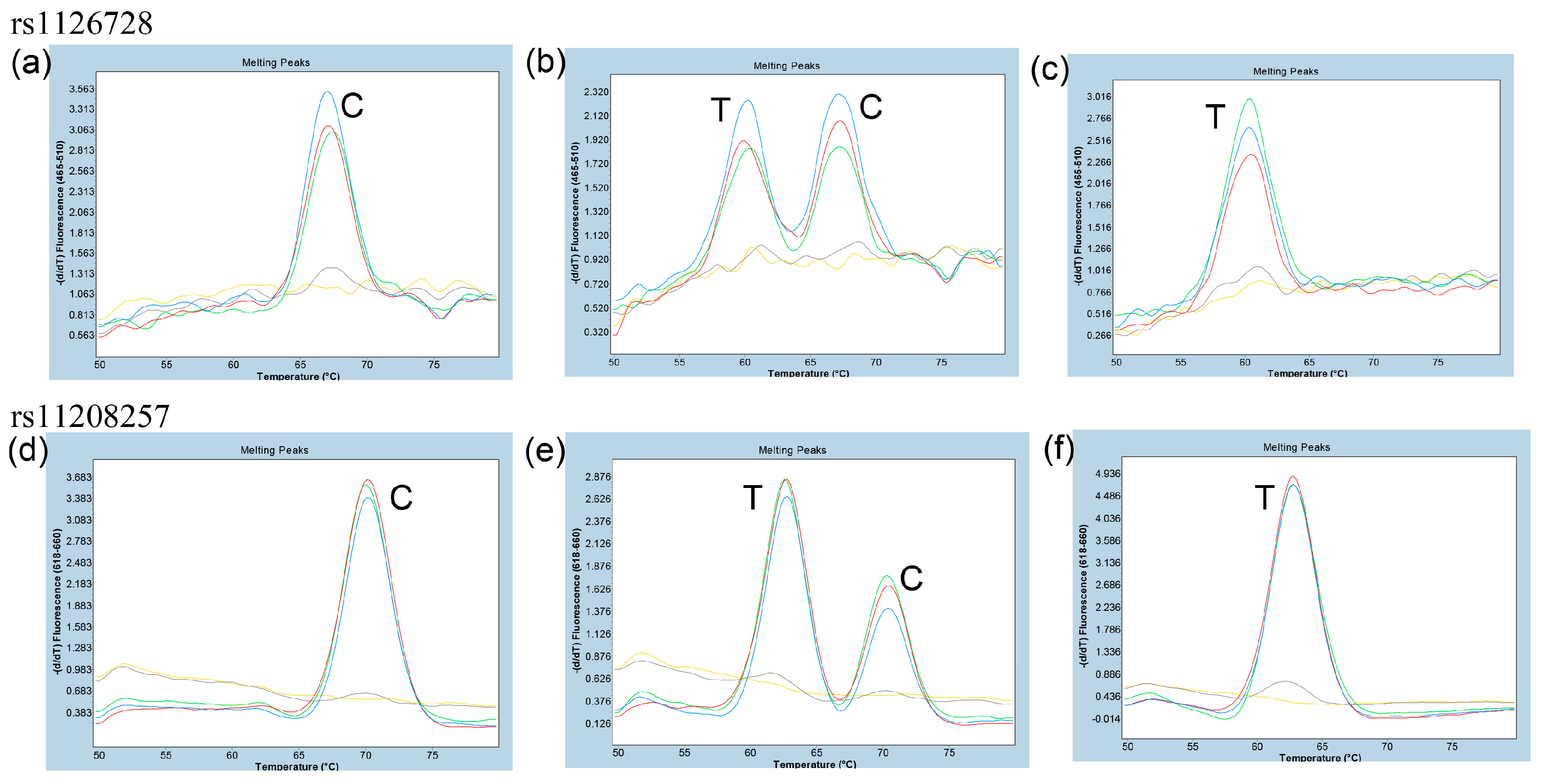
3.1. Determination of the Genotypes of rs1126728 and rs11208257 by Direct Sanger Sequencing of PCR Products
3.2. Determination of the Optimal Primer Concentration Ratio for Asymmetric PCR for Duplex FMCA of rs1126728 and rs11208257
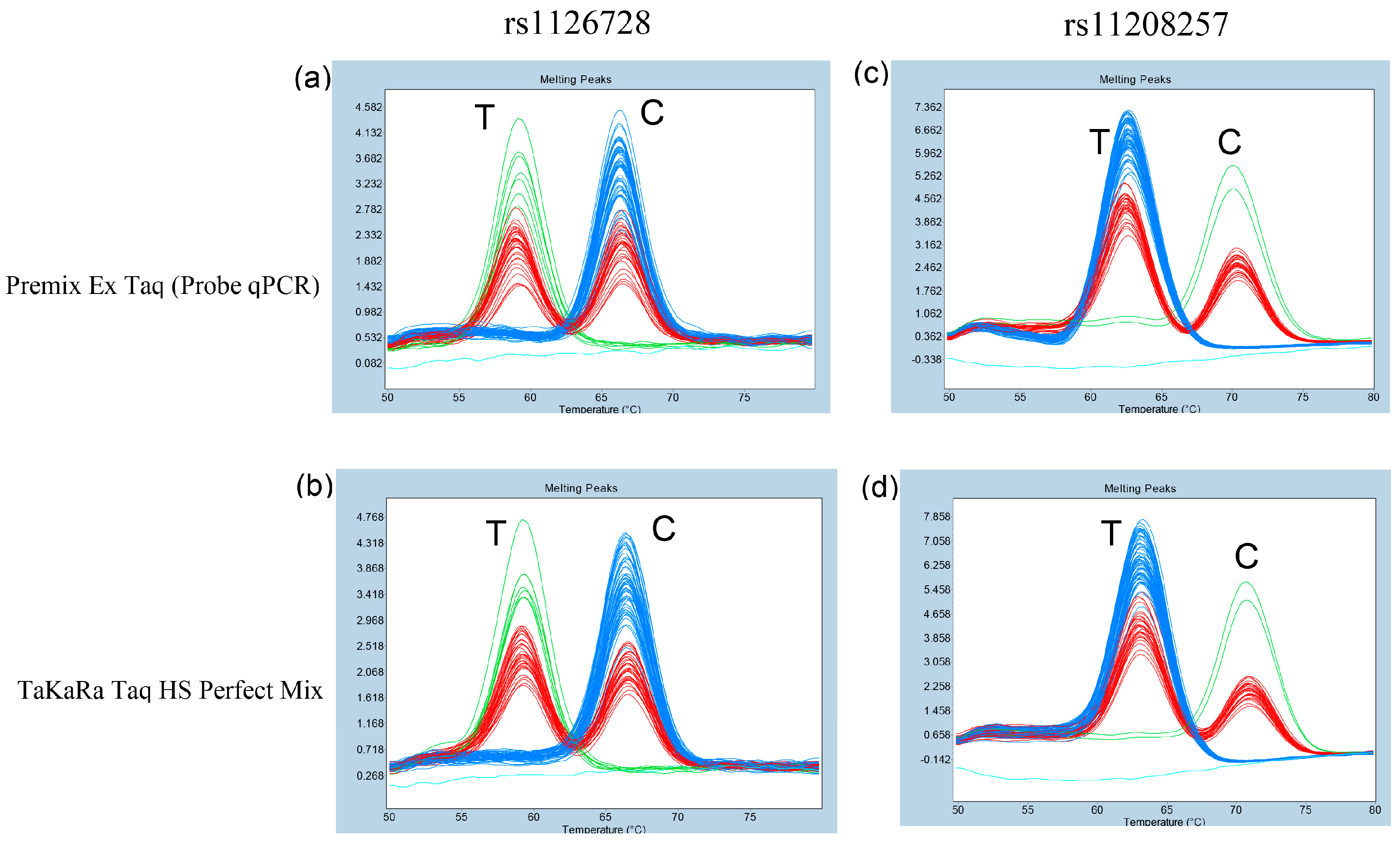
3.3. Comparison of Different DNA Polymerases in Probe-Based FMCA to Determine rs1126728 and rs11208257 Genotypes in PGM1
3.4. Genotyping of rs1126728 and rs11208257 in 95 Japanese Subjects Using Probe-Based FMCA
3.5. PGM1 Phenotypes Inferred by Genotyping Results of rs1126728 and rs11208257 in 95 Japanese Subjects
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Cy5 | cyanine 5 |
| FAM | 5-carboxyfluorescein |
| FMCA | fluorescence melting curve analysis |
| PCR-RFLP | PCR-restriction fragment length polymorphism |
| PCR-SSCP | PCR-single-strand conformation polymorphism |
| PMG | phosphoglucomutase |
| SNV | single-nucleotide variant |
| Tm | melting temperature |
References
- Muenks, A.G.; Stiers, K.M.; Beamer, L.J. Sequence-structure relationships, expression profiles, and disease-associated mutations in the paralogs of phosphoglucomutase 1. PLoS ONE 2017, 12, e0183563. [Google Scholar] [CrossRef]
- Spencer, N.; Hopkinson, D.A.; Harris, H. Phosphoglucomutase polymorphism in man. Nature 1964, 204, 742–745. [Google Scholar] [CrossRef]
- Hopkinson, D.A.; Harris, H. Rare phosphoglucomutase phenotypes. Ann. Hum. Genet. 1966, 30, 167–181. [Google Scholar] [CrossRef]
- Hopkinson, D.A.; Harris, H. A third phosphoglucomutase locus in man. Ann. Hum. Genet. 1968, 31, 359–367. [Google Scholar] [CrossRef]
- Cantu, J.M.; Ibarra, B. Phosphoglucomutase: Evidence for a new locus expressed in human milk. Science 1982, 216, 639–640. [Google Scholar] [CrossRef] [PubMed]
- Edwards, Y.H.; Putt, W.; Fox, M.; Ives, J.H. A novel human phosphoglucomutase (PGM5) maps to the centromeric region of chromosome 9. Genomics 1995, 30, 350–353. [Google Scholar] [CrossRef]
- McAlpine, P.J.; Hopkinson, D.A.; Harris, H. The relative activities attributable to the three phosphoglucomutase loci (PGM1, PGM2, PGM3) in human tissues. Ann. Hum. Genet. 1970, 34, 169–175. [Google Scholar] [CrossRef]
