Genotype Frequency of HLA-B*58:01 and Its Association with Paraclinical Characteristics and PSORS1C1 rs9263726 in Gout Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Paraclinical Characteristics Analysis of Subjects
2.3. DNA Extraction, PCR Direct Sequencing, and Genotype Analysis
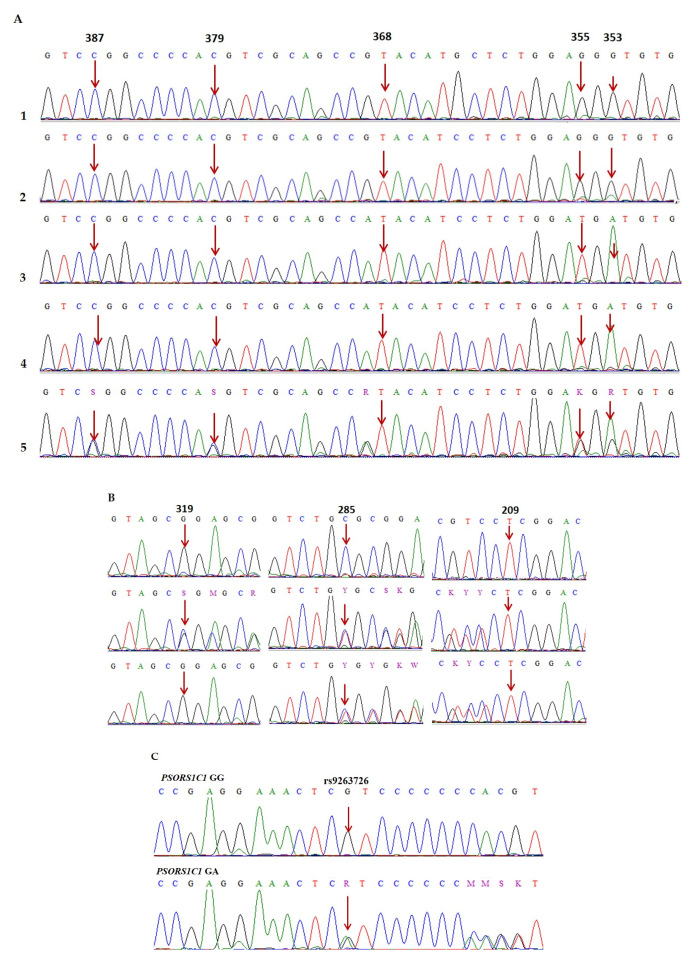
2.4. Method for Identifying the HLA-B*58 Allele and PSORS1C1 rs9263726 in Gout Patients
2.5. Statistical Analysis
3. Results
3.1. Age, Gender, and Paraclinical Characteristics of Subjects
3.2. Genotype and Allele Frequencies of HLA-B*58:01 and PSORS1C1 rs9263726
3.3. Association Between HLA-B*58:01 and Paraclinical Characteristics
3.4. Association Between HLA-B*58:01 and PSORS1C1 rs9263726
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Asghari, K.M.; Zahmatyar, M.; Seyedi, F.; Motamedi, A.; Zolfi, M.; Alamdary, S.J.; Fazlollahi, A.; Shamekh, A.; Mousavi, S.E.; Nejadghaderi, S.A.; et al. Gout: Global epidemiology, risk factors, comorbidities and complications: A narrative review. BMC Musculoskelet. Disord. 2024, 25, 1047. [Google Scholar] [CrossRef] [PubMed]
- Dean, L. Lesinurad Therapy and CYP2C9 genotype. In Medical Genetics Summaries; Pratt, V.M., Pirmohamed, M., Esquivel, B., Kane, M.S., Kattman, B.L., Malheiro, A.J., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2019; pp. 347–353. [Google Scholar]
- Dean, L.; Kane, M. Pegloticase therapy and G6PD genotype. In Medical Genetics Summaries; Pratt, V.M., Pirmohamed, M., Esquivel, B., Kane, M.S., Kattman, B.L., Malheiro, A.J., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2020; pp. 415–424. [Google Scholar]
- Dean, L.; Kane, M. Allopurinol therapy and HLA-B*58:01 genotype. In Medical Genetics Summaries; Pratt, V.M., Pirmohamed, M., Esquivel, B., Kane, M.S., Kattman, B.L., Malheiro, A.J., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2020; pp. 17–27. [Google Scholar]
- Dean, L.; Kane, M. Celecoxib therapy and CYP2C9 genotype. In Medical Genetics Summaries; Pratt, V.M., Pirmohamed, M., Esquivel, B., Kane, M.S., Kattman, B.L., Malheiro, A.J., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2021; pp. 141–150. [Google Scholar]
- Zhang, X.; Ma, H.; Hu, C.; Yu, B.; Ma, W.; Wu, Z.; Luo, X.; Zou, H.; Guan, M. Detection of HLA-B*58: 01 with TaqMan assay and its association with allopurinol-induced sCADR. Clin. Chem. Lab. Med. 2015, 53, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.-I.; Chung, W.-H.; Liou, L.-B.; Chu, C.-C.; Lin, M.; Huang, H.-P.; Lin, Y.-L.; Lan, J.-L.; Yang, L.-C.; Hong, H.-S.; et al. HLA-B*58:01 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc. Natl. Acad. Sci. USA 2005, 102, 4134–4139. [Google Scholar] [CrossRef] [PubMed]
- Jantararoungtong, T.; Chen, P.; Lin, P.-Y.; Tiamkao, S.; Khunarkornsiri, U.; Chucherd, P.; Konyoung, P.; Vannaprasaht, S.; Choonhakarn, C.; Pisuttimarn, P.; et al. Strong association between HLA-B*58:01 and allopurinol-induced Stevens–Johnson syndrome and toxic epidermal necrolysis in a Thai population. Pharmacogenet. Genom. 2009, 19, 704–709. [Google Scholar]
