Current Trends and Future Directions of Digital Pathology and Artificial Intelligence in Dermatopathology: A Scientometric-Based Review
Abstract
1. Introduction
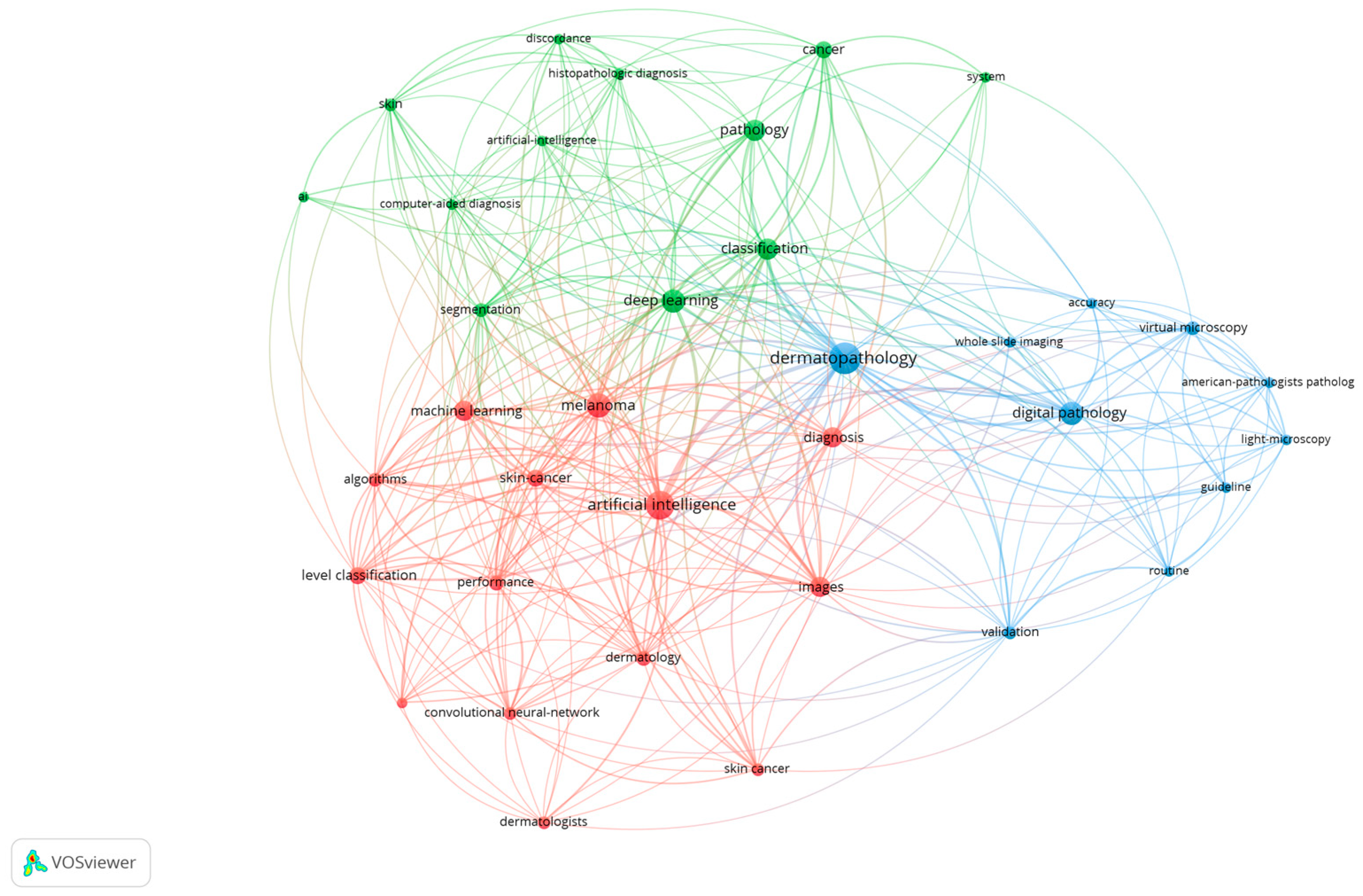
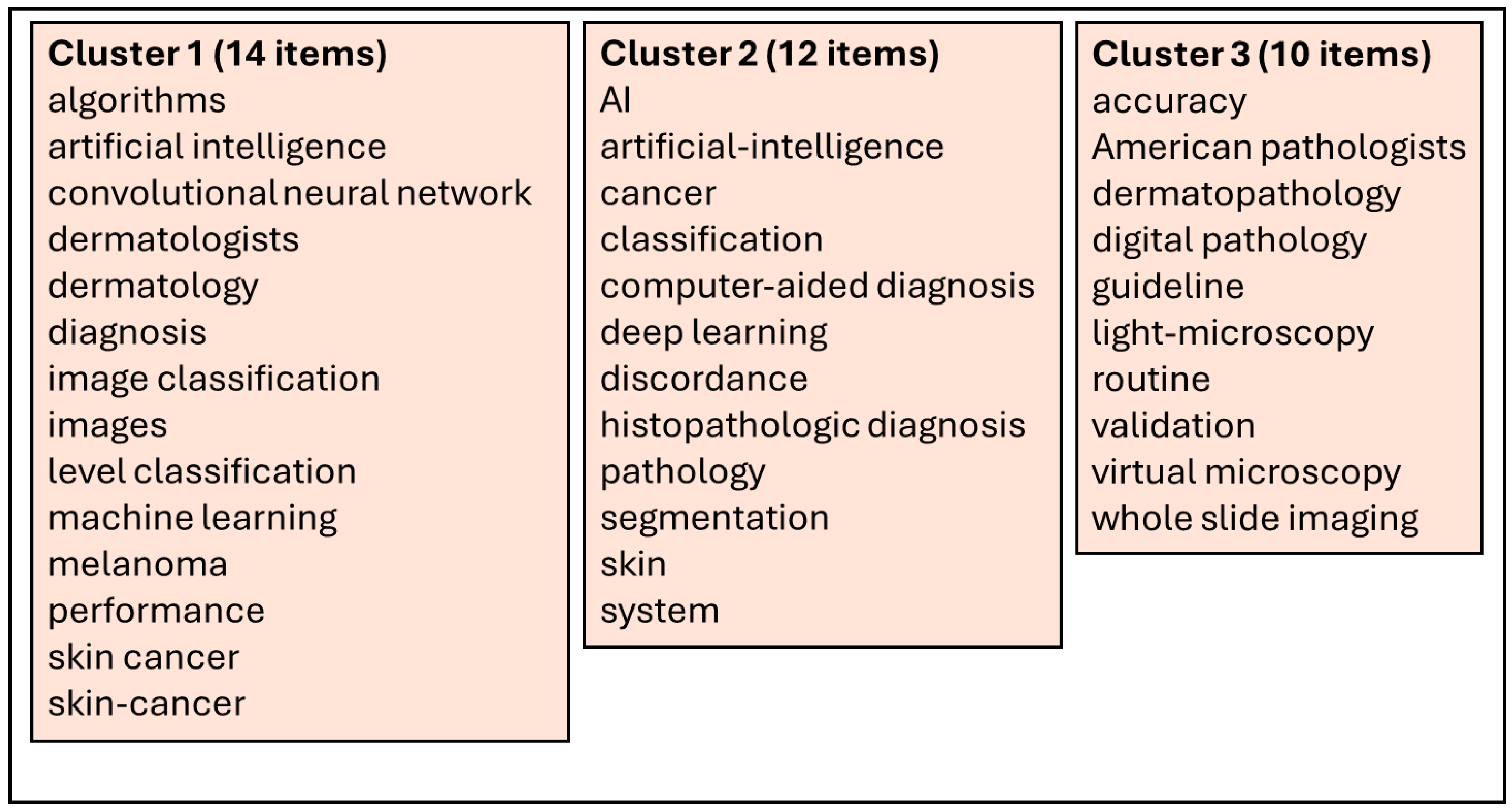
2. Methodology
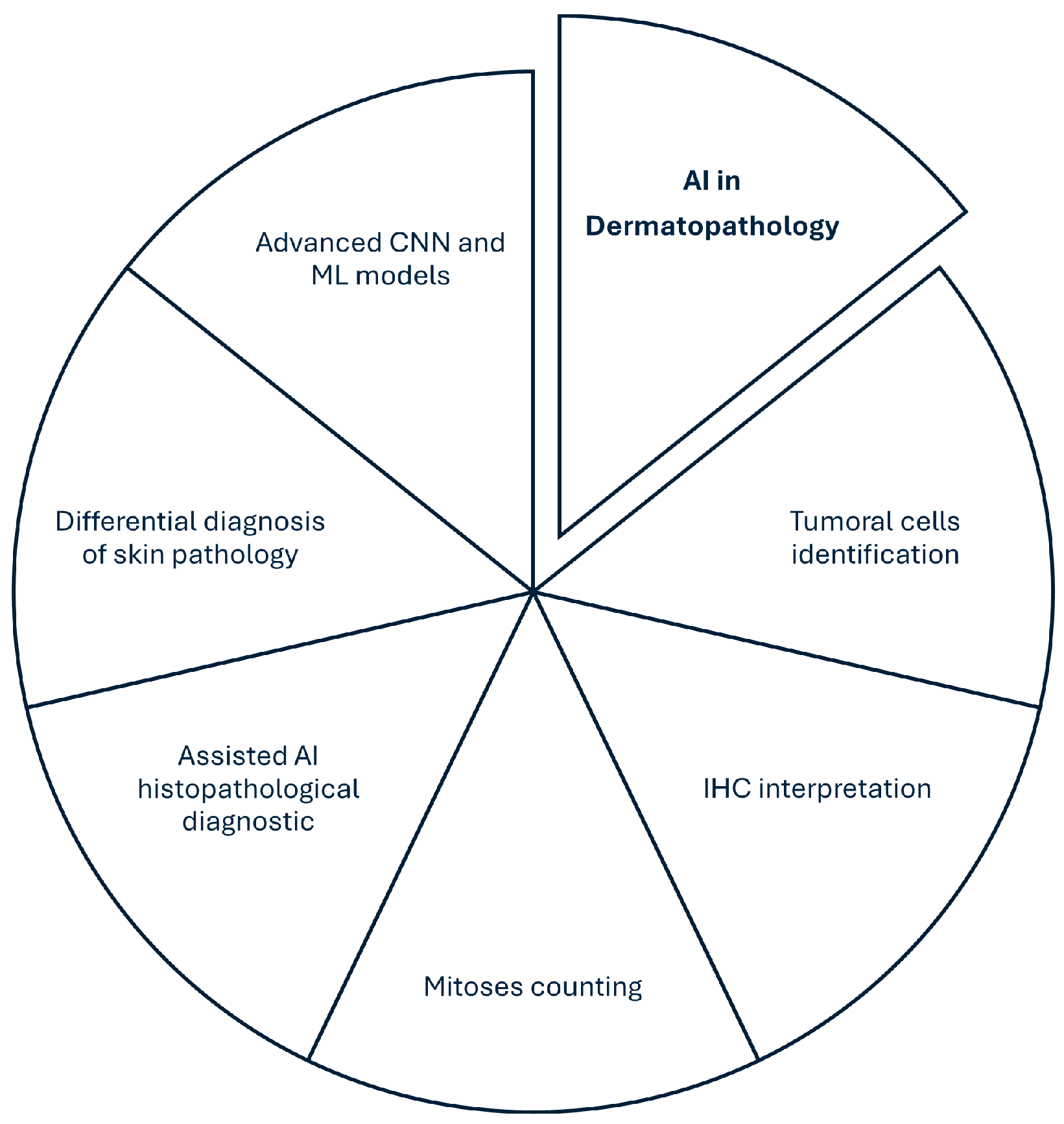
3. Research Direction 1—Advanced Algorithms and Artificial Intelligence for Diagnosis in Dermatopathology
4. Research Direction 2—Integration of Digital Pathology and Standardization in Dermatopathology Practice
5. Research Direction 3—Validation, Regulation, and Global Access Expansion Through Telepathology
6. Conclusions, Foundations and Challenges of Implementing DP and AI in Dermatopathology
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bruce, C.; Prassas, I.; Mokhtar, M.; Clarke, B.; Youssef, E.; Wang, C.; Yousef, G.M. Transforming Diagnostics: The Implementation of Digital Pathology in Clinical Laboratories. Histopathology 2024, 85, 207–214. [Google Scholar] [CrossRef]
- Betmouni, S. Diagnostic Digital Pathology Implementation: Learning from the Digital Health Experience. Digit. Health 2021, 7, 205520762110202. [Google Scholar] [CrossRef]
- Jahn, S.W.; Plass, M.; Moinfar, F. Digital Pathology: Advantages, Limitations and Emerging Perspectives. J. Clin. Med. 2020, 9, 3697. [Google Scholar] [CrossRef]
- Hanna, M.G.; Reuter, V.E.; Hameed, M.R.; Tan, L.K.; Chiang, S.; Sigel, C.; Hollmann, T.; Giri, D.; Samboy, J.; Moradel, C.; et al. Whole Slide Imaging Equivalency and Efficiency Study: Experience at a Large Academic Center. Mod. Pathol. 2019, 32, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Jansen, P.; Baguer, D.O.; Duschner, N.; Arrastia, J.L.; Schmidt, M.; Landsberg, J.; Wenzel, J.; Schadendorf, D.; Hadaschik, E.; Maass, P.; et al. Deep Learning Detection of Melanoma Metastases in Lymph Nodes. Eur. J. Cancer 2023, 188, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Chatziioannou, E.; Roßner, J.; Aung, T.N.; Rimm, D.L.; Niessner, H.; Keim, U.; Serna-Higuita, L.M.; Bonzheim, I.; Cuellar, L.K.; Westphal, D.; et al. Deep Learning-Based Scoring of Tumour-Infiltrating Lymphocytes Is Prognostic in Primary Melanoma and Predictive to PD-1 Checkpoint Inhibition in Melanoma Metastases. EBioMedicine 2023, 93, 104644. [Google Scholar] [CrossRef]
- Fatima, G.; Alhmadi, H.; Mahdi, A.A.; Hadi, N.; Fedacko, J.; Magomedova, A.; Parvez, S.; Raza, A.M. Transforming Diagnostics: A Comprehensive Review of Advances in Digital Pathology. Cureus 2024, 16, e71890. [Google Scholar] [CrossRef]
- Wen, Z.; Wang, S.; Yang, D.; Xie, Y.; Chen, M.; Bishop, J.; Xiao, G. Deep Learning in Digital Pathology for Personalized Treatment Plans of Cancer Patients. Semin. Diagn. Pathol. 2023, 40, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Koteluk, O.; Wartecki, A.; Mazurek, S.; Kołodziejczak, I.; Mackiewicz, A. How Do Machines Learn? Artificial Intelligence as a New Era in Medicine. J. Pers. Med. 2021, 11, 32. [Google Scholar] [CrossRef]
- Rizzo, P.C.; Caputo, A.; Maddalena, E.; Caldonazzi, N.; Girolami, I.; Dei Tos, A.P.; Scarpa, A.; Sbaraglia, M.; Brunelli, M.; Gobbo, S.; et al. Digital Pathology World Tour. Digit. Health 2023, 9, 20552076231194551. [Google Scholar] [CrossRef]
- Zarella, M.D.; Bowman, D.; Aeffner, F.; Farahani, N.; Xthona, A.; Absar, S.F.; Parwani, A.; Bui, M.; Hartman, D.J. A Practical Guide to Whole Slide Imaging: A White Paper from the Digital Pathology Association. Arch. Pathol. Lab. Med. 2019, 143, 222–234. [Google Scholar] [CrossRef]
- Cazzato, G.; Massaro, A.; Colagrande, A.; Lettini, T.; Cicco, S.; Parente, P.; Nacchiero, E.; Lospalluti, L.; Cascardi, E.; Giudice, G.; et al. Dermatopathology of Malignant Melanoma in the Era of Artificial Intelligence: A Single Institutional Experience. Diagnostics 2022, 12, 1972. [Google Scholar] [CrossRef]
