Bayesian Graphical Models for Multiscale Inference in Medical Image-Based Joint Degeneration Analysis
Abstract
1. Introduction
2. Methodology
3. Bayesian Inference in Medical Imaging
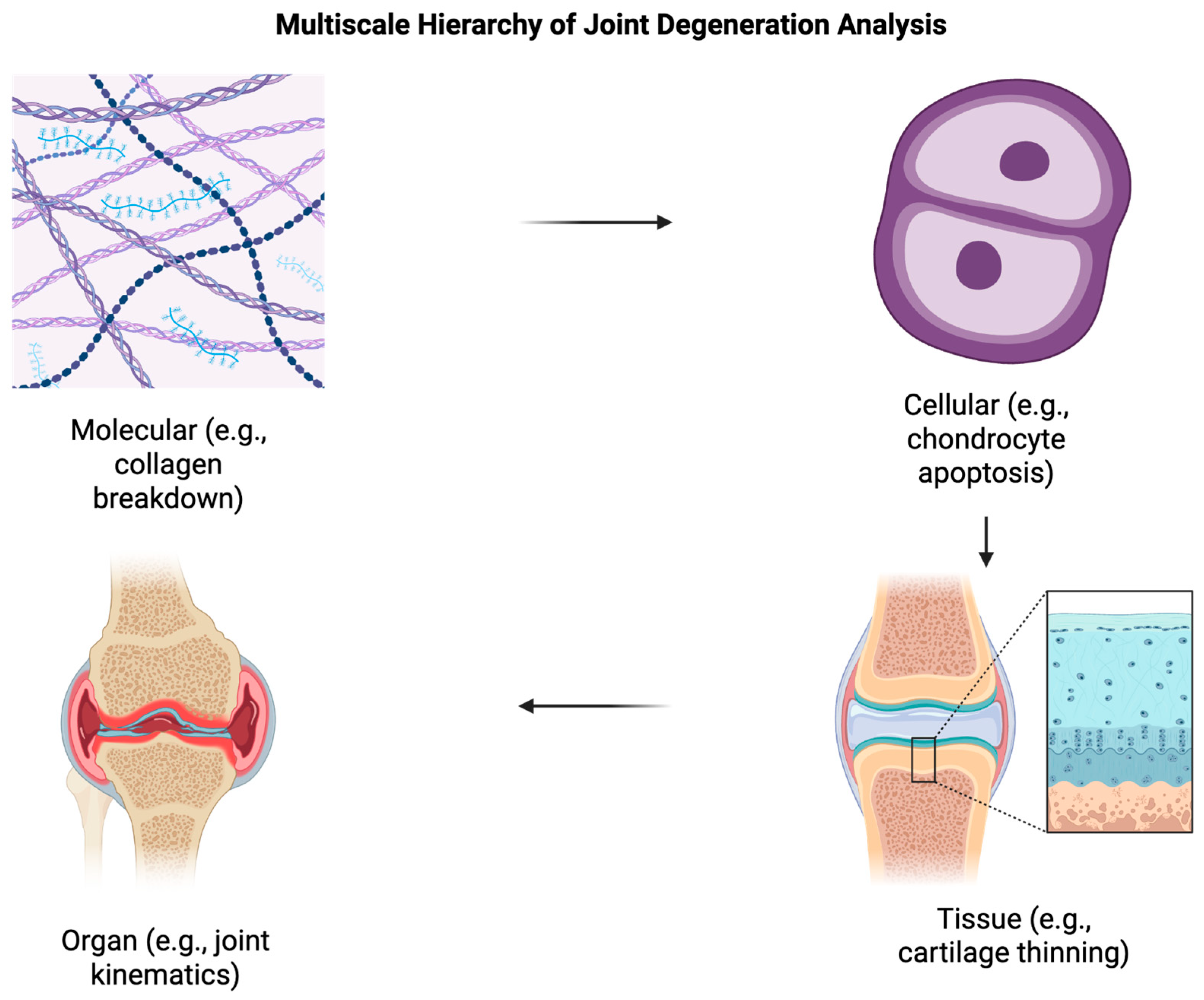
3.1. Multiscale Analysis
- Multi-resolution convolutional neural networks (MRCNNs): These architectures process images at different resolutions in parallel, fusing low-resolution context with high-resolution detail. For example, in knee osteoarthritis detection from MRI, MRCNNs achieved an AUC of 0.95 versus 0.91 for single-scale CNNs, with improved sensitivity for early-stage disease [25]. Pros: strong performance when both fine and coarse structures matter; cons: higher memory requirements and longer training times.
- Pyramid feature extraction: Using Gaussian or Laplacian pyramids, features are extracted at progressively downsampled resolutions. In cartilage lesion segmentation, pyramid-based U-Nets improved Dice coefficients by 3–5% over baseline U-Nets [26]. Pros: efficient capture of context at multiple scales; cons: potential loss of fine detail if too aggressively downsampled.
- Scale-invariant feature descriptors (e.g., SIFT, wavelet transforms): These approaches capture features robust to magnification changes, making them suitable for heterogeneous acquisition protocols. In bone microarchitecture assessment, wavelet-based texture analysis produced classification accuracies of 88–92%, outperforming single-scale texture descriptors by ~6% [27]. Pros: robustness to acquisition variability; cons: sometimes less effective for deep learning integration without adaptation.