- Tohkin, M.; Kaniwa, N.; Saito, Y.; Sugiyama, E.; Kurose, K.; Nishikawa, J.; Hasegawa, R.; Aihara, M.; Matsunaga, K.; Abe, M.; et al. A whole-genome association study of major determinants for allopurinol-related Stevens–Johnson syndrome and toxic epidermal necrolysis in Japanese patients. Pharmacogenom. J. 2013, 13, 60–69. [Google Scholar] [CrossRef]
- Kang, H.-R.; Jee, Y.K.; Kim, Y.-S.; Hwa, L.C.; Jung, J.-W.; Kim, S.H.; Park, H.-W.; Chang, Y.-S.; Jang, I.-J.; Cho, S.-H.; et al. Positive and negative associations of HLA class I alleles with allopurinol-induced SCARs in Koreans. Pharmacogenet. Genom. 2011, 21, 303–307. [Google Scholar] [CrossRef]
- Low, D.E.; Nurul-Aain, A.F.; Tan, W.C.; Tang, J.J.; Bakhtiar, M.F.; Murad, S.; Park, H.-W.; Chang, Y.-S.; Jang, I.-J.; Cho, S.-H. HLA-B* 58: 01 association in allopurinol-induced severe cutaneous adverse reactions: The implication of ethnicity and clinical phenotypes in multiethnic Malaysia. Pharmacogenet. Genom. 2020, 30, 153–160. [Google Scholar] [CrossRef]
- Tse, T.; Wu, B.; Vagholkar, S.; Willcock, S. Allopurinol for gout: Consider the case for limited HLA-B*58:01 screening. Aust. J. Gen. Pract. 2022, 51, 813–814. [Google Scholar] [CrossRef]
- Hoa, B.; Hang, N.; Kashiwase, K.; Ohashi, J.; Lien, L.; Horie, T.; Shojima, J.; Hijikata, M.; Sakurada, S.; Satake, M.; et al. HLA-A, -B, -C, -DRB1 and-DQB1 alleles and haplotypes in the Kinh population in Vietnam. Tissue Antigens. 2008, 71, 127–134. [Google Scholar] [CrossRef]
- Nguyen, D.V.; Vidal, C.; Chu, H.C.; van Nunen, S. Developing pharmacogenetic screening methods for an emergent country: Vietnam. World Allergy Organ. J. 2019, 12, 100037. [Google Scholar] [CrossRef]
- He, Y.; Hoskins, J.M.; Clark, S.; Campbell, N.H.; Wagner, K.; Motsinger-Reif, A.A.; McLeod, H.L. Accuracy of SNPs to predict risk of HLA alleles associated with drug-induced hypersensitivity events across racial groups. Pharmacogenomics 2015, 16, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Vidal, C.; Li, J.; Fulton, R.; Fernando, S. A polymorphism within the psoriasis susceptibility 1 candidate 1 (PSORS1C1) gene is not linked to HLA-B*58:01 in an Australian cohort. Drug Metab. Pharmacokinet. 2016, 31, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Saksit, N.; Nakkam, N.; Konyoung, P.; Khunarkornsiri, U.; Tassaneeyakul, W.; Chumworathayi, P.; Kanjanawart, S.; Sukasem, C.; Sangviroon, A.; Pattanacheewapull, O.; et al. Comparison between the HLA-B*58:01 Allele and Single-Nucleotide Polymorphisms in Chromosome 6 for Prediction of Allopurinol-Induced Severe Cutaneous Adverse Reactions. J. Immunol. Res. 2017, 2017, 2738784. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, S.; Zhang, J.; Zhang, Y.; Xue, L.; Miao, L. rs9263726 is a specific genetic marker for allopurinol-induced severe cutaneous adverse reactions in Chinese patients. J. Pers. Med. 2015, 12, 585–592. [Google Scholar] [CrossRef]