- Marletta, S.; Eccher, A.; Martelli, F.M.; Santonicco, N.; Girolami, I.; Scarpa, A.; Pagni, F.; L’Imperio, V.; Pantanowitz, L.; Gobbo, S.; et al. Artificial Intelligence–Based Algorithms for the Diagnosis of Prostate Cancer: A Systematic Review. Am. J. Clin. Pathol. 2024, 161, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.; Zaheer, S. Advancements in Pathology: Digital Transformation, Precision Medicine, and Beyond. J. Pathol. Inform. 2024, 16, 100408. [Google Scholar] [CrossRef]
- Yousif, M.; Hassell, L.; Pantanowitz, L. Impact of COVID-19 on the Adoption of Digital Pathology. In Digital Innovation for Healthcare in COVID-19 Pandemic; Elsevier eBooks; Elsevier: Amsterdam, The Netherlands, 2022; pp. 95–107. [Google Scholar] [CrossRef]
- Lujan, G.M.; Savage, J.; Shana’ah, A.; Yearsley, M.; Thomas, D.; Allenby, P.; Otero, J.; Limbach, A.L.; Cui, X.; Scarl, R.T.; et al. Digital Pathology Initiatives and Experience of a Large Academic Institution during the Coronavirus Disease 2019 (COVID-19) Pandemic. Arch. Pathol. Lab. Med. 2021, 145, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Hanna, M.G.; Reuter, V.E.; Ardon, O.; Kim, D.; Sirintrapun, S.J.; Schüffler, P.J.; Busam, K.J.; Sauter, J.L.; Brogi, E.; Tan, L.K.; et al. Validation of a Digital Pathology System Including Remote Review during the COVID-19 Pandemic. Mod. Pathol. 2020, 33, 2115–2127. [Google Scholar] [CrossRef] [PubMed]
- Browning, L.; Colling, R.; Rakha, E.; Rajpoot, N.; Rittscher, J.; James, J.A.; Salto-Tellez, M.; Snead, D.R.J.; Verrill, C. Digital Pathology and Artificial Intelligence Will Be Key to Supporting Clinical and Academic Cellular Pathology through COVID-19 and Future Crises: The PathLAKE Consortium Perspective. J. Clin. Pathol. 2020, 74, 443–447. [Google Scholar] [CrossRef]
- McGenity, C.; Clarke, E.L.; Jennings, C.; Matthews, G.; Cartlidge, C.; Freduah-Agyemang, H.; Stocken, D.D.; Treanor, D. Artificial Intelligence in Digital Pathology: A Systematic Review and Meta-Analysis of Diagnostic Test Accuracy. npj Digit. Med. 2024, 7, 114. [Google Scholar] [CrossRef]
- Wells, A.; Patel, S.; Lee, J.B.; Motaparthi, K. Artificial Intelligence in Dermatopathology: Diagnosis, Education, and Research. J. Cutan. Pathol. 2021, 48, 1061–1068. [Google Scholar] [CrossRef]
- Jartarkar, S.R. Artificial Intelligence: Its Role in Dermatopathology. Indian J. Dermatol. Venereol. Leprol. 2023, 89, 549–552. [Google Scholar] [CrossRef]
- Wei, M.L.; Tada, M.; So, A.; Torres, R. Artificial Intelligence and Skin Cancer. Front. Med. 2024, 11, 1331895. [Google Scholar] [CrossRef] [PubMed]
- Young, A.T.; Xiong, M.; Pfau, J.; Keiser, M.J.; Wei, M.L. Artificial Intelligence in Dermatology: A Primer. J. Investig. Dermatol. 2020, 140, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Maron, R.C.; Hekler, A.; Stenzinger, A.; Hauschild, A.; Weichenthal, M.; Tiemann, M.; Krahl, D.; Kutzner, H.; Utikal, J.S.; et al. Hidden Variables in Deep Learning Digital Pathology and Their Potential to Cause Batch Effects: Prediction Model Study. J. Med. Internet Res. 2021, 23, e23436. [Google Scholar] [CrossRef]
- Chan, S.; Reddy, V.; Myers, B.; Thibodeaux, Q.; Brownstone, N.; Liao, W. Machine Learning in Dermatology: Current Applications, Opportunities, and Limitations. Dermatol. Ther. 2020, 10, 365–386. [Google Scholar] [CrossRef]
- Shi, Z.; Zhu, J.; Yu, L.; Li, X.; Li, J.; Chen, H.; Chen, L. A Two-Stage End-To-End Deep Learning Framework for Pathologic Examination in Skin Tumor Diagnosis. Am. J. Pathol. 2023, 193, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Sauter, D.; Lodde, G.; Nensa, F.; Schadendorf, D.; Livingstone, E.; Kukuk, M. Validating Automatic Concept-Based Explanations for AI-Based Digital Histopathology. Sensors 2022, 22, 5346. [Google Scholar] [CrossRef]
- Martorell, A.; Martin-Gorgojo, A.; Rios-Vinuela, E.; Rueda-Carnero, J.M.; Alfageme, F.; Taberner, R. Artificial Intelligence in Dermatology: A Threat or an Opportunity? Actas Dermo-Sifiliogr. 2022, 113, 30–46. [Google Scholar] [CrossRef]
- Cazzato, G.; Rongioletti, F. Artificial Intelligence in Dermatopathology: Updates, Strengths, and Challenges. Clin. Dermatol. 2024, 42, 437–442. [Google Scholar] [CrossRef]