- Attention-based multiscale fusion: Self-attention mechanisms weight contributions from different scales adaptively. Applied to multimodal MRI for osteoarthritis progression prediction, attention-fusion models achieved AUCs of 0.97 and reduced false positives by 15% compared to unweighted fusion [28]. Pros: adaptive feature importance learning; cons: increased model complexity and training instability if not carefully regularized.
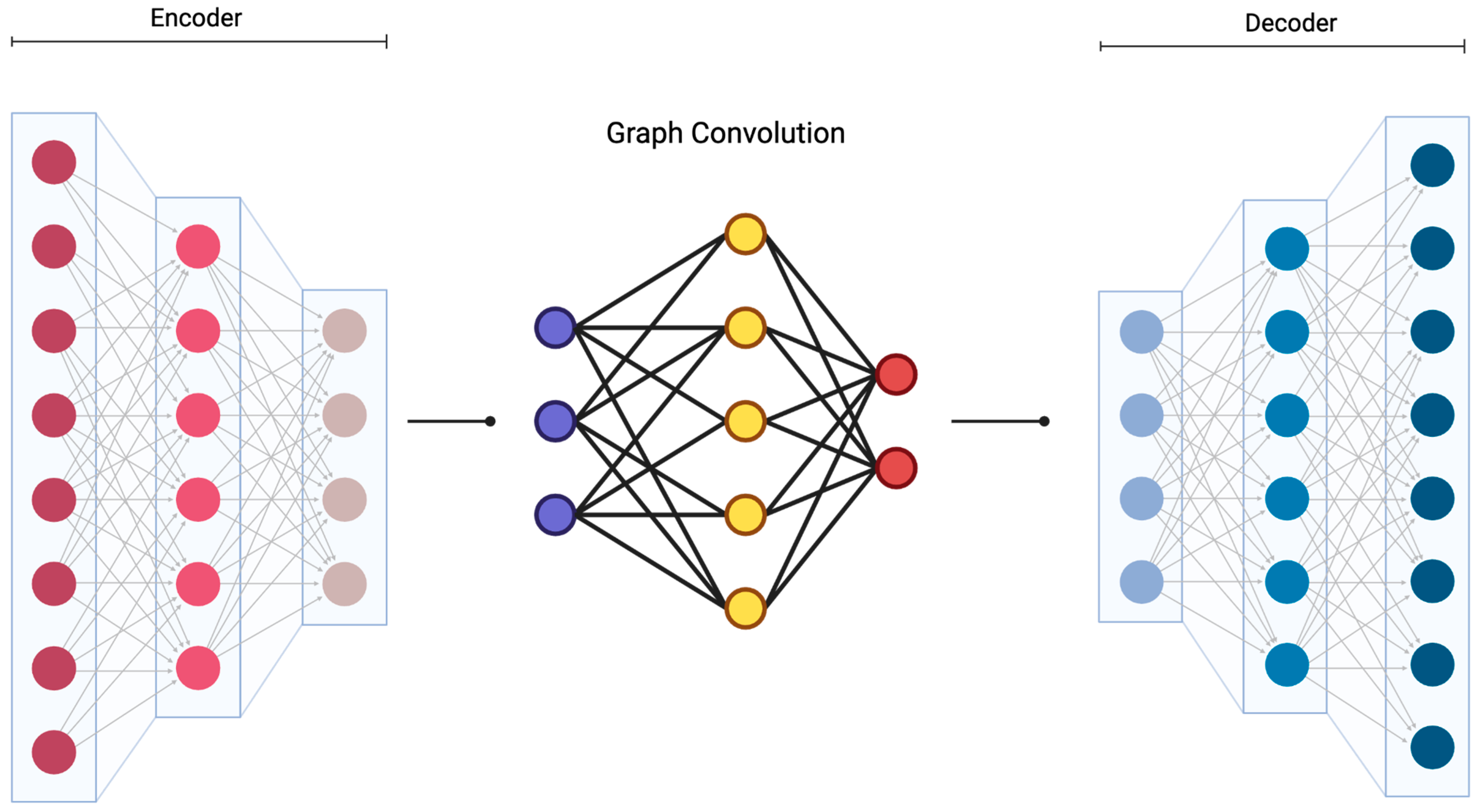
3.2. Probabilistic Graphical Models
3.3. Spatial-Temporal Modeling
3.4. Network Connectivity Analysis
3.5. Advanced Imaging Biomarkers and Quantitative Analysis
3.6. Quantitative MRI Techniques
3.7. Radiomics and Texture Analysis
3.8. Multimodal Integration Strategies
3.9. Uncertainty Quantification and Bayesian/Variational Inference Methods
3.10. Monte Carlo Methods
3.11. Model Selection and Validation
3.12. Diffusion Models for Medical Imaging
3.13. Bayesian Joint Diffusion Models
3.14. Clinical Translation and Validation
4. Discussion
4.1. Clinical Implementation Challenges
4.2. Potential Solutions for Widespread Implementation
4.3. Bayesion Graphical Models and Multimodal Imaging: Advancing Precision Medicine
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baum, T.; Joseph, G.B.; Karampinos, D.C.; Jungmann, P.M.; Link, T.M.; Bauer, J.S. Cartilage and meniscal T2 relaxation time as non-invasive biomarker for knee osteoarthritis and cartilage repair procedures. Osteoarthr. Cartil. 2013, 21, 1474–1484. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mihaljević, B.; Bielza, C.; Larrañaga, P. Bayesian networks for interpretable machine learning and optimization. Neurocomputing 2021, 456, 648–665. [Google Scholar] [CrossRef]
- Teng, H.L.; Wu, D.; Su, F.; Pedoia, V.; Souza, R.B.; Ma, C.B.; Li, X. Gait Characteristics Associated with a Greater Increase in Medial Knee Cartilage T1ρ and T2 Relaxation Times in Patients Undergoing Anterior Cruciate Ligament Reconstruction. Am. J. Sports Med. 2017, 45, 3262–3271. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Su, F.; Wu, D.; Pedoia, V.; Heitkamp, L.; Ma, C.B.; Souza, R.B.; Li, X. Frontal Plane Knee Mechanics and Early Cartilage Degeneration in People with Anterior Cruciate Ligament Reconstruction: A Longitudinal Study. Am. J. Sports Med. 2018, 46, 378–387. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, K.; Duka, B.; Xie, H.; Oathes, D.J.; Calhoun, V.; Zhang, Y. A dynamic graph convolutional neural network framework reveals new insights into connectome dysfunctions in ADHD. Neuroimage 2022, 246, 118774. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Paverd, H.; Zormpas-Petridis, K.; Clayton, H.; Burge, S.; Crispin-Ortuzar, M. Radiology and multi-scale data integration for precision oncology. npj Precis. Onc. 2024, 8, 158. [Google Scholar] [CrossRef] [PubMed]
- DuBois Bowman, F.; Caffo, B.; Bassett, S.S.; Kilts, C. A Bayesian hierarchical framework for spatial modeling of fMRI data. Neuroimage 2008, 39, 146–156. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rombach, R.; Blattmann, A.; Lorenz, D.; Esser, P.; Ommer, B. High-resolution image synthesis with latent diffusion models. arXiv 2022, arXiv:2112.10752v2. [Google Scholar] [CrossRef]
- Wu, R.; Guo, Y.; Chen, Y.; Zhang, J. Osteoarthritis burden and inequality from 1990 to 2021: A systematic analysis for the global burden of disease Study 2021. Sci. Rep. 2025, 15, 8305. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fusco, M.; Skaper, S.D.; Coaccioli, S.; Varrassi, G.; Paladini, A. Degenerative Joint Diseases and Neuroinflammation. Pain. Pr. 2017, 17, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Miller, D.J.; Urish, K.L. T2 map signal variation predicts symptomatic osteoarthritis progression: Data from the Osteoarthritis Initiative. Skelet. Radiol. 2016, 45, 909–913. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Urish, K.L.; Keffalas, M.G.; Durkin, J.R.; Miller, D.J.; Chu, C.R.; Mosher, T.J. T2 texture index of cartilage can predict early symptomatic OA progression: Data from the osteoarthritis initiative. Osteoarthr. Cartil. 2013, 21, 1550–1557. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gimenez, O.; Royle, A.; Kéry, M.; Nater, C.R. Ten quick tips to get you started with Bayesian statistics. PLoS Comput. Biol. 2025, 21, e1012898. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zohuri, B.; Rahmani, F.M.; Behgounia, F. Knowledge Is Power in Four Dimensions: Models to Forecast Future Paradigm; Academic Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Gaser, C.; Dahnke, R.; Thompson, P.M.; Kurth, F.; Luders, E. The Alzheimer’s Disease Neuroimaging Initiative. CAT: A computational anatomy toolbox for the analysis of structural MRI data. Gigascience 2024, 13, giae049. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ceritoglu, C.; Oishi, K.; Li, X.; Chou, M.C.; Younes, L.; Albert, M.; Lyketsos, C.; van Zijl, P.C.; Miller, M.I.; Mori, S. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. Neuroimage 2009, 47, 618–627. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bedini, L.; Benvenuti, L.; Salerno, E.; Tonazzini, A. A mixed-annealing algorithm for edge preserving image reconstruction using a limited number of projections. Signal Process. 1993, 32, 397–408. [Google Scholar] [CrossRef]
- Väärälä, A.; Casula, V.; Peuna, A.; Panfilov, E.; Mobasheri, A.; Haapea, M.; Lammentausta, E.; Nieminen, M.T. Predicting osteoarthritis onset and progression with 3D texture analysis of cartilage MRI DESS: 6-Year data from osteoarthritis initiative. J. Orthop. Res. 2022, 40, 2597–2608. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xie, Y.; Dan, Y.; Tao, H.; Wang, C.; Zhang, C.; Wang, Y.; Yang, J.; Yang, G.; Chen, S. Radiomics Feature Analysis of Cartilage and Subchondral Bone in Differentiating Knees Predisposed to Posttraumatic Osteoarthritis after Anterior Cruciate Ligament Reconstruction from Healthy Knees. Biomed. Res. Int. 2021, 2021, 4351499. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vervullens, S. Heterogeneity in Individuals with Knee Osteoarthritis Awaiting Total Knee Arthroplasty and Its Impact on Outcome from a Biopsychosocial Perspective. Ph.D. Thesis, Maastricht University, Maastricht, Netherlands, 2024. [Google Scholar] [CrossRef]
- Jiang, C.A.; Leong, T.Y.; Poh, K.L. PGMC: A framework for probabilistic graphic model combination. AMIA Annu. Symp. Proc. 2005, 2005, 370–374. [Google Scholar] [PubMed] [PubMed Central]
- Shin, D.A.; Lee, S.H.; Oh, S.; Yoo, C.; Yang, H.J.; Jeon, I.; Park, S.B. Probabilistic graphical modelling using Bayesian networks for predicting clinical outcome after posterior decompression in patients with degenerative cervical myelopathy. Ann. Med. 2023, 55, 2232999. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Altenbuchinger, M.; Weihs, A.; Quackenbush, J.; Grabe, H.J.; Zacharias, H.U. Gaussian and Mixed Graphical Models as (multi-)omics data analysis tools. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194418. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van der Velden, B.H.M.; Kuijf, H.J.; Gilhuijs, K.G.A.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, P.S.Q.; Lai, K.W.; Goh, S.L.; Hasikin, K.; Hum, Y.C.; Tee, Y.K.; Dhanalakshmi, S. Emergence of deep learning in knee osteoarthritis diagnosis. Comput. Intell. Neurosci. 2021, 2021, 4931437. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.C.W.; Chung, A.C.S. Large deformation diffeomorphic image registration with Laplacian pyramid networks. Lect. Notes Comput. Sci. 2020, 12263, 211–221. [Google Scholar] [CrossRef]
- Zheng, K.; Makrogiannis, S. Bone texture characterization for osteoporosis diagnosis using digital radiography. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2016, 2016, 1034–1037. [Google Scholar] [CrossRef]
- Panfilov, E.; Saarakkala, S.; Nieminen, M.T.; Tiulpin, A. End-to-end prediction of knee osteoarthritis progression with multimodal transformers. IEEE J. Biomed. Health Inform. 2025, 1–11. [Google Scholar] [CrossRef]
- Bomhals, B.; Cossement, L.; Maes, A.; Sathekge, M.; Mokoala, K.M.G.; Sathekge, C.; Ghysen, K.; Van de Wiele, C. Principal Component Analysis Applied to Radiomics Data: Added Value for Separating Benign from Malignant Solitary Pulmonary Nodules. J. Clin. Medicine. 2023, 12, 7731. [Google Scholar] [CrossRef]