- Dou, Y.; Peng, P.; Cai, C.; Ye, A.; Kong, L.; Zhang, R. HLA-B*58: 01 and rs9263726 have a linkage, but not absolute linkage disequilibrium in Han Chinese population. Drug Metab. Pharmacokinet. 2018, 33, 228–231. [Google Scholar] [CrossRef]
- González-Galarza, F.F.; Takeshita, L.Y.; Santos, E.J.; Kempson, F.; Maia, M.H.T.; Da Silva, A.L.S.; E Silva, A.L.T.; Ghattaoraya, G.S.; Alfirevic, A.; Jones, A.R.; et al. Allele frequency net 2015 update: New features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. J. Nucleic Acids Res. 2015, 43, D784–D788. [Google Scholar] [CrossRef]
- Nguyen, D.V.; Chu, H.C.; Vidal, C.; Anderson, J.; Nguyen, N.N.; Do, N.Q.T.; Tran, T.L.; Nguyen, T.N.; Nguyen, H.T.T.; Fulton, R.B.; et al. Gene expression profiling in allopurinol-induced severe cutaneous adverse reactions in Vietnamese. Pharmacogenomics 2020, 21, 985–994. [Google Scholar] [CrossRef]
- Singh, J.A.; Gaffo, A. Gout epidemiology and comorbidities. In Seminars in Arthritis and Rheumatism; Elsevier: Amsterdam, The Netherlands, 2020; pp. S11–S16. [Google Scholar]
- Timsans, J.; Palomäki, A.; Kauppi, M. Gout and hyperuricemia: A narrative review of their comorbidities and clinical implications. J. Clin. Med. 2024, 13, 7616. [Google Scholar] [CrossRef]
- Feig, D.I.; Kang, D.-H.; Johnson, R. Uric acid and cardiovascular risk. N. Engl. J. Med. 2008, 359, 1811–1821. [Google Scholar] [CrossRef]
- Jalal, D.I.; Chonchol, M.; Chen, W.; Targher, G. Uric acid as a target of therapy in CKD. Am. J. Kidney Dis. 2013, 61, 134–146. [Google Scholar] [CrossRef]
- Lowe, M.; Payton, A.; Verma, A.; Gemmell, I.; Worthington, J.; Hamilton, P.; Ollier, W.; Augustine, T.; Poulton, K. Human leukocyte antigen associations with renal function among ethnic minorities in the United Kingdom. HLA 2020, 96, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Malekmohammad, K.; Bezsonov, E.E.; Rafieian-Kopaei, M. Role of lipid accumulation and inflammation in atherosclerosis: Focus on molecular and cellular mechanisms. Front. Cardiovasc. Med. 2021, 8, 707529. [Google Scholar] [CrossRef] [PubMed]
- Supriami, K.; Puspitawati, I.; Mayasari, D.S.; Hartopo, A.B. Increased Platelet-derived Microparticles Counts is Correlated with Elevated Blood LDL Cholesterol in Acute Myocardial Infarction. Indones. Biomed. J. 2022, 14, 261–268. [Google Scholar] [CrossRef]
- Dao, H.H.; Harun-Or-Rashid, M.; Sakamoto, J. Body composition and metabolic syndrome in patients with primary gout in Vietnam. J. Rheumatol. 2010, 49, 2400–2407. [Google Scholar] [CrossRef][Green Version]