- Rezk, E.; Eltorki, M.; El-Dakhakhni, W. Leveraging Artificial Intelligence to Improve the Diversity of Dermatological Skin Color Pathology: Protocol for an Algorithm Development and Validation Study. JMIR Res. Protoc. 2022, 11, e34896. [Google Scholar] [CrossRef]
- Kriegsmann, K.; Lobers, F.; Zgorzelski, C.; Kriegsmann, J.; Janssen, C.; Meliss, R.R.; Muley, T.; Sack, U.; Steinbuss, G.; Kriegsmann, M. Deep Learning for the Detection of Anatomical Tissue Structures and Neoplasms of the Skin on Scanned Histopathological Tissue Sections. Front. Oncol. 2022, 12, 1022967, Erratum in Front. Oncol. 2023, 13, 1201237. [Google Scholar] [CrossRef]
- Amin, S.; Mori, T.; Itoh, T. A Validation Study of Whole Slide Imaging for Primary Diagnosis of Lymphoma. Pathol. Int. 2019, 69, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Siarov, J.; Siarov, A.; Kumar, D.; Paoli, J.; Molne, J.; Neittaanmaki, N. Deep Learning Model Shows Pathologist-Level Detection of Sentinel Node Metastasis of Melanoma and Intra-Nodal Nevi on Whole Slide Images. Front. Med. 2024, 11, 1418013. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, E.J.; Eliahiai, I.; Chaib, S.; Elmorabit, K.; Mouatakid, M.; Kharmoum, J.; Chraibi, M. The State of Telepathology in Africa in the Age of Digital Pathology Advancements: A Bibliometric Analysis and Literature Review. Cureus J. Med. Sci. 2024, 16, e63835. [Google Scholar] [CrossRef]
- Chong, Y.; Kim, D.C.; Jung, C.K.; Kim, D.-C.; Song, S.Y.; Joo, H.J.; Yi, S.-Y.; Medical Informatics Study Group of the Korean Society of Pathologists. Recommendations for Pathologic Practice Using Digital Pathology: Consensus Report of the Korean Society of Pathologists. J. Pathol. Transl. Med. 2020, 54, 437–452. [Google Scholar] [CrossRef]
- Grant, S.R.; Andrew, T.W.; Alvarez, E.V.; Huss, W.J.; Paragh, G. Diagnostic and Prognostic Deep Learning Applications for Histological Assessment of Cutaneous Melanoma. Cancers 2022, 14, 6231. [Google Scholar] [CrossRef]
- Cho, W.C.; Gill, P.; Aung, P.P.; Gu, J.; Nagarajan, P.; Ivan, D.; Curry, J.L.; Prieto, V.G.; Torres-Cabala, C.A. The Utility of Digital Pathology in Improving the Diagnostic Skills of Pathology Trainees in Commonly Encountered Pigmented Cutaneous Lesions during the COVID-19 Pandemic: A Single Academic Institution Experience. Ann. Diagn. Pathol. 2021, 54, 151807. [Google Scholar] [CrossRef]
- Mosquera-Zamudio, A.; Launet, L.; Tabatabaei, Z.; Parra-Medina, R.; Colomer, A.; Moll, O.; Monteagudo, C.; Janssen, E.; Naranjo, V. Deep Learning for Skin Melanocytic Tumors in Whole-Slide Images: A Systematic Review. Cancers 2023, 15, 42. [Google Scholar] [CrossRef]
- Sauter, D.; Lodde, G.; Nensa, F.; Schadendorf, D.; Livingstone, E.; Kukuk, M. Deep Learning in Computational Dermatopathology of Melanoma: A Technical Literature Review. Comput. Biol. Med. 2023, 163, 107083. [Google Scholar] [CrossRef]
- Cazzato, G.; Colagrande, A.; Cimmino, A.; Arezzo, F.; Loizzi, V.; Caporusso, C.; Marangio, M.; Foti, C.; Romita, P.; Lospalluti, L.; et al. Artificial Intelligence in Dermatopathology: New Insights and Perspectives. Dermatopathology 2021, 8, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Zemouri, R.; Devalland, C.; Valmary-Degano, S.; Zerhouni, N. Neural Network: A Future in Pathology? Ann. Pathol. 2019, 39, 119–129. [Google Scholar] [CrossRef]
- Arrastia, J.L.; Heilenktoetter, N.; Baguer, D.O.; Hauberg-Lotte, L.; Boskamp, T.; Hetzer, S.; Duschner, N.; Schaller, J.; Maass, P. Deeply Supervised UNet for Semantic Segmentation to Assist Dermatopathological Assessment of Basal Cell Carcinoma. J. Imaging 2021, 7, 71. [Google Scholar] [CrossRef]
- Rinck, D.; Dittmer, M.; Tinker, D.; Smith, K.; Heinecke, G. National Resident Survey in Dermatopathology: The Role of Slide Scanners in Resident Learning. J. Cutan. Pathol. 2023, 50, 1078–1082. [Google Scholar] [CrossRef]
- Decroos, F.; Springenberg, S.; Lang, T.; Paepper, M.; Zapf, A.; Metze, D.; Steinkraus, V.; Boeer-Auer, A. A Deep Learning Approach for Histopathological Diagnosis of Onychomycosis: Not Inferior to Analogue Diagnosis by Histopathologists. Acta Derm.-Venereol. 2021, 101, adv00532. [Google Scholar] [CrossRef]
