Active Microbiological Surveillance for Contrasting Multi-Drug-Resistant Pathogens: Comparison Between a Multiplex Real-Time PCR Method and Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and General Information
2.2. Culture-Based Method
2.3. Real-Time PCR Analysis
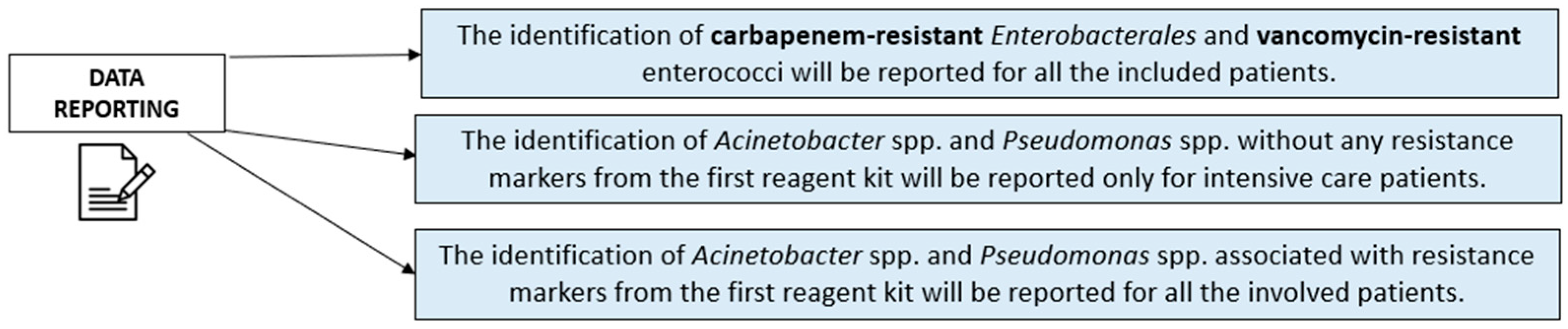
- The identification of carbapenem-resistant Enterobacterales, van genes, and blaCTX-M genes was reported for all the included patients, considering the high dissemination risk from gastrointestinal colonization among critical patients.
- The identification of Acinetobacter spp. and Pseudomonas spp. without any resistance markers from the first reagent kit was reported only for intensive care patients, especially in the case of at least three colonized anatomical sites, due to the risk of related respiratory (i.e., ventilator-associated pneumonia) or systemic infections.
- The identification of Acinetobacter spp. and Pseudomonas spp. associated with resistance markers from the first reagent kit was reported for all the involved patients to limit the resistant strains’ spread among all the wards.
