EndoNet: A Multiscale Deep Learning Framework for Multiple Gastrointestinal Disease Classification via Endoscopic Images
Abstract
1. Introduction
2. Literature Review
- Multi-model feature fusion was performed not only at the network level but also across different hierarchical layers (shallow and deep), enhancing feature diversity;
- A two-stage attribute optimization was applied (NNMF followed by mRMR), ensuring dimensionality reduction and relevance filtering before classification;
- The final decision-making step was handled using traditional ML classifiers, providing improved interpretability and adaptability;
- The system is designed to be lightweight, avoiding the computational burden of large pre-trained or transformer-based models, making it suitable for real-time diagnostic support.
3. Methodology
3.1. Feature Reduction and Selection Approaches
3.1.1. Non-Negative Matrix Factorization
3.1.2. Minimum Redundancy Maximum Relevance
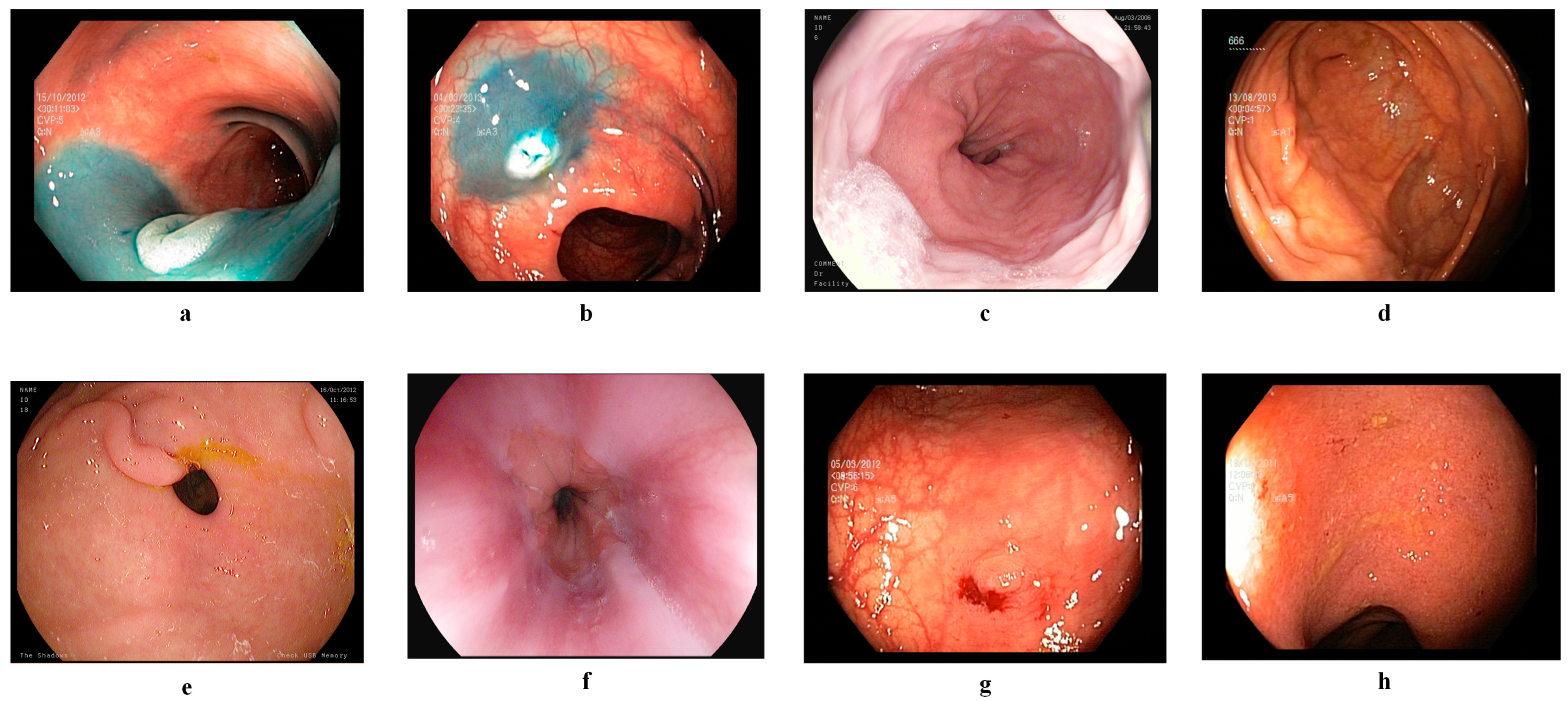
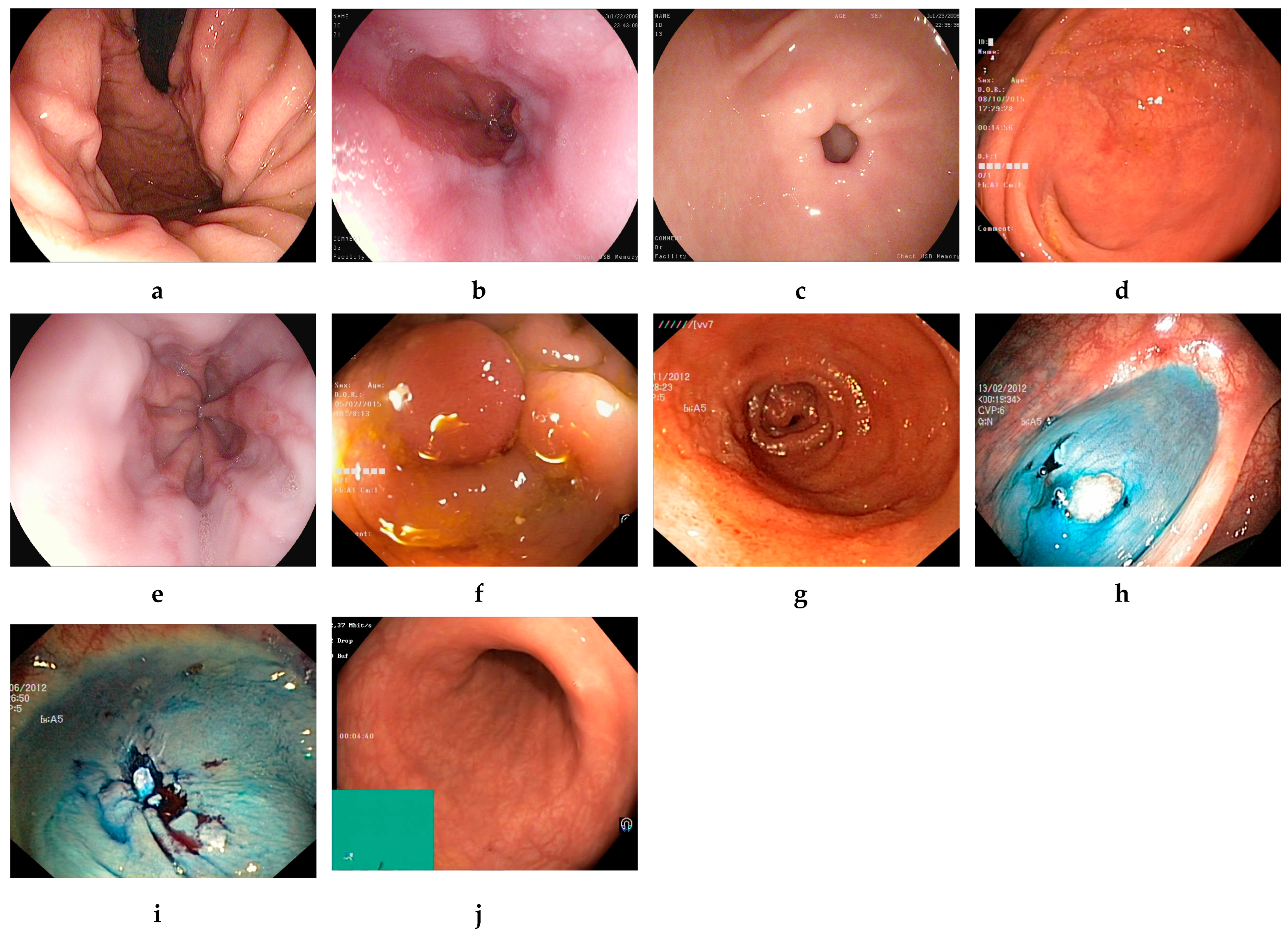