- Doeleman, T.; Hondelink, L.M.; Vermeer, M.H.; van Dijk, M.R.; Schrader, A.M.R. Artificial Intelligence in Digital Pathology of Cutaneous Lymphomas: A Review of the Current State and Future Perspectives. Semin. Cancer Biol. 2023, 94, 81–88. [Google Scholar] [CrossRef]
- Smith, H.; Blalock, T.; Stoff, B.K. Ethics of Artificial Intelligence-Assisted Image Interpretation in Dermatopathology. JAAD Int. 2025, 19, 56–57. [Google Scholar] [CrossRef] [PubMed]
- Cazzato, G.; Massaro, A.; Colagrande, A.; Trilli, I.; Ingravallo, G.; Casatta, N.; Lupo, C.; Ronchi, A.; Franco, R.; Maiorano, E.; et al. Artificial Intelligence Applied to a First Screening of Naevoid Melanoma: A New Use of Fast Random Forest Algorithm in Dermatopathology. Curr. Oncol. 2023, 30, 6066–6078. [Google Scholar] [CrossRef] [PubMed]
- Jartarkar, S.R.; Cockerell, C.J.; Patil, A.; Kassir, M.; Babaei, M.; Weidenthaler-Barth, B.; Grabbe, S.; Goldust, M. Artificial Intelligence in Dermatopathology. J. Cosmet. Dermatol. 2023, 22, 1163–1167. [Google Scholar] [CrossRef] [PubMed]
- Polesie, S.; McKee, P.H.; Gardner, J.M.; Gillstedt, M.; Siarov, J.; Neittaanmaki, N.; Paoli, J. Attitudes toward Artificial Intelligence within Dermatopathology: An International Online Survey. Front. Med. 2020, 7, 591952. [Google Scholar] [CrossRef]
- Sturm, B.; Creytens, D.; Smits, J.; Ooms, A.H.A.G.; Eijken, E.; Kurpershoek, E.; Küsters-Vandevelde, H.V.N.; Wauters, C.; Blokx, W.A.M.; van der Laak, J.A.W.M. Computer-Aided Assessment of Melanocytic Lesions by Means of a Mitosis Algorithm. Diagnostics 2022, 12, 436. [Google Scholar] [CrossRef]
- Ibraheim, M.K.; Gupta, R.; Gardner, J.M.; Elsensohn, A. Artificial Intelligence in Dermatopathology: An Analysis of Its Practical Application. Dermatopathology 2023, 10, 93–94. [Google Scholar] [CrossRef]
- Shah, A.; Wahood, S.; Guermazi, D.; Brem, C.E.; Saliba, E. Skin and Syntax: Large Language Models in Dermatopathology. Dermatopathology 2024, 11, 101–111. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, J.; Zhang, Q.; Chang, J.; Lu, D.; Fu, Y. Artificial Intelligence-Aided Recognition of Pathological Characteristics and Subtype Classification of Superficial Perivascular Dermatitis. Front. Med. 2021, 8, 696305. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, V.; Levine, L.; Solans, E.P.; Dola, S.; Chervony, L.; Polak, S. Performance of Automated Classification of Diagnostic Entities in Dermatopathology Validated on Multisite Data Representing the Real-World Variability of Pathology Workload. Arch. Pathol. Lab. Med. 2023, 147, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.M.; Lefevre, J.G.; Baxter, G.; Hamilton, N.A. Non-Melanoma Skin Cancer Segmentation for Histopathology Dataset. Data Brief 2021, 39, 107587. [Google Scholar] [CrossRef]
- Mitteldorf, C.; Tronnier, M. Dermatopathology–Current Status and Development in German-Speaking Dermatology. J. Dtsch. Dermatol. Ges. 2023, 21, 393–398. [Google Scholar] [CrossRef]
- Oh, Y.; Kim, H.M.; Hong, S.W.; Shin, E.; Kim, J.; Choi, Y.J. Digital Dermatopathology and Its Application to Mohs Micrographic Surgery. Yonsei Med. J. 2022, 63, S112–S114. [Google Scholar] [CrossRef]
- Gomolin, A.; Netchiporouk, E.; Gniadecki, R.; Litvinov, I.V. Artificial Intelligence Applications in Dermatology: Where Do We Stand? Front. Med. 2020, 7, 100. [Google Scholar] [CrossRef]
- Sokolov, K.; Shpudeiko, V. Dynamics of the Neural Network Accuracy in the Context of Modernization of the Algorithms of Skin Pathology Recognition. Indian J. Dermatol. 2022, 67, 312. [Google Scholar] [CrossRef] [PubMed]
- Higgins, A.D.; Dunn, R.J.; Malikzai, O.; Ahmadzai, M.; Gardner, J.M.; Stoff, B.K.; McMichael, J.R. Kaposi Sarcoma in Afghanistan: A Case Series from a Tertiary Referral Center. Dermatopathology 2022, 9, 258–270. [Google Scholar] [CrossRef]
- Bertram, C.A.; Stathonikos, N.; Donovan, T.A.; Bartel, A.; Fuchs-Baumgartinger, A.; Lipnik, K.; van Diest, P.J.; Bonsembiante, F.; Klopfleisch, R. Validation of Digital Microscopy: Review of Validation Methods and Sources of Bias. Vet. Pathol. 2022, 59, 26–38. [Google Scholar] [CrossRef]