2.4. Data Comparison and Statistical Analysis
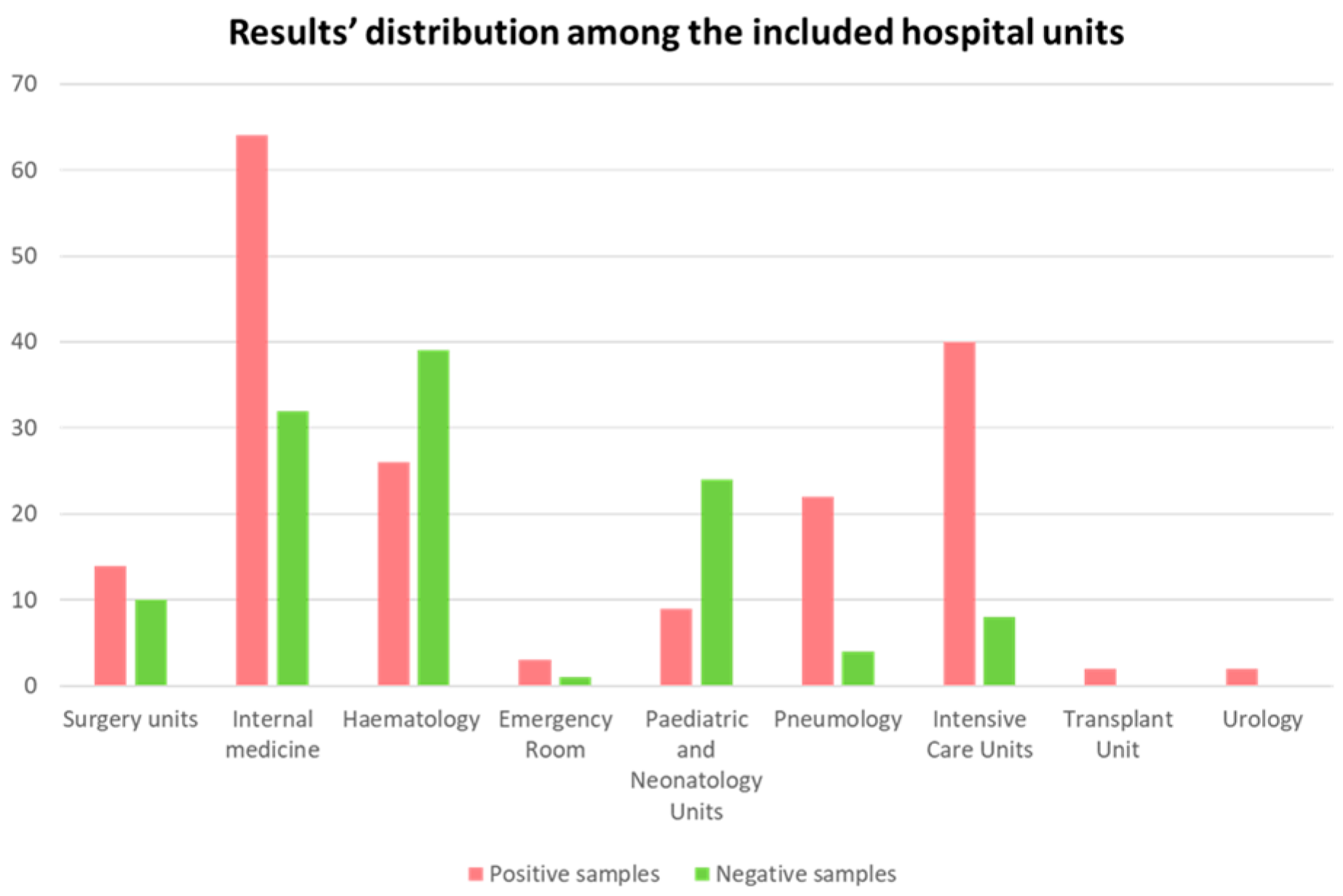
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mojica, M.F.; De La Cadena, E.; Correa, A.; Appel, T.M.; Pallares, C.J.; Villegas, M.V. Evaluation of Allplex™ Entero-DR assay for detection of antimicrobial resistance determinants from bacterial cultures. BMC Res. Notes 2020, 13, 154. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, M.; Tickler, I.A.; Al Deesi, Z.; AlFouzan, W.; Al Jabri, A.; Al Jindan, R.; Al Johani, S.; Alkahtani, S.A.; Al Kharusi, A.; Mokaddas, E.; et al. Molecular detection of carbapenem resistance genes in rectal swabs from patients in Gulf Cooperation Council hospitals. J. Hosp. Infect. 2021, 112, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Antimicrobial%20resistance%20surveillance%20in%20Europe%202023%20-%202021%20data.pdf (accessed on 25 May 2025).
- Available online: https://www.qualitasiciliassr.it/?q=rete-laboratori (accessed on 7 June 2025).