3.2. Dataset Description
3.2.1. Kvasir v2 Dataset
3.2.2. HyperKvasir Dataset
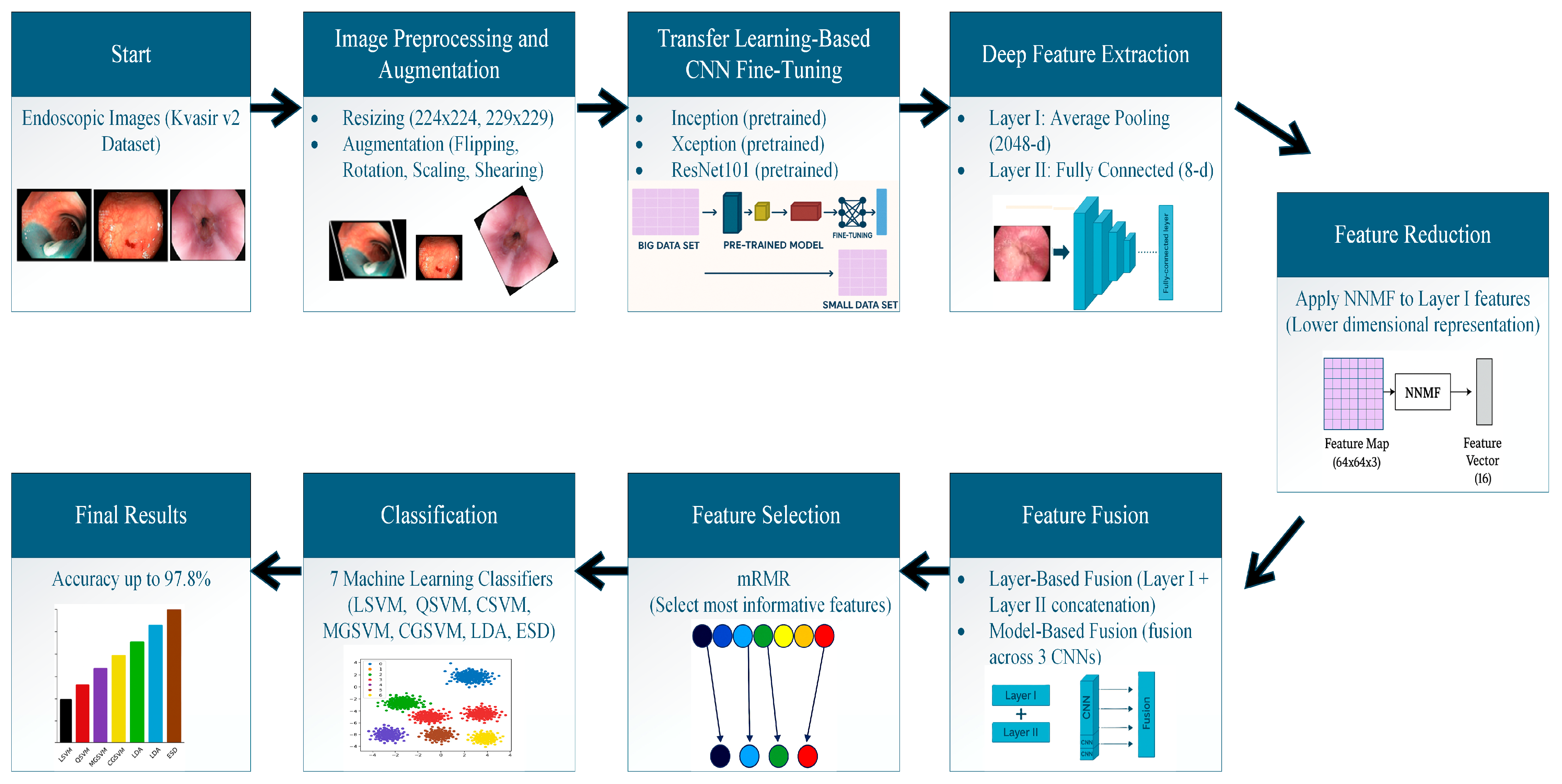
3.3. Proposed Framework
3.3.1. Image Preparation and Augmentation
3.3.2. DL Models Fine-Tuning and Re-Training
3.3.3. Deep Feature Extraction and Reduction
3.3.4. Feature Fusion and Selection
3.3.5. Multi-GI Disease Classification
4. Experimental Setup and Results
4.1. Performance Indicators
4.2. Experimental Setup
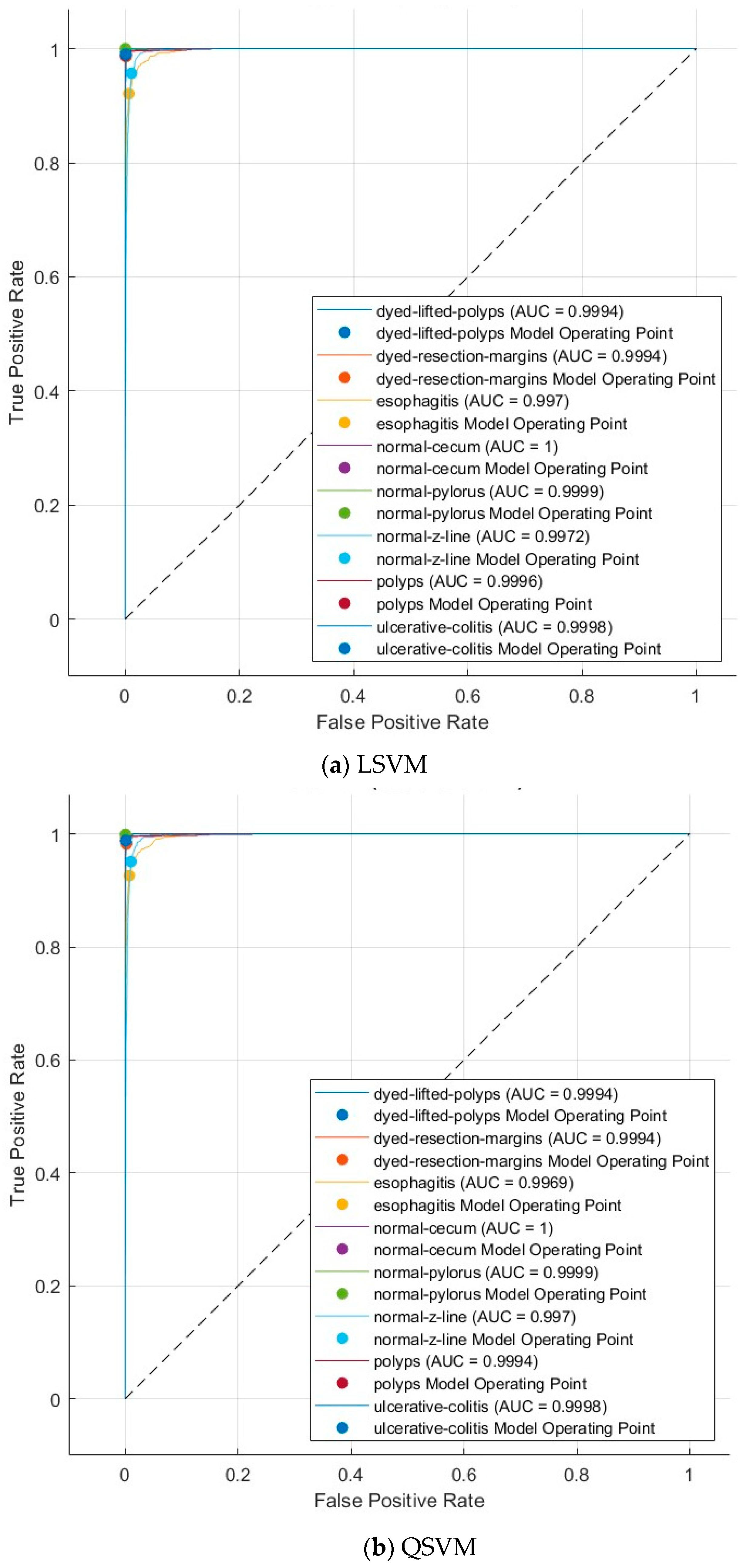
4.3. Results
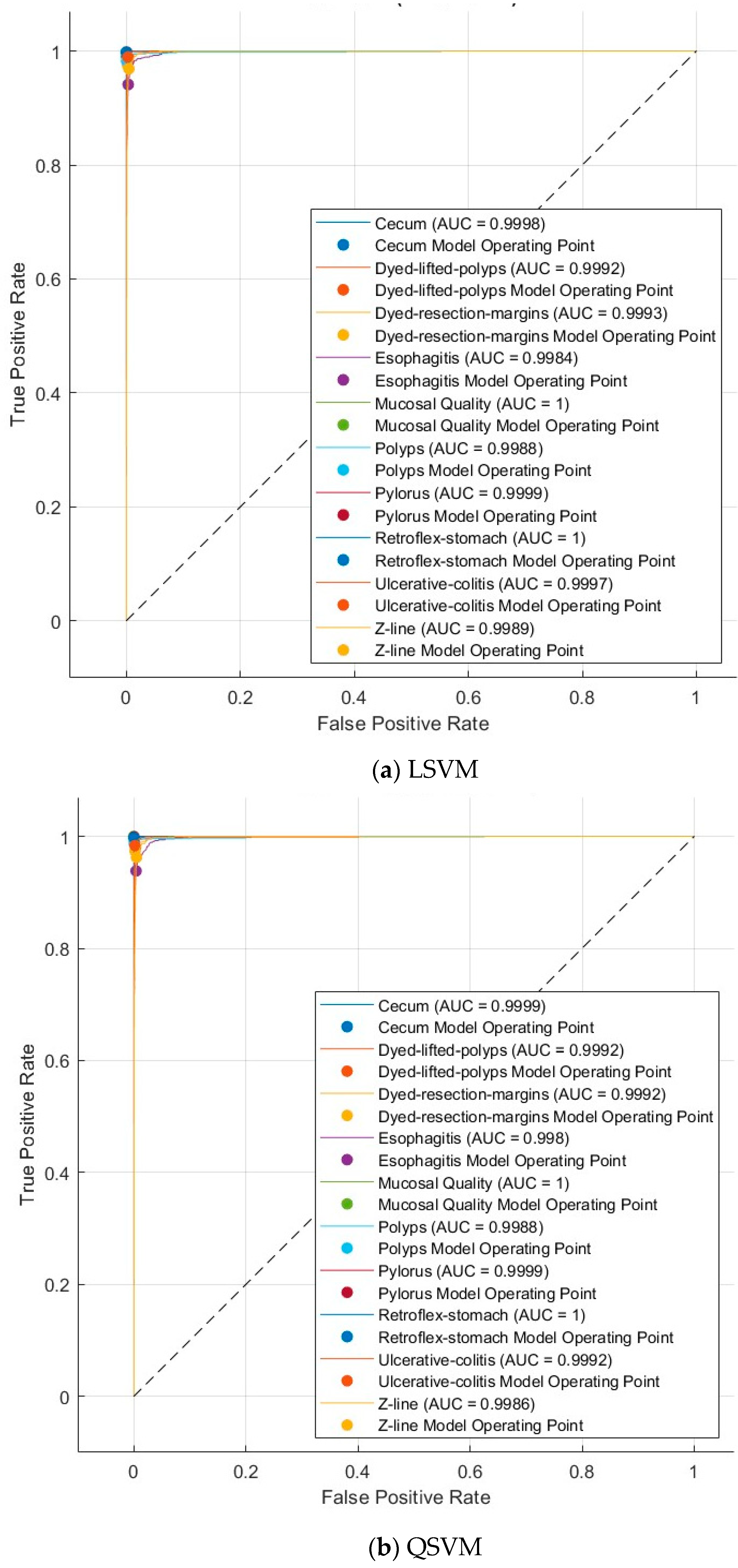
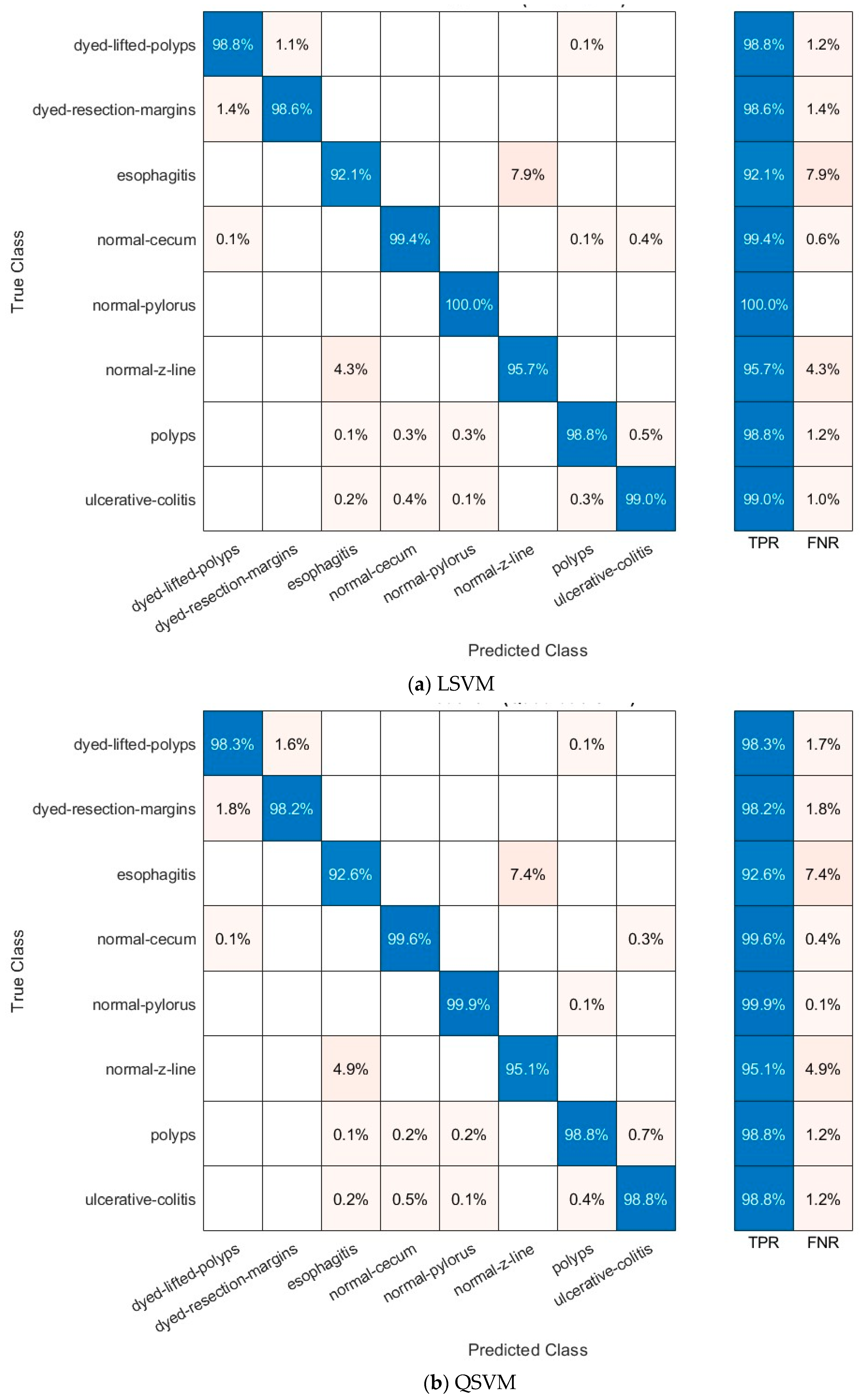
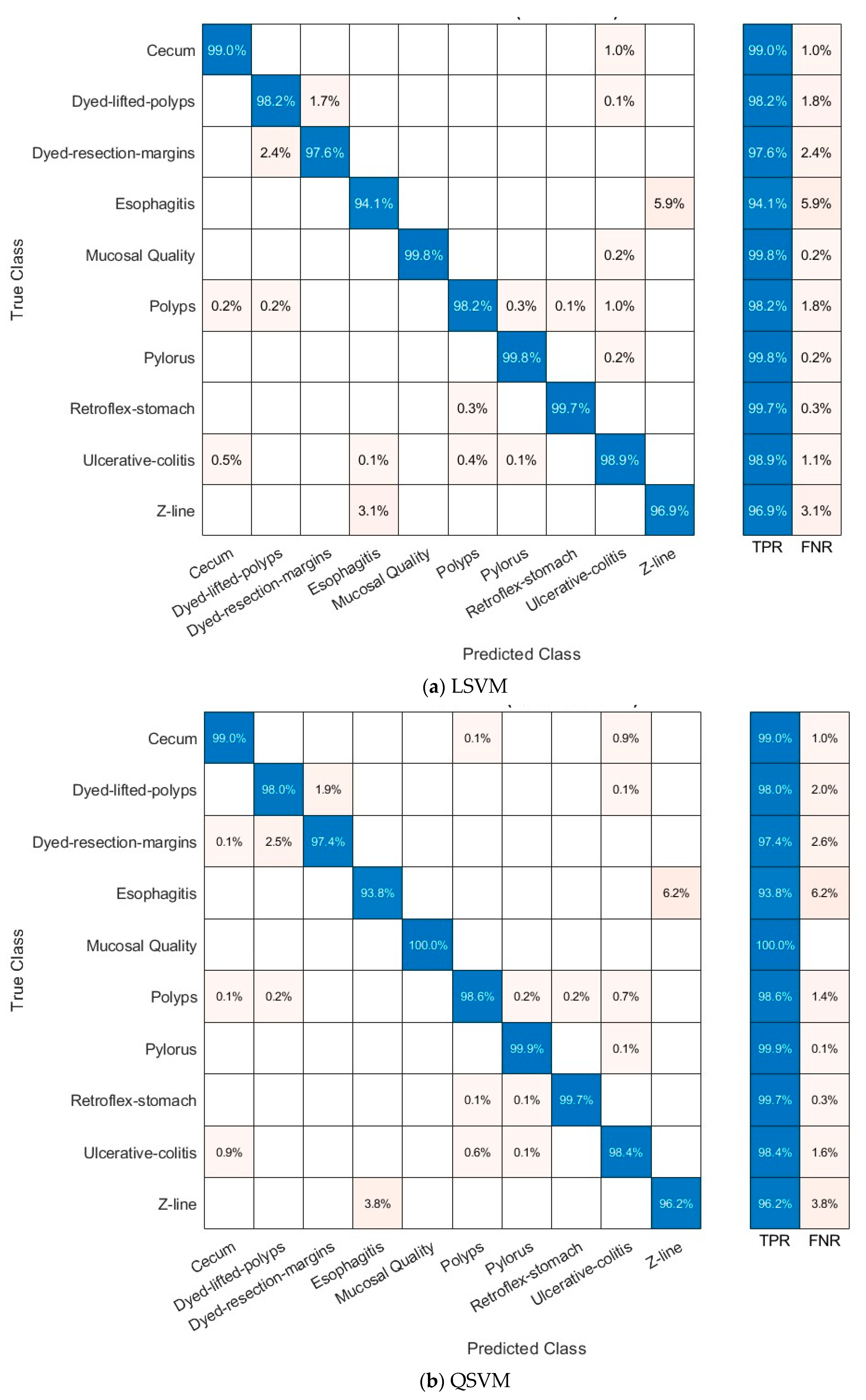
4.3.1. Multi-Layer Feature Extraction Results
4.3.2. Dimensionality Reduction Results
4.3.3. Deep Layer Fusion Level Results
4.3.4. Deep Networks Fusion Level Results
5. Discussion
5.1. State of the Art Comparison
5.2. Limitations and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shaheen, N.; Ransohoff, D.F. Gastroesophageal reflux, barrett esophagus, and esophageal cancer: Scientific review. JAMA 2002, 287, 1972–1981. [Google Scholar] [CrossRef] [PubMed]