- Khemani, B.; Patil, S.; Kotecha, K.; Tanwar, S. A review of graph neural networks: Concepts, architectures, techniques, challenges, datasets, applications, and future directions. J. Big Data 2024, 11, 18. [Google Scholar] [CrossRef]
- Gaggion, N.; Mansilla, L.; Mosquera, C.; Milone, D.H.; Ferrante, E. Improving Anatomical Plausibility in Medical Image Segmentation via Hybrid Graph Neural Networks: Applications to Chest X-Ray Analysis. IEEE Trans. Med. Imaging 2023, 42, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Gaggion, N.; Mosquera, C.; Mansilla, L.; Saidman, J.M.; Aineseder, M.; Milone, D.H.; Ferrante, E. CheXmask: A large-scale dataset of anatomical segmentation masks for multi-center chest x-ray images. Sci. Data 2024, 11, 511. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kikuchi, T.; Hanaoka, S.; Nakao, T.; Takenaga, T.; Nomura, Y.; Mori, H.; Yoshikawa, T. Synthesis of Hybrid Data Consisting of Chest Radiographs and Tabular Clinical Records Using Dual Generative Models for COVID-19 Positive Cases. J. Imaging Inf. Med. 2024, 37, 1217–1227. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- See, T.J.; Zhang, D.; Boley, M.; Chalmers, D.K. Graph Neural Network-Based Molecular Property Prediction with Patch Aggregation. J. Chem. Theory Comput. 2024, 20, 8886–8896. [Google Scholar] [CrossRef] [PubMed]
- Rahman, H.; Khan, A.R.; Sadiq, T.; Farooqi, A.H.; Khan, I.U.; Lim, W.H. A Systematic Literature Review of 3D Deep Learning Techniques in Computed Tomography Reconstruction. Tomography 2023, 9, 2158–2189. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bhattacharya, S.; Reddy Maddikunta, P.K.; Pham, Q.V.; Gadekallu, T.R.; Krishnan, S.S.R.; Chowdhary, C.L.; Alazab, M.; Jalil Piran, M. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey. Sustain. Cities Soc. 2021, 65, 102589. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, S.; Li, C.; Wang, R.; Liu, Z.; Wang, M.; Tan, H.; Wu, Y.; Liu, X.; Sun, H.; Yang, R.; et al. Annotation-efficient deep learning for automatic medical image segmentation. Nat. Commun. 2021, 12, 5915. [Google Scholar] [CrossRef]
- Jiang, J.; Chen, X.; Tian, G.; Liu, Y. ViG-UNet: Vision Graph Neural Networks for Medical Image Segmentation. arXiv 2023. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Yang, R.; Wu, Z.; Sun, L.; Zou, L. DRLSU-Net: Level set with U-Net for medical image segmentation. Digit. Signal Process. 2025, 157, 104884. [Google Scholar] [CrossRef]
- Liu, L.; Cheng, J.; Quan, Q.; Wu, F.-X.; Wang, Y.-P.; Wang, J. A survey on U-shaped networks in medical image segmentations. Neurocomputing 2020, 409, 244–258. [Google Scholar] [CrossRef]
- Amaral, A.V.R.; González, J.A.; Moraga, P. Spatio-temporal modeling of infectious diseases by integrating compartment and point process models. Stoch. Env. Res. Risk Assess. 2023, 37, 1519–1533. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mononen, M.E.; Tanska, P.; Isaksson, H.; Korhonen, R.K. A Novel Method to Simulate the Progression of Collagen Degeneration of Cartilage in the Knee: Data from the Osteoarthritis Initiative. Sci. Rep. 2016, 6, 21415. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Goldenstein, J.; Schooler, J.; Crane, J.C.; Ozhinsky, E.; Pialat, J.B.; Carballido-Gamio, J.; Majumdar, S. Prospective image registration for automated scan prescription of follow-up knee images in quantitative studies. Magn. Reson. Imaging 2011, 29, 693–700. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Dunn, T.C.; Lu, Y.; Jin, H.; Ries, M.D.; Majumdar, S. T2 relaxation time of cartilage at MR imaging: Comparison with severity of knee osteoarthritis. Radiology 2004, 232, 592–598. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Du, Y.; Guo, Y.; Calhoun, V.D. Aging brain shows joint declines in brain within-network connectivity and between-network connectivity: A large-sample study (n > 6000). Front. Aging Neurosci. 2023, 15, 1159054. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, J.; Ma, J.; Xi, H.; Li, J.; Zhu, J. Multi-scale graph harmonies: Unleashing U-Net’s potential for medical image segmentation through contrastive learning. Neural Netw. 2025, 182, 106914. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Schwartz, L.H.; Zhao, B. Automatic liver segmentation by integrating fully convolutional networks into active contour models. Med. Phys. 2019, 46, 4455–4469. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.; Karwowski, W. Graph Neural Networks in Brain Connectivity Studies: Methods, Challenges, and Future Directions. Brain Sci. 2024, 15, 17. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lu, H.-Y.; Li, Y.; Kaluvakolanu Thyagarajan, U.P.K.; Ma, K.-L. GNNAnatomy: Rethinking model-level explanations for graph neural networks. arXiv 2025, arXiv:2406.04548v3. [Google Scholar] [CrossRef]