- Jean, S.; Yarbrough, M.L.; Anderson, N.W.; Burnham, C.A. Culture of Rectal Swab Specimens for Enteric Bacterial Pathogens Decreases Time to Test Result While Preserving Assay Sensitivity Compared to Bulk Fecal Specimens. J. Clin. Microbiol. 2019, 57, e02077-18. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.C.; Cataldo, M.A.; Dancer, S.J.; De Angelis, G.; Falcone, M.; Frank, U.; Kahlmeter, G.; Pan, A.; Petrosillo, N.; Rodríguez-Baño, J.; et al. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant gram-negative bacteria in hospitalized patients. Clin. Microbiol. Infect. 2014, 20, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Song, S.A.; Lee, J.N.; Oh, M.; Jo, K.M.; Kim, H.J.; Lee, J.H.; Park, J.; Jang, H.J.; Kim, H.K.; et al. Clinical factors predicting persistent carriage of Klebsiella pneumoniae carbapenemase-producing carbapenem-resistant Enterobacteriaceae among patients with known carriage. J. Hosp. Infect. 2018, 99, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Adler, A.; Navon-Venezia, S.; Moran-Gilad, J.; Marcos, E.; Schwartz, D.; Carmeli, Y. Laboratory and clinical evaluation of screening agar plates for detection of carbapenem-resistant Enterobacteriaceae from surveillance rectal swabs. J. Clin. Microbiol. 2011, 49, 2239–2242. [Google Scholar] [CrossRef] [PubMed]
- Foschi, C.; Gaibani, P.; Lombardo, D.; Re, M.C.; Ambretti, S. Rectal screening for carbapenemase-producing Enterobacteriaceae: A proposed workflow. J. Glob. Antimicrob. Resist. 2020, 21, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.; Maugeri, G.; Bongiorno, D.; Migliorisi, G.; Stefani, S. Integrating an LFA Carbapenemase Detection System into the Laboratory Diagnostic Routine: Preliminary Data and Effectiveness Against Enzyme Variants. Diagnostics 2025, 15, 1434. [Google Scholar] [CrossRef] [PubMed]
- Del Bianco, F.; Morotti, M.; Zannoli, S.; Dirani, G.; Fantini, M.; Pedna, M.F.; Farabegoli, P.; Sambri, V. Comparison of Four Commercial Screening Assays for the Detection of blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA48 in Rectal Secretion Collected by Swabs. Microorganisms 2019, 7, 704. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.; Migliorisi, G.; Maugeri, G.; Bongiorno, D.; Bonomo, C.; Nicitra, E.; Scalia, G.; Stefani, S. The molecular detection of carbapenem markers with a two-levels amplification screening protocol: Epidemiological and resistome insights. Front. Microbiol. 2024, 15, 1346442. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bilavsky, E.; Schwaber, M.J.; Carmeli, Y. How to stem the tide of carbapenemase-producing Enterobacteriaceae: Proactive versus reactive strategies. Curr. Opin. Infect. Dis. 2010, 23, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 15.0. 2025. Available online: https://www.eucast.org (accessed on 15 May 2025).
- Loconsole, D.; Sallustio, A.; Sacco, D.; Santantonio, M.; Casulli, D.; Gatti, D.; Accogli, M.; Parisi, A.; Zagaria, R.; Colella, V.; et al. Genomic surveillance of carbapenem-resistant Klebsiella pneumoniae reveals a prolonged outbreak of extensively drug-resistant ST147 NDM-1 during the COVID-19 pandemic in the Apulia region (Southern Italy). J. Glob. Antimicrob. Resist. 2024, 36, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Pitout, J.D.D.; Dingle, T.C. Laboratory detection of carbapenemases among Gram-negative organisms. Clin. Microbiol. Rev. 2024, 37, e0005422. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yee, R.; Fisher, S.; Bergman, Y.; Chambers, K.K.; Tamma, P.D.; Carroll, K.C.; Simner, P.J. Combined selective culture and molecular methods for the detection of carbapenem-resistant organisms from fecal specimens. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2315–2321. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.K.; Shrestha, D.; Kunwar, A.J.; Thapa, S.; Shrestha, N.; Dhoubhadel, B.G.; Parry, C.M. The overlap of accessory virulence factors and multidrug resistance among clinical and surveillance Klebsiella pneumoniae isolates from a neonatal intensive care unit in Nepal: A single-centre experience in a resource-limited setting. Trop. Med. Health 2024, 52, 30. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maugeri, G.; Calvo, M.; Bongiorno, D.; Bivona, D.; Migliorisi, G.; Privitera, G.F.; Scalia, G.; Stefani, S. Sequencing Analysis of Invasive Carbapenem-Resistant Klebsiella pneumoniae Isolates Secondary to Gastrointestinal Colonization. Microorganisms 2025, 13, 89. [Google Scholar] [CrossRef] [PubMed]