- Laggis, C.W.; Bailey, E.E.; Novoa, R.; Stewart, C.L.; Stoff, B.; Wanat, K.A.; Barbieri, J.; Kovarik, C. Validation of Image Quality and Diagnostic Accuracy Using a Mobile Phone Camera Microscope Adaptor Compared with Glass Slide Review in Teledermatopathology. Am. J. Dermatopathol. 2020, 42, 349–353. [Google Scholar] [CrossRef]
- Al-Ali, F.; Polesie, S.; Paoli, J.; Aljasser, M.; Salah, L.A. Attitudes towards Artificial Intelligence among Dermatologists Working in Saudi Arabia. Dermatol. Pract. Concept. 2023, 13, e2023035. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.; Shaheed, A.; Patel, R. Artificial Intelligence in Dermoscopy: Enhancing Diagnosis to Distinguish Benign and Malignant Skin Lesions. Cureus J. Med. Sci. 2024, 16, e54656. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, D.; Kleczek, P.; Jaworek-Korjakowska, J.; Dyduch, G.; Gorgon, M. Semi-Supervised Nests of Melanocytes Segmentation Method Using Convolutional Autoencoders. Sensors 2020, 20, 1546. [Google Scholar] [CrossRef] [PubMed]
- Guiter, G.E.; Sapia, S.; Wright, I.A.; Hutchins, G.G.A.; Arayssi, T. Development of a Remote Online Collaborative Medical School Pathology Curriculum with Clinical Correlations, across Several International Sites, through the COVID-19 Pandemic. Med. Sci. Educ. 2021, 31, 549–556. [Google Scholar] [CrossRef]
- Ianni, J.D.; Soans, R.E.; Sankarapandian, S.; Chamarthi, R.V.; Ayyagari, D.; Olsen, T.G.; Bonham, M.J.; Stavish, C.C.; Motaparthi, K.; Cockerell, C.J.; et al. Tailored for Real-World: A Whole Slide Image Classification System Validated on Uncurated Multi-Site Data Emulating the Prospective Pathology Workload. Sci. Rep. 2020, 10, 3217. [Google Scholar] [CrossRef] [PubMed]
- Blocker, S.J.; Cook, J.; Everitt, J.I.; Austin, W.M.; Watts, T.L.; Mowery, Y.M. Automated Nuclear Segmentation in Head and Neck Squamous Cell Carcinoma Pathology Reveals Relationships between Cytometric Features and ESTIMATE Stromal and Immune Scores. Am. J. Pathol. 2022, 192, 1305–1320. [Google Scholar] [CrossRef]
- Meneveau, M.O.; Vavolizza, R.D.; Mohammad, A.; Kumar, P.; Manderfield, J.T.; Callahan, C.; Lynch, K.T.; Abbas, T.; Slingluff, C.L.; Bekiranov, S. A Step toward Personalized Surgical Decision Making Machine Learning Predicts 1 versus Numerous Melanoma Lymph Node Metastases Using RNA-Sequencing. Ann. Surg. 2023, 278, E589–E597. [Google Scholar] [CrossRef]
- Evans, H.; Kimani, P.K.; Hiller, L.; Tsang, Y.W.; Sah, S.; Gopalakrishnan, K.; Boyd, C.; Loughrey, M.B.; Kelly, P.J.; Boyle, D.P.; et al. What Factors Influence Cellular Pathologists’ Confidence in Case Reporting? Virchows Arch. 2025, 486, 1165–1173. [Google Scholar] [CrossRef]
- D’Alonzo, M.; Bozkurt, A.; Alessi-Fox, C.; Gill, M.; Brooks, D.H.; Rajadhyaksha, M.; Kose, K.; Dy, J.G. Semantic Segmentation of Reflectance Confocal Microscopy Mosaics of Pigmented Lesions Using Weak Labels. Sci. Rep. 2021, 11, 3679. [Google Scholar] [CrossRef]
- Brunye, T.T.; Drew, T.; Saikia, M.J.; Kerr, K.F.; Eguchi, M.M.; Lee, A.C.; May, C.; Elder, D.E.; Elmore, J.G. Melanoma in the Blink of an Eye: Pathologists’ Rapid Detection, Classification, and Localization of Skin Abnormalities. Vis. Cogn. 2021, 29, 386–400. [Google Scholar] [CrossRef]
- Koch, E.A.T.; Erdmann, M.; Berking, C.; Kiesewetter, F.; Kramer, R.; Schliep, S.; Heppt, M.V. Standardized Computer-Assisted Analysis of PRAME Immunoreactivity in Dysplastic Nevi and Superficial Spreading Melanomas. Int. J. Mol. Sci. 2023, 24, 6388. [Google Scholar] [CrossRef]
- Quiohilag, K.; Caie, P.; Oniscu, A.; Brenn, T.; Harrison, D. The Differential Expression of Micro-RNAs 21, 200c, 204, 205, and 211 in Benign, Dysplastic and Malignant Melanocytic Lesions and Critical Evaluation of Their Role as Diagnostic Biomarkers. Virchows Arch. 2020, 477, 121–130. [Google Scholar] [CrossRef]
- Ncube, B.; Mars, M.; Scott, R.E. The Need for a Telemedicine Strategy for Botswana? A Scoping Review and Situational Assessment. BMC Health Serv. Res. 2020, 20, 794. [Google Scholar] [CrossRef] [PubMed]