- Keicho, N.; Itoyama, S.; Kashiwase, K.; Phi, N.C.; Long, H.T.; Ha, L.D.; Van Ban, V.; Hoa, B.K.; Le Hang, N.T.; Hijikata, M.; et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum. Immunol. 2009, 70, 527–531. [Google Scholar] [CrossRef]
- Duc, D.M.; Hoang, L.L.G.; Nguyen, V.T.; Dang, T.N.; Nguyen, N.H.; Vu, H.A.; Mai, T.P. High-Resolution HLA Typing of HLA-A, -B, -C, -DRB1, and -DQB1 in Kinh Vietnamese by Using Next-Generation Sequencing. Front. Genet. 2020, 11, 383. [Google Scholar]
- Smith, A.; Baumgartner, K.; Bositis, C. Cirrhosis: Diagnosis and management. Am. Fam. Physician 2019, 100, 759–770. [Google Scholar]
- Hoang, Y.T.T.; Nguyen, Y.T.; Vu, L.T.; Bui, H.T.T.; Nguyen, Q.V.; Vu, N.P.; Nguyen, T.D.; Nguyen, H.H. Association of ADH1B rs1229984, ADH1C rs698, and ALDH2 rs671 with Alcohol abuse and Alcoholic Cirrhosis in People Living in Northeast Vietnam. Asian Pac. J. Cancer Prev. 2023, 24, 2073. [Google Scholar] [CrossRef]
- Hoang, Y.T.T.; Nguyen, Y.T.; Nguyen, H.D.; Le, A.T.P.; Bui, H.T.T.; Vu, N.P. Single nucleotide polymorphisms of ADH1B, ADH1C and ALDH2 genes in 235 people living in thai nguyen province of Vietnam. Asian Pac. J. Cancer Prev. 2022, 23, 4243. [Google Scholar] [CrossRef]
- Thuong, N.V.; Chinh, N.T.; Ly, T.T.; Hoa, N.P. The nutritional practice of gout patients at Duc Giang general hopital, in 2020. Vietnam. Med. J. 2023, 531, 94–98. (In Vietnamese) [Google Scholar]
- Thuy, N.T.; Thuy, N.T.P.; Trang, T.T.H.; Thyy, N.T.T. Clinical manifastations and laboratory charactoristics of gout and some related factor at Nge An general friendship hospital. Vietnam. Med. J. 2024, 544, 72–73. (In Vietnamese) [Google Scholar]
- Cross, M.; Ong, K.L.; Culbreth, G.T.; Steinmetz, J.D.; Cousin, E.; Lenox, H.; Kopec, J.A.; Haile, L.M.; Brooks, P.M.; Kopansky-Giles, D.R.; et al. Global, regional, and national burden of gout, 1990–2020, and projections to 2050: A systematic analysis of the Global Burden of Disease Study 2021. J. Lancet Rheumatol. 2024, 6, e507–e517. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Pandya, B.J.; Choi, H.K. Prevalence of gout and hyperuricemia in the US general population: The National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheum. 2011, 63, 3136–3141. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-F.; Grainge, M.J.; Zhang, W.; Doherty, M. Global epidemiology of gout: Prevalence, incidence and risk factors. J. Nat. Rev. Rheumatol. 2015, 11, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.L.; Prior, J.A.; Belcher, J.; Hay, C.A.; Mallen, C.D.; Roddy, E. Gender-specific risk factors for gout: A systematic review of cohort studies. J. Adv. Rheumatol. 2019, 59, 24. [Google Scholar] [CrossRef]
- Choi, H.K.; Atkinson, K.; Karlson, E.W.; Willett, W.; Curhan, G. Alcohol intake and risk of incident gout in men: A prospective study. J. Lancet Rheumatol. 2004, 363, 1277–1281. [Google Scholar] [CrossRef]