| Statistical Parameter | Percentage |
|---|---|
| Sensitivity | 100% |
| Specificity | 90.4% |
| Positive predictive value | 52.7% |
| Negative predictive value | 100% |
| Statistical Parameter | Percentage |
|---|---|
| Sensitivity | 100% |
| Specificity | 62.2% |
| Positive predictive value | 59.3% |
| Negative predictive value | 100% |
| Statistical Parameter | Percentage |
|---|---|
| Sensitivity | 100% |
| Specificity | 74.2% |
| Positive predictive value | 22.6% |
| Negative predictive value | 100% |
| Resistance Markers | Times of Detection (%) * | Culture Result (%) ** |
|---|---|---|
| blaCTX-M | 51 (29.8) | 5 (9.8%) |
| vanA | 22 (12.9) | 10 (45.4%) |
| blaCTX-M + vanA | 22 (12.9) | 10 VRE (45.4%); 1 case with VRE and CTX-M-producing strains (4.5%) |
| vanB | 16 (9.3) | 0 |
| blaNDM + blaOXA-48 + blaCTX-M | 7 (4.1) | 5 (71.4%) |
| blaNDM + blaOXA-48 + blaCTX-M + vanA | 6 (3.5) | 5 (83.3%) |
| blaNDM + blaKPC + blaOXA-48 + blaCTX-M + vanA | 5 (2.9) | 0 |
| blaKPC + blaCTX-M + vanA | 5 (2.9) | 1 VRE (20%); 1 CTX-M-producing strain (20%) |
| blaKPC | 4 (2.3) | 4 (100%) |
| blaKPC + blaCTX-M | 4 (2.3) | 4 KPC-producing strains (100%) |
| blaKPC + vanA | 4 (2.3) | 2 VRE (50%); 1 KPC-producing strain (25%); 1 case with KPC-producing and VRE strains (25%) |
| blaCTX-M + vanB | 3 (1.7) | 0 |
| vanA + vanB | 3 (1.7) | 1 VRE (33.3%) |
| blaKPC + blaCTX-M + vanB | 2 (1.2) | 1 KPC-producing strain (50%) |
| blaNDM + blaOXA-48 + blaCTX-M + blaVIM | 2 (1.2) | 1 CTX-M-producing strain (50%) |
| blaNDM + blaCTX-M + vanA | 2 (1.2) | 1 NDM-producing strain (50%) |
| vanA + vanB + blaCTX-M | 2 (1.2) | 1 CTX-M-producing strain (50%) |
| blaVIM | 2 (1.2) | 0 |
| blaKPC + blaCTX-M + vanA + blaVIM | 1 (0.6) | 0 |
| blaNDM + blaCTX-M + vanB | 1 (0.6) | 0 |
| blaNDM + blaKPC + blaCTX-M + vanA | 1 (0.6) | 0 |
| blaNDM + blaKPC + blaOXA-48 + blaCTX-M | 1 (0.6) | 1 CTX-M-producing strain (50%) |
| blaNDM | 1 (0.6) | 1 NDM-producing strain (100%) |
| blaNDM + blaOXA-48 + blaCTX-M + vanB | 1 (0.6) | 1 CTX-M-producing strain (50%) |
| blaKPC + blaCTX-M + blaOXA-48 | 1 (0.6) | 0 |
| blaOXA-48 + blaCTX-M + vanA | 1 (0.6) | 1 OXA-48-producing strain (100%) |
| blaVIM + vanB | 1 (0.6) | 1 VIM-producing strain (100%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maugeri, G.; Calvo, M.; Scalia, G.; Stefani, S. Active Microbiological Surveillance for Contrasting Multi-Drug-Resistant Pathogens: Comparison Between a Multiplex Real-Time PCR Method and Culture. Diagnostics 2025, 15, 2128. https://doi.org/10.3390/diagnostics15172128
Maugeri G, Calvo M, Scalia G, Stefani S. Active Microbiological Surveillance for Contrasting Multi-Drug-Resistant Pathogens: Comparison Between a Multiplex Real-Time PCR Method and Culture. Diagnostics. 2025; 15(17):2128. https://doi.org/10.3390/diagnostics15172128
Chicago/Turabian StyleMaugeri, Gaetano, Maddalena Calvo, Guido Scalia, and Stefania Stefani. 2025. "Active Microbiological Surveillance for Contrasting Multi-Drug-Resistant Pathogens: Comparison Between a Multiplex Real-Time PCR Method and Culture" Diagnostics 15, no. 17: 2128. https://doi.org/10.3390/diagnostics15172128
APA StyleMaugeri, G., Calvo, M., Scalia, G., & Stefani, S. (2025). Active Microbiological Surveillance for Contrasting Multi-Drug-Resistant Pathogens: Comparison Between a Multiplex Real-Time PCR Method and Culture. Diagnostics, 15(17), 2128. https://doi.org/10.3390/diagnostics15172128