- van Eck, N.J.; Waltman, L. Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef]
- Flores, J.; Misra, R.; Shah, B.; Williams, Y.; Haghighat, B.; Miranda, G.; Jani, P.; Frasier, K. Artificial Intelligence and Machine Learning Transforming Dermatopathology with Diagnosis and Predictive Analytics. Dermis 2025, 5, 29. [Google Scholar] [CrossRef]
- Zia, S.; Yildiz-Aktas, I.Z.; Zia, F.; Parwani, A.V. An Update on Applications of Digital Pathology: Primary Diagnosis; Telepathology, Education and Research. Diagn. Pathol. 2025, 20, 17. [Google Scholar] [CrossRef]
- Elmore, J.G.; Eguchi, M.M.; Barnhill, R.L.; Reisch, L.M.; Elder, D.E.; Piepkorn, M.W.; Brunyé, T.T.; Radick, A.C.; Shucard, H.L.; Knezevich, S.R.; et al. Effect of Prior Diagnoses on Dermatopathologists’ Interpretations of Melanocytic Lesions. JAMA Dermatol. 2022, 158, 1040. [Google Scholar] [CrossRef]
- Faghihi, A.; Fathollahi, M.; Rajabi, R. Diagnosis of Skin Cancer Using VGG16 and VGG19 Based Transfer Learning Models. Multimed. Tools Appl. 2023, 83, 57495–57510. [Google Scholar] [CrossRef]
- Crowley, R.S.; Tseytlin, E.; Jukic, D. ReportTutor—An Intelligent Tutoring System That Uses a Natural Language Interface. AMIA Annu. Symp. Proc. 2025, 2005, 171. [Google Scholar]
- Tran, M.; Schmidle, P.; Guo, R.R.; Wagner, S.J.; Koch, V.; Lupperger, V.; Novotny, B.; Murphree, D.H.; Hardway, H.D.; D’Amato, M.; et al. Generating Dermatopathology Reports from Gigapixel Whole Slide Images with HistoGPT. Nat. Commun. 2025, 16, 4886. [Google Scholar] [CrossRef]
- Lalmalani, R.M.; Lim, C.X.Y.; Oh, C.C. Artificial Intelligence in Dermatopathology: A Systematic Review. Clin. Exp. Dermatol. 2024, 50, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Kamal, R.; AlSamhori, J.F.; Noel, A.; Qaqish, L.N.; Jaber, L.A.; Abujudeh, R.; Hathal, M.; Mohammed, A.Y.; Nashwan, A.J. Transforming Dermatopathology with AI: Addressing Bias, Enhancing Interpretability, and Shaping Future Diagnostics. Dermatol. Rev. 2025, 6, e70018. [Google Scholar] [CrossRef]
- Varnosfaderani, S.M.; Forouzanfar, M. The Role of AI in Hospitals and Clinics: Transforming Healthcare in the 21st Century. Bioengineering 2024, 11, 337. [Google Scholar] [CrossRef]
- Hellmeier, F.; Brosien, K.; Eickhoff, C.; Meyer, A. Beyond One-Time Validation: A Framework for Adaptive Validation of Prognostic and Diagnostic AI-Based Medical Devices. arXiv 2024, arXiv:2409.04794. [Google Scholar] [CrossRef]
- Laohawetwanit, T.; Gonzalez, R.S.; Bychkov, A. Learning at a Distance: Results of an International Survey on the Adoption of Virtual Conferences and Whole Slide Imaging by Pathologists. J. Clin. Pathol. 2024, 77, 632–638. [Google Scholar] [CrossRef]
- Rohr, J.M.; Ginnebaugh, K.; Tuthill, M.; Pimentel, J.; Markin, R. Real-Time Telepathology Is Substantially Equivalent to In-Person Intraoperative Frozen Section Diagnosis. Arch. Pathol. Lab. Med. 2024, 148, 68–73. [Google Scholar] [CrossRef]
- Matthews, G.A.; McGenity, C.; Bansal, D.; Treanor, D. Public Evidence on AI Products for Digital Pathology. npj Digit. Med. 2024, 7, 300. [Google Scholar] [CrossRef]
- McGraw, D.; Mandl, K.D. Privacy Protections to Encourage Use of Health-Relevant Digital Data in a Learning Health System. npj Digit. Med. 2021, 4, 2. [Google Scholar] [CrossRef]
- Jonnagaddala, J.; Wong, Z.S.-Y. Privacy Preserving Strategies for Electronic Health Records in the Era of Large Language Models. npj Digit. Med. 2025, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Charrière, K.; Pazart, L. Clinical Evidence Requirements according to the IVDR 2017/746: Practical Tools and References for Underpinning Clinical Evidence of IVD-MDs. Clin. Chem. Lab. Med. 2023, 61, 1150–1157. [Google Scholar] [CrossRef]
- Schwen, L.O.; Kiehl, T.-R.; Carvalho, R.; Zerbe, N.; Homeyer, A. Digitization of Pathology Labs: A Review of Lessons Learned. Lab. Investig. 2023, 103, 100244. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.E.; Zhang, J.; Huang, H.-C.; Yamashita, R.; Keim-Malpass, J.; Simko, J.P.; DeVries, S.; Morgan, T.M.; Souhami, L.; Dobelbower, M.C.; et al. External Validation of a Digital Pathology-Based Multimodal Artificial Intelligence Architecture in the NRG/RTOG 9902 Phase 3 Trial. Eur. Urol. Oncol. 2024, 7, 1024–1033. [Google Scholar] [CrossRef]