- Peery, A.F.; Murphy, C.C.; Anderson, C.; Jensen, E.T.; Deutsch-Link, S.; Egberg, M.D.; Lund, J.L.; Subramaniam, D.; Dellon, E.S.; Sperber, A.D. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the united States: Update 2024. Gastroenterology 2025, 168, 1000–1024. [Google Scholar] [CrossRef]
- Leggett, C.L.; Wang, K.K. Computer-aided diagnosis in GI endoscopy: Looking into the future. Gastrointest. Endosc. 2016, 84, 842–844. [Google Scholar] [CrossRef]
- Bang, C.S.; Lee, J.J.; Baik, G.H. Computer-aided diagnosis of esophageal cancer and neoplasms in endoscopic images: A systematic review and meta-analysis of diagnostic test accuracy. Gastrointest. Endosc. 2021, 93, 1006–1015.e1013. [Google Scholar] [CrossRef]
- Mori, Y.; Kudo, S.-E.; Mohmed, H.E.N.; Misawa, M.; Ogata, N.; Itoh, H.; Oda, M.; Mori, K. Artificial intelligence and upper gastrointestinal endoscopy: Current status and future perspective. Dig. Endosc. 2019, 31, 378–388. [Google Scholar] [CrossRef]
- Aslan, M.F. A robust semantic lung segmentation study for CNN-based COVID-19 diagnosis. Chemom. Intell. Lab. Syst. 2022, 231, 104695. [Google Scholar] [CrossRef] [PubMed]
- Aslan, M.F.; Sabanci, K.; Ropelewska, E. A CNN-Based Solution for Breast Cancer Detection With Blood Analysis Data: Numeric to Image. In Proceedings of the 2021 29th Signal Processing and Communications Applications Conference (SIU), Istanbul, Turkey, 9–11 June 2021; pp. 1–4. [Google Scholar]
- Min, J.K.; Kwak, M.S.; Cha, J.M. Overview of Deep Learning in Gastrointestinal Endoscopy. Gut Liver 2019, 13, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Penrice, D.D.; Rattan, P.; Simonetto, D.A. Artificial Intelligence and the Future of Gastroenterology and Hepatology. Gastro Hep Adv. 2022, 1, 581–595. [Google Scholar] [CrossRef]
- Alayba, A.M.; Senan, E.M.; Alshudukhi, J.S. Enhancing early detection of Alzheimer’s disease through hybrid models based on feature fusion of multi-CNN and handcrafted features. Sci. Rep. 2024, 14, 31203. [Google Scholar] [CrossRef]
- Pan, Z.; Wang, J.; Shen, Z.; Chen, X.; Li, M. Multi-layer convolutional features concatenation with semantic feature selector for vein recognition. IEEE Access 2019, 7, 90608–90619. [Google Scholar] [CrossRef]
- Ravì, D.; Wong, C.; Deligianni, F.; Berthelot, M.; Andreu-Perez, J.; Lo, B.; Yang, G.Z. Deep Learning for Health Informatics. IEEE J. Biomed. Health Inform. 2017, 21, 4–21. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Radovic, M.; Ghalwash, M.; Filipovic, N.; Obradovic, Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinform. 2017, 18, 9. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; de Lange, T.; Johansen, D.; Spampinato, C.; Dang-Nguyen, D.-T.; Lux, M.; Schmidt, P.T. Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection. In Proceedings of the 8th ACM on Multimedia Systems Conference, Taipei, Taiwan, 20–23 June 2017; pp. 164–169. [Google Scholar]
- Kvasir v2. Available online: https://www.kaggle.com/datasets/plhalvorsen/kvasir-v2-a-gastrointestinal-tract-dataset (accessed on 12 June 2025).