- del Río, E. Thick or Thin? Implications of Cartilage Architecture for Osteoarthritis Risk in Sedentary Lifestyles. Biomedicines 2025, 13, 1650. [Google Scholar] [CrossRef]
- Ou, J.; Zhang, J.; Alswadeh, M.; Zhu, Z.; Tang, J.; Sang, H.; Lu, K. Advancing osteoarthritis research: The role of AI in clinical, imaging and omics fields. Bone Res. 2025, 13, 48. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lavalle, S.; Scapaticci, R.; Masiello, E.; Salerno, V.M.; Cuocolo, R.; Cannella, R.; Botteghi, M.; Orro, A.; Saggini, R.; Donati Zeppa, S.; et al. Beyond the Surface: Nutritional Interventions Integrated with Diagnostic Imaging Tools to Target and Preserve Cartilage Integrity: A Narrative Review. Biomedicines 2025, 13, 570. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fan, X.; Sun, A.R.; Young, R.S.E.; Afara, I.O.; Hamilton, B.R.; Ong, L.J.Y.; Crawford, R.; Prasadam, I. Spatial analysis of the osteoarthritis microenvironment: Techniques, insights, and applications. Bone Res. 2024, 12, 7. [Google Scholar] [CrossRef]
- Brakel, B.A.; Sussman, M.S.; Majeed, H.; Teitel, J.; Man, C.; Rayner, T.; Weiss, R.; Moineddin, R.; Blanchette, V.; Doria, A.S. T2 mapping magnetic resonance imaging of cartilage in hemophilia. Res. Pr. Thromb. Haemost. 2023, 7, 102182. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Müller-Franzes, G.; Nolte, T.; Ciba, M.; Schock, J.; Khader, F.; Prescher, A.; Wilms, L.M.; Kuhl, C.; Nebelung, S.; Truhn, D. Fast, Accurate, and Robust T2 Mapping of Articular Cartilage by Neural Networks. Diagnostics 2022, 12, 688. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Kim, J.; Yang, M.; Ok, A.H.; Zbýň, Š.; Link, T.M.; Majumdar, S.; Ma, C.B.; Spindler, K.P.; Winalski, C.S. Cartilage compositional MRI—A narrative review of technical development and clinical applications over the past three decades. Skelet. Radiol. 2024, 53, 1761–1781. [Google Scholar] [CrossRef] [PubMed]
- Kajabi, A.W.; Zbýň, Š.; Smith, J.S.; Hedayati, E.; Knutsen, K.; Tollefson, L.V.; Homan, M.; Abbasguliyev, H.; Takahashi, T.; Metzger, G.J.; et al. Seven tesla knee MRI T2*-mapping detects intrasubstance meniscus degeneration in patients with posterior root tears. Radiol. Adv. 2024, 1, umae005. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Das, T.; Roos, J.C.P.; Patterson, A.J.; Graves, M.J.; Murthy, R. T2-relaxation mapping and fat fraction assessment to objectively quantify clinical activity in thyroid eye disease: An initial feasibility study. Eye 2019, 33, 235–243. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Imamura, R.; Teramoto, A.; Murahashi, Y.; Okada, Y.; Okimura, S.; Akatsuka, Y.; Watanabe, K.; Yamashita, T. Ultra-Short Echo Time-MRI T2* Mapping of Articular Cartilage Layers Is Associated with Histological Early Degeneration. Cartilage 2025, 16, 118–124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hu, Y.; Chen, X.; Wang, S.; Jing, Y.; Su, J. Subchondral bone microenvironment in osteoarthritis and pain. Bone Res. 2021, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Peng, C.; Wang, G.; Deng, C.; Zhang, Z.; Liu, X. Cartilage T2 mapping-based radiomics in knee osteoarthritis research: Status, progress and future outlook. Eur. J. Radiol. 2024, 181, 111826. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, W.; Liu, D.; Chen, P.; Li, P.; Li, F.; Yuan, W.; Wang, S.; Chen, C.; Chen, Q.; et al. Radiomics analysis using magnetic resonance imaging of bone marrow edema for diagnosing knee osteoarthritis. Front. Bioeng. Biotechnol. 2024, 12, 1368188. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alkhatatbeh, T.; Alkhatatbeh, A.; Guo, Q.; Chen, J.; Song, J.; Qin, X.; Wei, W. Interpretable machine learning and radiomics in hip MRI diagnostics: Comparing ONFH and OA predictions to experts. Front. Immunol. 2025, 16, 1532248. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bilgin, E. Current application, possibilities, and challenges of artificial intelligence in the management of rheumatoid arthritis, axial spondyloarthritis, and psoriatic arthritis. Ther. Adv. Musculoskelet. Dis. 2025, 17, 1759720X251343579. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Paladugu, P.; Kumar, R.; Hage, T.; Vaja, S.; Sekhar, T.; Weisberg, S.; Sporn, K.; Waisberg, E.; Ong, J.; Vadhera, A.S.; et al. Leveraging lower body negative pressure for enhanced outcomes in orthopedic arthroplasty—Insights from NASA’s bone health research. Life Sci. Space Res. 2025, 46, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Xuan, A.; Chen, H.; Chen, T.; Li, J.; Lu, S.; Fan, T.; Zeng, D.; Wen, Z.; Ma, J.; Hunter, D.; et al. The application of machine learning in early diagnosis of osteoarthritis: A narrative review. Ther. Adv. Musculoskelet. Dis. 2023, 15, 1759720X231158198. [Google Scholar] [CrossRef]
- Lin, L.; Shi, W.; Ye, J.; Li, J. Multisource Single-Cell Data Integration by MAW Barycenter for Gaussian Mixture Models. Biometrics 2023, 79, 866–877. [Google Scholar] [CrossRef]
- Wu, D.; Ma, T.; Ceritoglu, C.; Li, Y.; Chotiyanonta, J.; Hou, Z.; Hsu, J.; Xu, X.; Brown, T.; Miller, M.I.; et al. Resource atlases for multi-atlas brain segmentations with multiple ontology levels based on T1-weighted MRI. Neuroimage 2016, 125, 120–130. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gao, S.; Zhou, H.; Gao, Y.; Zhuang, X. BayeSeg: Bayesian modeling for medical image segmentation with interpretable generalizability. Med. Image Anal. 2023, 89, 102889. [Google Scholar] [CrossRef]
- Raunig, D.L.; Pennello, G.A.; Delfino, J.G.; Buckler, A.J.; Hall, T.J.; Guimaraes, A.R.; Wang, X.; Huang, E.P.; Barnhart, H.X.; deSouza, N.; et al. Multiparametric Quantitative Imaging Biomarker as a Multivariate Descriptor of Health: A Roadmap. Acad. Radiol. 2023, 30, 159–182. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sel, K.; Hawkins-Daarud, A.; Chaudhuri, A.; Osman, D.; Bahai, A.; Paydarfar, D.; Willcox, K.; Chung, C.; Jafari, R. Survey and perspective on verification, validation, and uncertainty quantification of digital twins for precision medicine. npj Digit. Med. 2025, 8, 40. [Google Scholar] [CrossRef]
- Honarmandi, P.; Duong, T.C.; Ghoreishi, S.F.; Allaire, D.; Arroyave, R. Bayesian uncertainty quantification and information fusion in CALPHAD-based thermodynamic modeling. Acta Mater. 2019, 164, 636–647. [Google Scholar] [CrossRef]
- Maceda, E.; Hector, E.C.; Lenzi, A.; Reich, B.J. A variational neural Bayes framework for inference on intractable posterior distributions. arXiv 2024, arXiv:2404.10899. [Google Scholar] [CrossRef]
- Jeyaraman, M.; Jeyaraman, N.; Nallakumarasamy, A.; Ramasubramanian, S.; Muthu, S. Insights of cartilage imaging in cartilage regeneration. World J. Orthop. 2025, 16, 106416. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Xu, K.; Yang, X. Variational image registration with learned prior using multi-stage VAEs. Comput. Biol. Med. 2024, 178, 108785. [Google Scholar] [CrossRef] [PubMed]
- Hamra, G.; MacLehose, R.; Richardson, D. Markov chain Monte Carlo: An introduction for epidemiologists. Int. J. Epidemiol. 2013, 42, 627–634. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kalyanasundaram, G.; Feng, J.E.; Congiusta, F.; Iorio, R.; DiCaprio, M.; Anoushiravani, A.A. Treating Hepatitis C Before Total Knee Arthroplasty is Cost-Effective: A Markov Analysis. J. Arthroplast. 2024, 39, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Campbell, H.; Gustafson, P. Bayes Factors and Posterior Estimation: Two Sides of the Very Same Coin. Am. Stat. 2022, 77, 248–258. [Google Scholar] [CrossRef]
- Nafi, A.A.N.; Hossain, M.A.; Rifat, R.H.; Zaman, M.M.U.; Ahsan, M.M.; Raman, S. Diffusion-based approaches in medical image generation and analysis. arXiv 2024, arXiv:2412.16860. [Google Scholar] [CrossRef]