- Kuhnl, P.; Schmidtmann, U.; Spielmann, W. Evidence for two additional common alleles at the PGM1 locus (phosphoglucomutase-E.C.: 2.7.5.1). A comparison by three different techniques. Hum. Genet. 1977, 35, 219–223. [Google Scholar] [CrossRef] [PubMed]
- March, R.E.; Putt, W.; Hollyoake, M.; Ives, J.H.; Lovegrove, J.U.; Hopkinson, D.A.; Edwards, Y.H.; Whitehouse, D.B. The classical human phosphoglucomutase (PGM1) isozyme polymorphism is generated by intragenic recombination. Proc. Natl. Acad. Sci. USA 1993, 90, 10730–10733. [Google Scholar] [CrossRef]
- Takahashi, N.; Neel, J.V. Intragenic recombination at the human phosphoglucomutase 1 locus: Predictions fulfilled. Proc. Natl. Acad. Sci. USA 1993, 90, 10725–10729. [Google Scholar] [CrossRef] [PubMed]
- Scacchi, R.; Corbo, R.M.; Palmarino, R.; Sacco, G.; Arnone, M.; Lucarelli, P. Human phosphoglucomutase locus 1: Red cell enzymatic activities associated with common isoelectric focusing phenotypes. Hum. Hered. 1983, 33, 218–222. [Google Scholar] [CrossRef]
- Lee, Y.; Stiers, K.M.; Kain, B.N.; Beamer, L.J. Compromised catalysis and potential folding defects in in vitro studies of missense mutants associated with hereditary phosphoglucomutase 1 deficiency. J. Biol. Chem. 2014, 289, 32010–32019. [Google Scholar] [CrossRef]
- Gururaj, A.; Barnes, C.J.; Vadlamudi, R.K.; Kumar, R. Regulation of phosphoglucomutase 1 phosphorylation and activity by a signaling kinase. Oncogene 2004, 23, 8118–8127. [Google Scholar] [CrossRef]
- Gloria-Bottini, F.; Magrini, A.; Antonacci, E.; La Torre, M.; Di Renzo, L.; De Lorenzo, A.; Bergamaschi, A.; Bottini, E. Phosphoglucomutase genetic polymorphism and body mass. Am. J. Med. Sci. 2007, 334, 421–425. [Google Scholar] [CrossRef]
- Gloria-Bottini, F.; Lucarini, N.; Palmarino, R.; La Torre, M.; Nicotra, M.; Borgiani, P.; Cosmi, E.; Bottini, E. Phosphoglucomutase genetic polymorphism of newborns. Am. J. Hum. Biol. 2001, 13, 9–14. [Google Scholar] [CrossRef]