- Warraich, H.J.; Tazbaz, T.; Califf, R.M. FDA Perspective on the Regulation of Artificial Intelligence in Health Care and Biomedicine. JAMA 2024, 333, 241–247. [Google Scholar] [CrossRef]
- McKay, F.; Williams, B.J.; Prestwich, G.; Bansal, D.; Hallowell, N.; Treanor, D. The Ethical Challenges of Artificial Intelligence-Driven Digital Pathology. J. Pathol. Clin. Res. 2022, 8, 209–216. [Google Scholar] [CrossRef]
- Chauhan, C.; Gullapalli, R.R. Ethics of AI in Pathology: Current Paradigms and Emerging Issues. Am. J. Pathol. 2021, 191, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- BJackson, R.; Rashidi, H.H.; Lennerz, J.K.; de Baca, M.E. Ethical and Regulatory Perspectives on Generative Artificial Intelligence in Pathology. Arch. Pathol. Lab. Med. 2024, 149, 123–129. [Google Scholar] [CrossRef]
- Bessen, J.L.; Alexander, M.; Foroughi, O.; Brathwaite, R.; Baser, E.; Lee, L.C.; Perez, O.; Gustavsen, G. Perspectives on Reducing Barriers to the Adoption of Digital and Computational Pathology Technology by Clinical Labs. Diagnostics 2025, 15, 794. [Google Scholar] [CrossRef]
- Clunie, D.A. DICOM Format and Protocol Standardization—A Core Requirement for Digital Pathology Success. Toxicol. Pathol. 2020, 49, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Senel, E.; Bas, Y. Evolution of Telepathology: A Comprehensive Analysis of Global Telepathology Literature between 1986 and 2017. Turk. J. Pathol. 2020, 36, 218–226. [Google Scholar] [CrossRef]
- Battazza, A.; Brasileiro, F.C.d.S.; Tasaka, A.C.; Bulla, C.; Ximenes, P.P.; Hosomi, J.E.; da Silva, P.F.; da Silva, L.F.; de Moura, F.B.C.; Rocha, N.S. Integrating Telepathology and Digital Pathology with Artificial Intelligence: An Inevitable Future. Vet. World 2024, 17, 1667–1671. [Google Scholar] [CrossRef] [PubMed]
- Archila, L.R.; Smith, L.; Sihvo, H.-K.; Westerling-Bui, T.; Koponen, V.; O’Sullivan, D.M.; Camila, M.; Alexander, E.E.; Wang, Y.; Sivasubramaniam, P.; et al. Development and Technical Validation of an Artificial Intelligence Model for Quantitative Analysis of Histopathologic Features of Eosinophilic Esophagitis. J. Pathol. Inform. 2022, 13, 100144. [Google Scholar] [CrossRef]
- Lord, S.J.; Horvath, A.R.; Sandberg, S.; Monaghan, P.J.; M Cobbaert, C.; Reim, M.; Tolios, A.; Mueller, R.; Bossuyt, P.M. Is This Test Fit-For-Purpose? Principles and a Checklist for Evaluating the Clinical Performance of a Test in the New Era of in Vitro Diagnostic (IVD) Regulation. Crit. Rev. Clin. Lab. Sci. 2025, 62, 182–197. [Google Scholar] [CrossRef]
- Song, A.H.; Jaume, G.; Williamson, D.F.K.; Lu, M.; Vaidya, A.; Miller, T.R.; Mahmood, F. Artificial Intelligence for Digital and Computational Pathology. Nat. Rev. Bioeng. 2023, 1, 930–949. [Google Scholar] [CrossRef]
- Herington, J.; McCradden, M.D.; Creel, K.; Boellaard, R.; Jones, E.; Jha, A.K.; Rahmim, A.; Scott, P.J.H.; Sunderland, J.; Wahl, R.L.; et al. Ethical Considerations for Artificial Intelligence in Medical Imaging: Data Collection, Development, and Evaluation. J. Nucl. Med. 2023, 64, 1848–1854. [Google Scholar] [CrossRef] [PubMed]
- Kothari, K.; Damoi, J.O.; Zeizafoun, N.; Asiimwe, P.; Glerum, K.; Bakaleke, M.B.; Giibwa, A.; Umphlett, M.; Marin, M.L.; Zhang, L.P. Increasing Access to Pathology Services in Low- and Middle-Income Countries through Innovative Use of Telepathology. Surg. Endosc. Other Interv. Tech. 2023, 37, 7206–7211. [Google Scholar] [CrossRef] [PubMed]



| Reference | Methodology/Technology | Application/Usage |
|---|---|---|
| [20,29,51,77] | AI pre-screening, CNNs | Highlighting regions of interest, preliminary slide interpretation, tumor lesion classification. |
| [26] | Attention Graph Gated Network + EfficientNetB6 | End-to-end DL framework for multiple skin tumors; patch-wise and slide-wise classification. |