- Neogi, T.; Chen, C.; Niu, J.; Chaisson, C.; Hunter, D.J.; Zhang, Y. Alcohol quantity and type on risk of recurrent gout attacks: An internet-based case-crossover study. J Am. J. Med. 2014, 127, 311–318. [Google Scholar] [CrossRef]
- Choi, H.K.; Curhan, G. Independent impact of gout on mortality and risk for coronary heart disease. J. Circ. 2007, 116, 894–900. [Google Scholar] [CrossRef]
- Luu, N.B.; Nguyen, T.T. Consumption of Alcohol Beverages in Viet Nam—Some National Investigation Results; National Economics University Publishing Company: Hanoi, Vietnam, 2018; p. 62. [Google Scholar]
- Tayefi, M.; Hassanian, S.M.; Maftouh, M.; Moohebati, M.; Bahrami, A.; Parizadeh, S.M.; Mahdizadeh, A.; Ghazizadeh, H.; Bazeli, J.; Heidari-Bakavoli, A.; et al. Relationship between platelet count and platelet width distribution and serum uric acid concentrations in patients with untreated essential hypertension. J. BioFactors 2018, 44, 532–538. [Google Scholar] [CrossRef]
- Nishida, Y. Relation between creatinine and uric acid excretion. J. Ann. Rheum. Dis. 1992, 51, 101–102. [Google Scholar] [CrossRef]
- Ephraim, R.K.; Awuku, Y.A.; Numekevor, P.; Botchway, F.; Adoba, P.; Dadzie, E.K.; Abrefa, C.A.; Abaka-Yawson, A. Serum Uric acid is a better indicator of kidney impairment than serum uric acid to creatine ratio; a cross sectional study of type 2 diabetes mellitus patients. J. Diabetes Metab. Disord. 2021, 20, 313–320. [Google Scholar] [CrossRef]
- Kaniawati, M.; Wijaya, A.; Susanto, A. The Correlations Between Concentrations of Myeloperoxidase, Serum Amyloid-A Protein and Scretory Phospolipase A-2 with Proinflammatory HDL in Healthy Male Person. Indones. Biomed. J. 2009, 1, 53–60. [Google Scholar] [CrossRef]
- Pirro, M.; Siepi, D.; Lupattelli, G.; Roscini, A.R.; Schillaci, G.; Gemelli, F.; Vaudo, G.; Marchesi, S.; Pasqualini, L.; Mannarino, E. Plasma C-reactive protein in subjects with hypo/hyperalphalipoproteinemias. Metabolism 2003, 52, 432–436. [Google Scholar] [CrossRef]
- Jung, E.; Kong, S.Y.; Ro, Y.S.; Ryu, H.H.; Shin, S.D. Serum cholesterol levels and risk of cardiovascular death: A systematic review and a dose-response meta-analysis of prospective cohort studies. Int. J. Environ. Res. Public Health 2022, 19, 8272. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, D.; Qin, L. Lipid profile and prognosis in patients with coronary heart disease: A meta-analysis of prospective cohort studies. BMC Cardiovasc. Disord. 2021, 21, 69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jin, L.; Wu, Z.; Ma, W.; Chen, Y.; Chen, G.; Wang, L.; Guan, M. Clinical evaluation of a substitute of HLA-B* 58: 01 in different Chinese ethnic groups. J. Genet. Mol. Biol. 2018, 41, 578–584. [Google Scholar] [CrossRef] [PubMed]
| Gene Region | Forward Primer (5′–3′) a | Reverse Primer (5′–3′) b | Fragment Size (bp) |
|---|---|---|---|
| HLA-B exon 2 | CAGTTCTAAAGTCCCCACGCAC | GATCTCGGACCCGGAGACTC | 613 |