- Inshaw, J.R.J.; Sidore, C.; Cucca, F.; Stefana, M.I.; Crouch, D.J.M.; McCarthy, M.I.; Mahajan, A.; Todd, J.A. Analysis of overlapping genetic association in type 1 and type 2 diabetes. Diabetologia 2021, 64, 1342–1347. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.P.; Kamarthy, P.; Balakrishna, S.; Manu, M.; Ramaswamy, S. Association between phosphoglucomutase-1 gene y420h polymorphism and type 2 diabetes mellitus: A Case-control study. Arch. Med. Health Sci. 2021, 9, 225. [Google Scholar] [CrossRef]
- El Housni, H.; Heimann, P.; Parma, J.; Vassart, G. Single-nucleotide polymorphism genotyping by melting analysis of dual-labeled probes: Examples using factor V Leiden and prothrombin 20210A mutations. Clin. Chem. 2003, 49, 1669–1672. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Liu, Z.; Liao, Y.; Chen, X.; Zhang, Y.; Li, Q. Multiplex fluorescence melting curve analysis for mutation detection with dual-labeled, self-quenched probes. PLoS ONE 2011, 6, e19206. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, D.; Li, B.; Xie, J.; Liu, J.; Zhang, Z. Single nucleotide polymorphism genotyping of ALDH2 gene based on asymmetric PCR and fluorescent probe-mediated melting curves. Anal. Biochem. 2021, 642, 114509. [Google Scholar] [CrossRef]
- Soejima, M.; Koda, Y. Detection of five common variants of ABO gene by a triplex probe-based fluorescence-melting-curve-analysis. Anal. Biochem. 2022, 648, 114668. [Google Scholar] [CrossRef]
- Soejima, M.; Koda, Y. Estimation of Lewis blood group status by fluorescence melting curve analysis in simultaneous genotyping of c.385A>T and fusion gene in FUT2 and c.59T>G and c.314C>T in FUT3. Diagnostics 2023, 13, 931. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Soejima, M.; Koda, Y. FUT2 polymorphism in Latin American populations. Clin. Chim. Acta 2020, 505, 1–5. [Google Scholar] [CrossRef]
- Conte, F.; Morava, E.; Bakar, N.A.; Wortmann, S.B.; Poerink, A.J.; Grunewald, S.; Crushell, E.; Al-Gazali, L.; de Vries, M.C.; Morkrid, L.; et al. Phosphoglucomutase-1 deficiency: Early presentation, metabolic management and detection in neonatal blood spots. Mol. Genet. Metab. 2020, 131, 135–146. [Google Scholar] [CrossRef]
- Solin, M.L.; Kaartinen, M. Allelic polymorphism of mouse Igh-J locus, which encodes immunoglobulin heavy chain joining (JH) segments. Immunogenetics 1992, 36, 306–313. [Google Scholar] [CrossRef]