| [29,83] | AutoML | Integration of AI in EMR systems by clinicians without technical expertise. |
| [31] | EfficientNetV2-S | WSI of 386 skin tumors; 98.7% accuracy, confusion between melanoma and BCC/cSCC. |
| [33] | CNN (U-Net, parent–child layers) | Differentiation of nodal metastasis (NM) vs. intranodal nevus (INN); high sensitivity/specificity. |
| [45] | AI for cutaneous lymphoma | Subclassification, biomarker identification, prognostic prediction. |
| [47] | Fast Random Forest | Pre-screening nevoid melanoma on WSI pixel clusters. |
| [80,84] | CNNs | WSI classification of melanocytic nevi vs. melanoma; ~95% accuracy. |
| [50] | CNN-based mitosis detection | Applied to melanocytic lesions; improved mitosis identification but false positives. |
| [80,84] | ResNet (Microsoft), VGG-19 (Oxford) | >9.9 M histology patches; melanoma vs. nevi classification with high accuracy. |
| [81] | ReportTutor (NLP model) | Automated report generation, promoting standardization. |
| [82] | HistoGPT (Generative AI) | Generating pathology reports/images, aiding education and diagnostics. |
| [85] | Explainable AI (XAI) | Improving transparency, mitigating “black-box” risks in clinical adoption. |
| Reference | Methodology/Technology | Application/Usage |
|---|---|---|
| [19] | Whole Slide Imaging (WSI), telepathology | Transition from optical to digital microscopy; secure cloud infrastructure for primary diagnosis. |
| [48,83] | Digital collections and archives | Education, annotation, standardization, dermatopathology teaching. |
| [67,89,90,91,92] | Validation studies, IVD software | Local and international validation of DP platforms; regulatory approval concerns. |
| [84] | Integration of WSI with molecular data | Early detection of aggressive melanoma; therapeutic decision-making. |
| [87,88] | Cloud systems + telepathology | International collaboration, remote consultation, second opinions, optimized human resources. |
| [89,92,93,94,95] | FDA/CE regulation and ISO protocols | Classification of DP/AI as medical devices; risk-based validation. |
| [89] | Registry of AI/DP products (Europe) | Tracking validation and certification of AI-based DP software. |
| [96,97,98] | Low-cost DP (microscope camera, cloud) | Implementation in resource-limited settings. |
| [99,100] | DICOM standards for pathology | Standardization and interoperability for WSI images. |
| [101,102] | Telepathology in collaborative networks | Multicenter research, biomarker validation, second-opinion services. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cocuz, I.G.; Niculescu, R.; Popelea, M.-C.; Cocuz, M.E.; Sabău, A.-H.; Tinca, A.-C.; Cozac-Szoke, A.R.; Chiorean, D.M.; Budin, C.E.; Cotoi, O.S. Current Trends and Future Directions of Digital Pathology and Artificial Intelligence in Dermatopathology: A Scientometric-Based Review. Diagnostics 2025, 15, 2196. https://doi.org/10.3390/diagnostics15172196
Cocuz IG, Niculescu R, Popelea M-C, Cocuz ME, Sabău A-H, Tinca A-C, Cozac-Szoke AR, Chiorean DM, Budin CE, Cotoi OS. Current Trends and Future Directions of Digital Pathology and Artificial Intelligence in Dermatopathology: A Scientometric-Based Review. Diagnostics. 2025; 15(17):2196. https://doi.org/10.3390/diagnostics15172196
Chicago/Turabian StyleCocuz, Iuliu Gabriel, Raluca Niculescu, Maria-Cătălina Popelea, Maria Elena Cocuz, Adrian-Horațiu Sabău, Andreea-Cătălina Tinca, Andreea Raluca Cozac-Szoke, Diana Maria Chiorean, Corina Eugenia Budin, and Ovidiu Simion Cotoi. 2025. "Current Trends and Future Directions of Digital Pathology and Artificial Intelligence in Dermatopathology: A Scientometric-Based Review" Diagnostics 15, no. 17: 2196. https://doi.org/10.3390/diagnostics15172196
APA StyleCocuz, I. G., Niculescu, R., Popelea, M.-C., Cocuz, M. E., Sabău, A.-H., Tinca, A.-C., Cozac-Szoke, A. R., Chiorean, D. M., Budin, C. E., & Cotoi, O. S. (2025). Current Trends and Future Directions of Digital Pathology and Artificial Intelligence in Dermatopathology: A Scientometric-Based Review. Diagnostics, 15(17), 2196. https://doi.org/10.3390/diagnostics15172196