- Özbay, F.A.; Özbay, E. Brain tumor detection with mRMR-based multimodal fusion of deep learning from MR images using Grad-CAM. Iran J. Comput. Sci. 2023, 6, 245–259. [Google Scholar] [CrossRef]
- Wang, G.; Lauri, F.; Hassani, A.H.E. Feature Selection by mRMR Method for Heart Disease Diagnosis. IEEE Access 2022, 10, 100786–100796. [Google Scholar] [CrossRef]
- Sarvamangala, D.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2022, 15, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O.; Aslan, M.F.; Sabanci, K. A framework for lung and colon cancer diagnosis via lightweight deep learning models and transformation methods. Diagnostics 2022, 12, 2926. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. Lung and Colon Cancer Classification Using Multiscale Deep Features Integration of Compact Convolutional Neural Networks and Feature Selection. Technologies 2025, 13, 54. [Google Scholar] [CrossRef]
- Attallah, O. Cervical cancer diagnosis based on multi-domain features using deep learning enhanced by handcrafted descriptors. Appl. Sci. 2023, 13, 1916. [Google Scholar] [CrossRef]
- Attallah, O. A Hybrid Trio-Deep Feature Fusion Model for Improved Skin Cancer Classification: Merging Dermoscopic and DCT Images. Technologies 2024, 12, 190. [Google Scholar] [CrossRef]
- Attallah, O. Skin cancer classification leveraging multi-directional compact convolutional neural network ensembles and gabor wavelets. Sci. Rep. 2024, 14, 20637. [Google Scholar] [CrossRef]
- Siddiqui, S.; Akram, T.; Ashraf, I.; Raza, M.; Khan, M.A.; Damaševičius, R. CG-Net: A novel CNN framework for gastrointestinal tract diseases classification. Int. J. Imaging Syst. Technol. 2024, 34, e23081. [Google Scholar] [CrossRef]
- Siddiqui, S.; Khan, J.A.; Algamdi, S. Deep ensemble learning for gastrointestinal diagnosis using endoscopic image classification. PeerJ Comput. Sci. 2025, 11, e2809. [Google Scholar] [CrossRef]
- Werner, J.; Gerum, C.; Nick, J.; Floch, M.L.; Brinkmann, F.; Hampe, J.; Bringmann, O. Enhanced Anomaly Detection for Capsule Endoscopy Using Ensemble Learning Strategies. arXiv 2025. [Google Scholar] [CrossRef]
- Dermyer, P.; Kalra, A.; Schwartz, M. Endodino: A foundation model for gi endoscopy. arXiv 2025. [Google Scholar] [CrossRef]
- Sivari, E.; Bostanci, E.; Guzel, M.S.; Acici, K.; Asuroglu, T.; Ercelebi Ayyildiz, T. A new approach for gastrointestinal tract findings detection and classification: Deep learning-based hybrid stacking ensemble models. Diagnostics 2023, 13, 720. [Google Scholar] [CrossRef]
- Gunasekaran, H.; Ramalakshmi, K.; Swaminathan, D.K.; Mazzara, M. GIT-Net: An ensemble deep learning-based GI tract classification of endoscopic images. Bioengineering 2023, 10, 809. [Google Scholar] [CrossRef] [PubMed]
- Lumini, A.; Nanni, L.; Maguolo, G. Deep ensembles based on stochastic activations for semantic segmentation. Signals 2021, 2, 820–833. [Google Scholar] [CrossRef]
- Khan, M.A.; Sahar, N.; Khan, W.Z.; Alhaisoni, M.; Tariq, U.; Zayyan, M.H.; Kim, Y.J.; Chang, B. GestroNet: A framework of saliency estimation and optimal deep learning features based gastrointestinal diseases detection and classification. Diagnostics 2022, 12, 2718. [Google Scholar] [CrossRef]
- Naz, J.; Sharif, M.I.; Sharif, M.I.; Kadry, S.; Rauf, H.T.; Ragab, A.E. A comparative analysis of optimization algorithms for gastrointestinal abnormalities recognition and classification based on ensemble XcepNet23 and ResNet18 features. Biomedicines 2023, 11, 1723. [Google Scholar] [CrossRef]
- El-Ghany, S.A.; Mahmood, M.A.; Abd El-Aziz, A. An Accurate Deep Learning-Based Computer-Aided Diagnosis System for Gastrointestinal Disease Detection Using Wireless Capsule Endoscopy Image Analysis. Appl. Sci. 2024, 14, 10243. [Google Scholar] [CrossRef]
- Tsai, C.M.; Lee, J.-D. Dynamic Ensemble Learning with Gradient-Weighted Class Activation Mapping for Enhanced Gastrointestinal Disease Classification. Electronics 2025, 14, 305. [Google Scholar] [CrossRef]
- Janutėnas, L.; Šešok, D. Perspective Transformation and Viewpoint Attention Enhancement for Generative Adversarial Networks in Endoscopic Image Augmentation. Appl. Sci. 2025, 15, 5655. [Google Scholar] [CrossRef]