| HLA-B exon 3 | AGGCGC GTTTACCCGGTTTC | CATTCAACGGAGGGCGACATTC | 495 |
| PSORS1C1 exon 3 | CTAGCTTTGTCCTCAGGCCAAC | AGAAGGTGCATCTGGCTCACC | 265 |
| Age (year) | Gender | Total | |
|---|---|---|---|
| Male | Female | ||
| ≤40 | 35 (100.0%) | 0 (0.0%) | 35 (26.3%) |
| 41 ≤ 59 | 54 (96.4%) | 2 (3.6%) | 56 (42.1%) |
| ≥60 | 35 (83.3%) | 7 (16.7%) | 42 (31.6%) |
| Total | 124 (93.2%) | 9 (6.8%) | 133 (100.0%) |
| Average age | 51.44 ± 14.59 | 70.33 ± 10.64 | 52.71 ± 15.09 |
| p value | <0.001 | ||
| Paraclinical Characteristics | Gout Patients (N = 133) | Male Gout Patients (N = 124) | Female Gout Patients (N = 9) | |||
|---|---|---|---|---|---|---|
| r | p Value | r | p Value | r | p Value | |
| RBC (1012/L) | −0.0006 | 0.946 | −0.033 | 0.719 | 0.389 | 0.301 |
| HGB (g/L) | −0.064 | 0.465 | −0.101 | 0.265 | 0.325 | 0.393 |
| HCT (%) | −0.061 | 0.487 | −0.107 | 0.238 | 0.406 | 0.278 |
| WBC (109/L) | 0.013 | 0.881 | −0.006 | 0.945 | 0.331 | 0.384 |
| NE (%) | −0.027 | 0.757 | −0.058 | 0.526 | 0.223 | 0.564 |
| LYM (%) | 0.074 | 0.396 | 0.063 | 0.487 | 0.359 | 0.342 |
| PLT (1012/L) | 0.174 | 0.045 | 0.202 | 0.024 | −0.110 | 0.778 |
| Glu (mmol/L) | −0.038 | 0.663 | −0.063 | 0.487 | 0.572 | 0.107 |
| Ure (µmol/L) | 0.159 | 0.067 | 0.217 | 0.016 | −0.330 | 0.386 |
| Cre (µmol/L) | 0.195 | 0.025 | 0.215 | 0.017 | −0.001 | 0.997 |
| TC (mmol/L) | −0.045 | 0.603 | −0.087 | 0.337 | 0.605 | 0.084 |
| TG (mmol/L) | −0.079 | 0.366 | −0.101 | 0.263 | 0.298 | 0.436 |
| HDL-C (mmol/L) | 0.025 | 0.772 | −0.046 | 0.612 | 0.885 | 0.002 |
| LDL-C (mmol/L) | −0.034 | 0.701 | −0.040 | 0.656 | 0.213 | 0.582 |
| Gene | Polymorphism | Nucleotide Change | Genotypes and Alleles | N and n | Frequencies (%) |
|---|---|---|---|---|---|
| HLA-B exons 2 and 3 | c.209A, 285A>G, 319G>C, 353C>T, 355C>A, 368A, 379G>C, 387G>C | *X/*X | 117 | 87.97 | |
| *X/*58:01 | 16 | 12.03 | |||
| *58:01/*58:01 | 0 | 0 | |||
| *X | 249 | 93.98 | |||
| *58:01 | 16 | 6.02 | |||
| PSORS1C1 exon 3 | rs9263726 | c.1418G>A | GG | 112 | 84.21 |
| GA | 21 | 15.79 | |||
| AA | 0 | 0 | |||
| G | 245 | 92.11 | |||
| A | 21 | 7.89 |
| Paraclinical Characteristics | Genotypes | Gout Patients (N = 133) | Male Gout Patients (N = 124) | ||
|---|---|---|---|---|---|
| Mean ± SD | p Value | Mean ± SD | p Value | ||
| RBC (1012/L) | *X/*X | 5.120 ± 1.356 | 0.979 | 5.1982 ± 1.37058 | 0.845 |
| *X/*58:01 | 5.128 ± 0.949 | 5.1288 ± 0.94898 | |||
| HGB (g/L) | *X/*X | 138.11 ± 18.348 | 0.305 | 139.51 ± 18.160 | 0.458 |
| *X/*58:01 | 143.06 ± 15.303 | 143.06 ± 15.303 | |||
| HCT (%) | *X/*X | 41.188 ± 4.857 | 0.102 | 41.597 ± 4.730 | 0.178 |
| *X/*58:01 | 43.344 ± 5.316 | 43.344 ± 5.316 | |||
| WBC (109/L) | *X/*X | 9.649 ± 3.036 | 0.018 | 9.617 ± 2.963 | 0.018 |
| *X/*58:01 | 12.351 ± 9.275 | 12.351 ± 9.275 | |||
| NE (%) | *X/*X | 48.277 ± 27.121 | 0.632 | 50.023 ± 26.214 | 0.811 |
| *X/*58:01 | 51.669 ± 21.459 | 51.669 ± 21.459 | |||