- Higuchi, R.; von Beroldingen, C.H.; Sensabaugh, G.F.; Erlich, H.A. DNA typing from single hairs. Nature 1988, 332, 543–546. [Google Scholar] [CrossRef]
- Suzuki, Y.; Orita, M.; Shiraishi, M.; Hayashi, K.; Sekiya, T. Detection of ras gene mutations in human lung cancers by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene 1990, 5, 1037–1043. [Google Scholar]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Fei, Z.; Xiao, P. Methods to improve the accuracy of next-generation sequencing. Front. Bioeng. Biotechnol. 2023, 11, 982111. [Google Scholar] [CrossRef] [PubMed]
- Hippman, C.; Nislow, C. Pharmacogenomic testing: Clinical evidence and implementation challenges. J. Pers. Med. 2019, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.G.; Connell, C.R.; Bloch, W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993, 21, 3761–3766. [Google Scholar] [CrossRef]
- Lee, K.; Park, H.C.; An, S.; Ahn, E.R.; Lee, Y.H.; Kim, M.J.; Lee, E.J.; Park, J.S.; Jung, J.W.; Lim, S. A new method for ABO genotyping using fluorescence melting curve analysis based on peptide nucleic acid probes. Leg. Med. 2015, 17, 334–339. [Google Scholar] [CrossRef]
- Yu, Z.; Shang, Z.; Huang, Q.; Wu, H.; Patil, S. Rapid Detection of SLCO1B1 Polymorphisms Using Duplex Fluorescence Melting Curve Analysis: Implications for Personalized Drug Dosing in Clinical Settings. Drug Des. Devel. Ther. 2024, 18, 4889–4899. [Google Scholar] [CrossRef] [PubMed]
- Soejima, M.; Koda, Y. Simultaneous genotyping of three nonsynonymous SNVs, rs1042602, rs1426654, and rs16891982 involved in skin pigmentation by fluorescent probe-based melting curve analysis. Hum. Mutat. 2025, 2025, 3468799. [Google Scholar] [CrossRef] [PubMed]

| SNV | Genotype | Observed Number | Allele Frequency | Expected Number | Chi-Square |
|---|---|---|---|---|---|
| C/C | 55 | C = 0.7526 T = 0.2474 | 53.8132 | Χ2 = 0.4278 p = 0.50 | |
| rs1126728 | C/T | 33 | 35.3737 | ||
| T/T | 7 | 5.8132 | |||
| T/T | 64 | T = 0.826 C = 0.173 | 64.8658 | Χ2 = 0.3831 p = 0.50 | |
| rs11208257 | C/T | 29 | 27.2684 | ||
| C/C | 2 | 2.8658 |
| rs1126728 | rs11208257 | Inferred Phenotype | Number of Subjects | ||||
|---|---|---|---|---|---|---|---|