- Khan, M.A.; Shafiq, U.; Hamza, A.; Mirza, A.M.; Baili, J.; AlHammadi, D.A.; Cho, H.-C.; Chang, B. A novel network-level fused deep learning architecture with shallow neural network classifier for gastrointestinal cancer classification from wireless capsule endoscopy images. BMC Med. Inform. Decis. Mak. 2025, 25, 150. [Google Scholar] [CrossRef] [PubMed]
- Kamble, A.; Bandodkar, V.; Dharmadhikary, S.; Anand, V.; Sanki, P.K.; Wu, M.X.; Jana, B. Enhanced Multi-Class Classification of Gastrointestinal Endoscopic Images with Interpretable Deep Learning Model. arXiv 2025, arXiv:2503.00780. [Google Scholar]
- Mohapatra, S.; Nayak, J.; Mishra, M.; Pati, G.K.; Naik, B.; Swarnkar, T. Wavelet transform and deep convolutional neural network-based smart healthcare system for gastrointestinal disease detection. Interdiscip. Sci. Comput. Life Sci. 2021, 13, 212–228. [Google Scholar] [CrossRef]
- Attallah, O.; Sharkas, M. GASTRO-CADx: A three stages framework for diagnosing gastrointestinal diseases. PeerJ Comput. Sci. 2021, 7, e423. [Google Scholar] [CrossRef]
- Khan, Z.F.; Ramzan, M.; Raza, M.; Khan, M.A.; Iqbal, K.; Kim, T.; Cha, J.-H. Deep convolutional neural networks for accurate classification of gastrointestinal tract syndromes. Comput. Mater. Contin. 2024, 78, 1207–1225. [Google Scholar] [CrossRef]
- Cogan, T.; Cogan, M.; Tamil, L. MAPGI: Accurate identification of anatomical landmarks and diseased tissue in gastrointestinal tract using deep learning. Comput. Biol. Med. 2019, 111, 103351. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.W.; Browne, M.; Langville, A.N.; Pauca, V.P.; Plemmons, R.J. Algorithms and applications for approximate nonnegative matrix factorization. Comput. Stat. Data Anal. 2007, 52, 155–173. [Google Scholar] [CrossRef]
- Févotte, C.; Idier, J. Algorithms for nonnegative matrix factorization with the β-divergence. Neural Comput. 2011, 23, 2421–2456. [Google Scholar] [CrossRef]
- Gillis, N. The why and how of nonnegative matrix factorization. arXiv 2014, arXiv:1401.5226. [Google Scholar] [PubMed]
- Ershadi, M.M.; Seifi, A. Applications of dynamic feature selection and clustering methods to medical diagnosis. Appl. Soft Comput. 2022, 126, 109293. [Google Scholar] [CrossRef]
- Borgli, H.; Thambawita, V.; Smedsrud, P.H.; Hicks, S.; Jha, D.; Eskeland, S.L.; Randel, K.R.; Pogorelov, K.; Lux, M.; Nguyen, D.T.D. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci. Data 2020, 7, 283. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Behbood, V.; Hao, P.; Zuo, H.; Xue, S.; Zhang, G. Transfer learning using computational intelligence: A survey. Knowl.-Based Syst. 2015, 80, 14–23. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Gjestang, H.L.; Hicks, S.A.; Thambawita, V.; Halvorsen, P.; Riegler, M.A. A self-learning teacher-student framework for gastrointestinal image classification. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 539–544. [Google Scholar]
- Yue, G.; Wei, P.; Liu, Y.; Luo, Y.; Du, J.; Wang, T. Automated endoscopic image classification via deep neural network with class imbalance loss. IEEE Trans. Instrum. Meas. 2023, 72, 5010611. [Google Scholar] [CrossRef]
| Augmentation Method | Range of Augmentation |
|---|---|
| Flipping in X and Y orientations | −40 to 40 |
| Rotation in X and Y orientations | 50% probability |
| Scaling in X and Y orientations | 0.5 to 2 |
| Shearing in X and Y orientations | −50 to 50 |
| Model | LSVM | QSVM | CSVM | MGSVM | CGSVM | LDA | ESD |
|---|---|---|---|---|---|---|---|
| Kvasir v2 Dataset | |||||||
| Inception | 96.0 | 96.1 | 95.8 | 95.7 | 95.6 | 95.6 | 95.5 |
| ResNet101 | 96.5 | 96.3 | 96.3 | 95.7 | 95.8 | 95.8 | 96.1 |
| Xception | 96.2 | 96.2 | 96.2 | 95.7 | 96.0 | 95.5 | 95.8 |
| HyperKvasir Dataset | |||||||
| Inception | 96.8 | 96.9 | 96.9 | 96.7 | 96.5 | 96.3 | 96.6 |
| ResNet101 | 96.9 | 96.8 | 96.8 | 95.9 | 96.2 | 96.3 | 96.6 |
| Xception | 97.5 | 97.4 | 97.3 | 97.0 | 97.3 | 97.1 | 97.2 |
| Model | LSVM | QSVM | CSVM | MGSVM | CGSVM | LDA | ESD |
|---|---|---|---|---|---|---|---|
| Kvasir v2 Dataset | |||||||
| Inception | 96.1 | 95.9 | 94.7 | 95.7 | 95.8 | 95.3 | 95.0 |
| ResNet101 | 96.2 | 95.9 | 95.2 | 96.0 | 96.0 | 95.3 | 95.0 |
| Xception | 96.4 | 96.0 | 95.2 | 96.0 | 96.1 | 95.9 | 95.5 |