- Fernandez, V.; Pinaya, W.H.L.; Borges, P.; Graham, M.S.; Tudosiu, P.D.; Vercauteren, T.; Cardoso, M.J. Generating multi-pathological and multi-modal images and labels for brain MRI. Med. Image Anal. 2024, 97, 103278. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mojiri Forooshani, P.; Biparva, M.; Ntiri, E.E.; Ramirez, J.; Boone, L.; Holmes, M.F.; Adamo, S.; Gao, F.; Ozzoude, M.; Scott, C.J.M.; et al. Deep Bayesian networks for uncertainty estimation and adversarial resistance of white matter hyperintensity segmentation. Hum. Brain Mapp. 2022, 43, 2089–2108. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Friedrich, P.; Wolleb, J.; Bieder, F.; Durrer, A.; Cattin, P.C. WDM: 3D Wavelet Diffusion Models for High-Resolution Medical Image Synthesis. In Deep Generative Models; Springer: Cham, Switzerland, 2024; pp. 11–21. [Google Scholar] [CrossRef]
- Khader, F.; Müller-Franzes, G.; Tayebi Arasteh, S.; Han, T.; Haarburger, C.; Schulze-Hagen, M.; Schad, P.; Engelhardt, S.; Baeßler, B.; Foersch, S.; et al. Denoising diffusion probabilistic models for 3D medical image generation. Sci. Rep. 2023, 13, 7303. [Google Scholar] [CrossRef]
- Wolleb, J.; Sandkühler, R.; Bieder, F.; Valmaggia, P.; Cattin, P.C. Diffusion Models for Implicit Image Segmentation Ensembles. arXiv 2021, arXiv:2112.03145. [Google Scholar] [CrossRef]
- Li, W.; Zhang, J.; Heng, P.A.; Gu, L. Comprehensive Generative Replay for Task-Incremental Segmentation with Concurrent Appearance and Semantic Forgetting. In Medical Image Computing and Computer Assisted Intervention (MICCAI) 2024; Springer Nature: Cham, Switzerland, 2024. [Google Scholar]
- Zhang, T.; Zhang, Q.; Wei, J.; Dai, Q.; Muratovic, D.; Zhang, W.; Diwan, A.; Gu, Z. Nanoparticle-enabled molecular imaging diagnosis of osteoarthritis. Mater. Today Bio 2025, 33, 101952. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Konz, N.; Chen, Y.; Dong, H.; Mazurowski, M.A. Anatomically-Controllable Medical Image Generation with Segmentation-Guided Diffusion Models. arXiv 2024, arXiv:2402.05210. [Google Scholar]
- Chen, B.; Thandiackal, K.; Pati, P.; Goksel, O. Generative appearance replay for continual unsupervised domain adaptation. Med. Image Anal. 2023, 89, 102924. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Mirando, A.J.; Cofer, G.; Qi, Y.; Hilton, M.J.; Johnson, G.A. Characterization complex collagen fiber architecture in knee joint using high-resolution diffusion imaging. Magn. Reson. Med. 2020, 84, 908–919. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dorjsembe, Z.; Pao, H.K.; Odonchimed, S.; Xiao, F. Conditional Diffusion Models for Semantic 3D Brain MRI Synthesis. IEEE J. Biomed. Health Inf. 2024, 28, 4084–4093. [Google Scholar] [CrossRef] [PubMed]
- Chartsias, A.; Joyce, T.; Papanastasiou, G.; Semple, S.; Williams, M.; Newby, D.E.; Dharmakumar, R.; Tsaftaris, S.A. Disentangled representation learning in cardiac image analysis. Med. Image Anal. 2019, 58, 101535. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chartsias, A.; Glocker, B.; Rueckert, D. Adversarial Image Synthesis for Unpaired Multi-Modal Cardiac Data. In Simulation and Synthesis in Medical Imaging; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Mead, K.; Cross, T.; Roger, G.; Sabharwal, R.; Singh, S.; Giannotti, N. MRI deep learning models for assisted diagnosis of knee pathologies: A systematic review. Eur. Radiol. 2025, 35, 2457–2469. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2018; Volume 11045, pp. 3–11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khader, F.; Mueller-Franzes, G.; Tayebi Arasteh, S.; Han, T.; Haarburger, C.; Schulze-Hagen, M.; Schad, P.; Engelhardt, S.; Baeßler, B.; Foersch, S.; et al. Medical Diffusion: Denoising Diffusion Probabilistic Models for 3D Medical Image Generation. arXiv 2022, arXiv:2211.03364. [Google Scholar]