| LYM (%) | *X/*X | 19.097 ± 13.672 | 0.877 | 20.054 ± 13.565 | 0.908 |
| *X/*58:01 | 19.646 ± 10.084 | 19.646 ± 10.084 | |||
| PLT (1012/L) | *X/*X | 274.315 ± 74.725 | 0.757 | 274.646 ± 72.420 | 0.764 |
| *X/*58:01 | 280.366 ± 59.102 | 280.366 ± 59.102 | |||
| Glu (mmol/L) | *X/*X | 6.313 ± 2.855 | 0.288 | 6.352 ± 2.943 | 0.327 |
| *X/*58:01 | 7.098 ± 1.805 | 7.098 ± 1.805 | |||
| Ure (µmol/L) | *X/*X | 6.542 ± 3.337 | 0.228 | 6.353 ± 3.249 | 0.313 |
| *X/*58:01 | 5.513 ± 1.682 | 5.513 ± 1.682 | |||
| Cre (µmol/L) | *X/*X | 104.171 ± 36.676 | 0.535 | 103.316 ± 36.799 | 0.598 |
| *X/*58:01 | 98.339 ± 19.912 | 98.339 ± 9.912 | |||
| Uri (µmol/L) | *X/*X | 522.868 ± 92.849 | 0.788 | 524.669 ± 93.033 | 0.733 |
| *X/*58:01 | 516.348 ± 74.586 | 516.348 ± 74.586 | |||
| TC (mmol/L) | *X/*X | 5.102 ± 0.999 | 0.813 | 5.080 ± 0.991 | 0.874 |
| *X/*58:01 | 5.034 ± 1.525 | 5.034 ± 1.525 | |||
| TG (mmol/L) | *X/*X | 2.792 ± 1.680 | 0.927 | 2.837 ± 1.719 | 0.991 |
| *X/*58:01 | 2.832 ± 1.258 | 2.832 ± 1.258 | |||
| HDL-C (mmol/L) | *X/*X | 1.295 ± 0.328 | 0.216 | 1.279 ± 0.313 | 0.275 |
| *X/*58:01 | 1.189 ± 0.274 | 1.189 ± 0.274 | |||
| LDL-C (mmol/L) | *X/*X | 2.818 ± 0.837 | 0.157 | 2.807 ± 0.859 | 0.183 |
| *X/*58:01 | 2.494 ± 0.978 | 2.494 ± 0.978 | |||
| Genotype | N | (%) | |
|---|---|---|---|
| HLA-B | PSORS1C1 (rs9263726) | 133 | 100 |
| *X/*X | GG | 106 | 79.7 |
| *X/*X | GA | 11 | 8.27 |
| *X/*58:01 | GG | 6 | 4.51 |
| *X/*58:01 | GA | 10 | 7.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, H.T.; Bui, H.T.; Hoang, Y.T.T.; Hoang, M.H.; Ngo, M.D.; Nguyen, M.H.; Nguyen, T.T.T.; Ngo, N.T.; Nguyen, Q.V. Genotype Frequency of HLA-B*58:01 and Its Association with Paraclinical Characteristics and PSORS1C1 rs9263726 in Gout Patients. Diagnostics 2025, 15, 2114. https://doi.org/10.3390/diagnostics15162114
Nguyen HT, Bui HT, Hoang YTT, Hoang MH, Ngo MD, Nguyen MH, Nguyen TTT, Ngo NT, Nguyen QV. Genotype Frequency of HLA-B*58:01 and Its Association with Paraclinical Characteristics and PSORS1C1 rs9263726 in Gout Patients. Diagnostics. 2025; 15(16):2114. https://doi.org/10.3390/diagnostics15162114
Chicago/Turabian StyleNguyen, Hien Thu, Ha Thi Bui, Yen Thi Thu Hoang, My Ha Hoang, Manh Duc Ngo, Mai Hoang Nguyen, Thuy Thi Thanh Nguyen, Nhuan Tien Ngo, and Quang Viet Nguyen. 2025. "Genotype Frequency of HLA-B*58:01 and Its Association with Paraclinical Characteristics and PSORS1C1 rs9263726 in Gout Patients" Diagnostics 15, no. 16: 2114. https://doi.org/10.3390/diagnostics15162114
APA StyleNguyen, H. T., Bui, H. T., Hoang, Y. T. T., Hoang, M. H., Ngo, M. D., Nguyen, M. H., Nguyen, T. T. T., Ngo, N. T., & Nguyen, Q. V. (2025). Genotype Frequency of HLA-B*58:01 and Its Association with Paraclinical Characteristics and PSORS1C1 rs9263726 in Gout Patients. Diagnostics, 15(16), 2114. https://doi.org/10.3390/diagnostics15162114