| Melting Temperature | Genotype | Inferred Phenotype | Melting Temperature | Genotype | Inferred Phenotype | ||
| High | C/C | 1/1 | High | C/C | −/− | 1−/1− | 1 |
| Double | C/T | +/− | 1+/1− | 15 | |||
| Low | T/T | +/+ | 1+/1+ | 39 | |||
| Double | C/T | 1/2 | High | C/C | −/− | 1−/2− | 1 |
| Double | C/T | +/− | 1+/2− or 1−/2+ | 14 | |||
| Low | T/T | +/+ | 1+/2+ | 18 | |||
| Low | T/T | 2/2 | High | C/C | −/− | 2−/2− | 0 |
| Double | C/T | +/− | 2+/2− | 0 | |||
| Low | T/T | +/+ | 2+/2+ | 7 | |||
| Single-Probe FMCA | Double-Probe FMCA | Direct Sanger Sequencing | |
|---|---|---|---|
| Time | About × 2 of double-probe FMCA | Fast (approximately one hour) | Time consuming |
| Cost | About × 2 of double-probe FMCA | Inexpensive | Use of costly reagent |
| Post-PCR procedures | Unnecessary | Unnecessary | Necessary |
| Risk of carry-over contaminations | Low probability | Low probability | Relatively high probability |
| Reproducibility | High | High | High |
| Accuracy | Accurate | Accurate | Most accurate |
| Multiple assays | Impossible | Possible | Impossible |
| Throughput | About half of double-probe FMCA | Relatively high | Low |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soejima, M.; Koda, Y. Duplex Probe-Based Fluorescence Melting Curve Analysis for Simultaneous Genotyping of rs1126728 and rs11208257 in the Phosphoglucomutase-1 Gene. Diagnostics 2025, 15, 2345. https://doi.org/10.3390/diagnostics15182345
Soejima M, Koda Y. Duplex Probe-Based Fluorescence Melting Curve Analysis for Simultaneous Genotyping of rs1126728 and rs11208257 in the Phosphoglucomutase-1 Gene. Diagnostics. 2025; 15(18):2345. https://doi.org/10.3390/diagnostics15182345
Chicago/Turabian StyleSoejima, Mikiko, and Yoshiro Koda. 2025. "Duplex Probe-Based Fluorescence Melting Curve Analysis for Simultaneous Genotyping of rs1126728 and rs11208257 in the Phosphoglucomutase-1 Gene" Diagnostics 15, no. 18: 2345. https://doi.org/10.3390/diagnostics15182345
APA StyleSoejima, M., & Koda, Y. (2025). Duplex Probe-Based Fluorescence Melting Curve Analysis for Simultaneous Genotyping of rs1126728 and rs11208257 in the Phosphoglucomutase-1 Gene. Diagnostics, 15(18), 2345. https://doi.org/10.3390/diagnostics15182345