- Nie, D.; Trullo, R.; Lian, J.; Petitjean, C.; Ruan, S.; Wang, Q.; Shen, D. Medical Image Synthesis with Context-Aware Generative Adversarial Networks. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2017; Springer: Cham, Switzerland, 2018; pp. 417–425. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Halilaj, E.; Le, Y.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Modeling and predicting osteoarthritis progression: Data from the osteoarthritis initiative. Osteoarthr. Cartil. 2018, 26, 1643–1650. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kendall, A.; Gal, Y. What Uncertainties Do We Need in Bayesian Deep Learning for Computer Vision? In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Wang, S.; Summers, R.M. Machine learning and radiology. Med. Image Anal. 2012, 16, 933–951. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Damschroder, L.J.; Aron, D.C.; Keith, R.E.; Kirsh, S.R.; Alexander, J.A.; Lowery, J.C. Fostering implementation of health services research findings into practice: A consolidated framework for advancing implementation science. Implement. Sci. 2009, 4, 50. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cook, D.A.; Dupras, D.M. A practical guide to developing effective web-based learning. J. Gen. Intern. Med. 2004, 19, 698–707. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Smith, J.J. Medical Imaging: The Basics of FDA Regulation. 1 August 2006. Available online: https://www.mddionline.com/radiological/medical-imaging-the-basics-of-fda-regulation?utm_ (accessed on 15 August 2025).
- Parikh, R.B.; Teeple, S.; Navathe, A.S. Addressing Bias in Artificial Intelligence in Health Care. J. Am. Med. Assoc. 2019, 322, 2377–2378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Mass spectrometry-based metabolomics: Applications to biomarker and metabolic pathway research. Biomed. Chromatogr. 2016, 30, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. Biology: The big challenges of big data. Nature 2013, 498, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Tarhan, S.; Unlu, Z. Magnetic resonance imaging and ultrasonographic evaluation of the patients with knee osteoarthritis: A comparative study. Clin. Rheumatol. 2003, 22, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Cherry, S.R. Multimodality imaging: Beyond PET/CT and SPECT/CT. Semin. Nucl. Med. 2009, 39, 348–353. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Samek, W.; Wiegand, T.; Müller, K.R. Explainable Artificial Intelligence: Understanding, Visualizing and Interpreting Deep Learning Models. arXiv 2017, arXiv:1708.08296. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The future of digital health with federated learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, S.X.; Dhanaliwala, A.H.; Rudie, J.D.; Rauschecker, A.M.; Roberts-Wolfe, D.; Haddawy, P.; Kahn, C.E., Jr. Bayesian Networks in Radiology. Radiol. Artif. Intell. 2023, 5, e210187. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Harpaldas, H.; Arumugam, S.; Campillo Rodriguez, C.; Kumar, B.A.; Shi, V.; Sia, S.K. Point-of-care diagnostics: Recent developments in a pandemic age. Lab A Chip 2021, 21, 4517–4548. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sheng, B.; Huang, L.; Wang, X.; Zhuang, J.; Tang, L.; Deng, C.; Zhang, Y. Identification of Knee Osteoarthritis Based on Bayesian Network: Pilot Study. JMIR Med. Inf. 2019, 7, e13562. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, R.; Marla, K.; Ravi, P.; Sporn, K.; Srinivas, R.; Vaja, S.; Ngo, A.; Tavakkoli, A. Bayesian Graphical Models for Multiscale Inference in Medical Image-Based Joint Degeneration Analysis. Diagnostics 2025, 15, 2295. https://doi.org/10.3390/diagnostics15182295
Kumar R, Marla K, Ravi P, Sporn K, Srinivas R, Vaja S, Ngo A, Tavakkoli A. Bayesian Graphical Models for Multiscale Inference in Medical Image-Based Joint Degeneration Analysis. Diagnostics. 2025; 15(18):2295. https://doi.org/10.3390/diagnostics15182295
Chicago/Turabian StyleKumar, Rahul, Kiran Marla, Puja Ravi, Kyle Sporn, Rohit Srinivas, Swapna Vaja, Alex Ngo, and Alireza Tavakkoli. 2025. "Bayesian Graphical Models for Multiscale Inference in Medical Image-Based Joint Degeneration Analysis" Diagnostics 15, no. 18: 2295. https://doi.org/10.3390/diagnostics15182295
APA StyleKumar, R., Marla, K., Ravi, P., Sporn, K., Srinivas, R., Vaja, S., Ngo, A., & Tavakkoli, A. (2025). Bayesian Graphical Models for Multiscale Inference in Medical Image-Based Joint Degeneration Analysis. Diagnostics, 15(18), 2295. https://doi.org/10.3390/diagnostics15182295