| HyperKvasir Dataset | |||||||
| Inception | 97.0 | 96.9 | 96.0 | 96.8 | 96.3 | 96.1 | 95.7 |
| ResNet101 | 97.1 | 96.8 | 96.1 | 96.9 | 96.6 | 96.2 | 96.8 |
| Xception | 97.7 | 97.3 | 96.4 | 97.3 | 97.5 | 97.6 | 97.5 |
| # Features | LSVM | QSVM | CSVM | MGSVM | CGSVM | LDA | ESD |
|---|---|---|---|---|---|---|---|
| Inception | |||||||
| 10 | 95.0 | 94.8 | 94.0 | 94.7 | 94.3 | 92.1 | 90.9 |
| 20 | 95.6 | 95.2 | 94.4 | 95.0 | 95.4 | 94.3 | 93.8 |
| 30 | 95.4 | 95.3 | 94.6 | 95.1 | 95.1 | 94.5 | 94.4 |
| 40 | 95.8 | 95.2 | 94.9 | 95.1 | 95.1 | 94.2 | 93.9 |
| 50 | 95.5 | 95.4 | 94.8 | 95.0 | 95.2 | 94.7 | 94.7 |
| ResNet101 | |||||||
| 10 | 94.6 | 94.3 | 93.5 | 94.1 | 93.9 | 92.0 | 90.4 |
| 20 | 95.2 | 94.4 | 93.5 | 94.4 | 94.6 | 93.0 | 92.5 |
| 30 | 95.2 | 95.1 | 94.2 | 94.7 | 95.0 | 94.1 | 94.0 |
| 40 | 95.6 | 95.0 | 94.5 | 94.6 | 95.1 | 94.3 | 94.0 |
| 50 | 95.2 | 95.2 | 94.7 | 94.7 | 94.9 | 94.4 | 94.1 |
| Xception | |||||||
| 10 | 95.1 | 94.7 | 93.5 | 94.5 | 94.7 | 93.9 | 93.9 |
| 20 | 95.2 | 94.7 | 93.8 | 94.5 | 95.1 | 94.3 | 93.9 |
| 30 | 95.3 | 95.0 | 94.1 | 94.5 | 95.0 | 94.8 | 94.2 |
| 40 | 95.3 | 95.3 | 94.5 | 94.8 | 95.1 | 94.5 | 94.1 |
| 50 | 95.2 | 95.1 | 94.4 | 94.6 | 95.0 | 94.5 | 94.6 |
| # Features | LSVM | QSVM | CSVM | MGSVM | CGSVM | LDA | ESD |
|---|---|---|---|---|---|---|---|
| Inception | |||||||
| 10 | 95.3 | 95.2 | 94.4 | 95.4 | 95.0 | 91.3 | 90.8 |
| 20 | 96.7 | 96.2 | 95.6 | 96.1 | 96.3 | 94.8 | 95.1 |
| 30 | 96.6 | 96.2 | 95.7 | 96.3 | 96.2 | 96.0 | 95.7 |
| 40 | 96.5 | 96.3 | 95.9 | 95.9 | 96.5 | 96.1 | 95.8 |
| 50 | 96.7 | 96.4 | 95.8 | 96.1 | 96.6 | 96.0 | 95.8 |
| ResNet101 | |||||||
| 10 | 93.7 | 93.4 | 92.6 | 93.2 | 92.8 | 88.5 | 87.1 |
| 20 | 96.0 | 95.6 | 94.9 | 94.9 | 95.1 | 94.3 | 93.7 |
| 30 | 96.1 | 95.7 | 94.9 | 94.5 | 95.2 | 94.6 | 93.5 |
| 40 | 96.2 | 95.9 | 95.5 | 94.8 | 95.6 | 94.8 | 94.0 |
| 50 | 96.1 | 95.9 | 95.2 | 94.3 | 95.0 | 95.3 | 94.5 |
| Xception | |||||||
| 10 | 96.8 | 96.3 | 95.5 | 96.1 | 96.5 | 96.3 | 96.0 |
| 20 | 97.0 | 96.5 | 95.6 | 95.9 | 96.6 | 96.3 | 95.7 |
| 30 | 97.1 | 96.5 | 96.0 | 95.8 | 96.6 | 96.7 | 96.3 |
| 40 | 96.9 | 96.4 | 95.8 | 95.6 | 96.5 | 96.8 | 96.5 |
| 50 | 96.9 | 96.7 | 95.8 | 95.8 | 96.5 | 97.0 | 96.6 |
| Features | Size | LSVM | QSVM | CSVM | MGSVM | CGSVM | LDA | ESD |
|---|---|---|---|---|---|---|---|---|
| Inception | ||||||||
| Pool | 2048 | 96.0 | 96.1 | 95.8 | 95.7 | 95.6 | 95.6 | 95.5 |
| Pool_NNMF | 40 | 95.8 | 95.2 | 94.9 | 95.1 | 95.1 | 94.2 | 93.9 |
| FC | 8 | 96.1 | 95.9 | 94.7 | 95.7 | 95.8 | 95.3 | 95.0 |
| Combined (Pool_NNMF + FC) | 48 | 96.7 | 96.3 | 95.7 | 96.1 | 96.0 | 95.5 | 95.4 |
| ResNet101 | ||||||||
| Pool | 2048 | 96.5 | 96.3 | 96.3 | 95.7 | 95.8 | 95.8 | 96.1 |
| Pool_NNMF | 40 | 95.6 | 95.0 | 94.5 | 94.6 | 95.1 | 94.3 | 94.0 |
| FC | 8 | 96.2 | 95.9 | 95.2 | 96.0 | 96.0 | 95.3 | 95.0 |
| Combined (Pool_NNMF + FC) | 48 | 96.7 | 96.4 | 95.5 | 95.9 | 96.4 | 95.8 | 96.0 |
| Xception | ||||||||
| Pool | 2048 | 96.2 | 96.2 | 96.2 | 95.7 | 96.0 | 95.5 | 95.8 |
| Pool_NNMF | 40 | 95.3 | 95.3 | 94.5 | 94.8 | 95.1 | 94.5 | 94.1 |
| FC | 8 | 96.4 | 96.0 | 95.2 | 96.0 | 96.1 | 95.9 | 95.5 |
| Combined (Pool_NNMF + FC) | 48 | 96.5 | 96.3 | 95.7 | 95.7 | 96.3 | 95.8 | 96.1 |
| Features | Size | LSVM | QSVM | CSVM | MGSVM | CGSVM | LDA | ESD |
|---|---|---|---|---|---|---|---|---|
| Inception | ||||||||
| Pool | 2048 | 96.8 | 96.9 | 96.9 | 96.7 | 96.5 | 96.3 | 96.6 |
| Pool_NNMF | 50 | 96.7 | 96.4 | 95.8 | 96.1 | 96.6 | 96.0 | 95.8 |
| FC | 10 | 97.0 | 96.9 | 96.0 | 96.8 | 96.3 | 96.1 | 95.7 |
| Combined (Pool_NNMF + FC) | 60 | 97.3 | 97.3 | 96.9 | 96.8 | 97.1 | 96.8 | 96.9 |
| ResNet101 | ||||||||
| Pool | 2048 | 96.9 | 96.8 | 96.8 | 95.9 | 96.2 | 96.3 | 96.6 |
| Pool_NNMF | 40 | 96.2 | 95.9 | 95.5 | 94.8 | 95.6 | 94.8 | 94.0 |
| FC | 10 | 97.1 | 96.8 | 96.1 | 96.9 | 96.6 | 96.2 | 96.8 |
| Combined (Pool_NNMF + FC) | 50 | 97.6 | 97.3 | 96.8 | 96.1 | 96.7 | 96.3 | 96.3 |
| Xception | ||||||||
| Pool | 2048 | 97.5 | 97.4 | 97.3 | 97.0 | 97.3 | 97.1 | 97.2 |
| Pool_NNMF | 50 | 96.9 | 96.7 | 95.8 | 95.8 | 96.5 | 97.0 | 96.6 |
| FC | 10 | 97.7 | 97.3 | 96.4 | 97.3 | 97.5 | 97.6 | 97.5 |
| Combined (Pool_NNMF + FC) | 60 | 97.7 | 97.6 | 97.2 | 96.8 | 97.5 | 97.6 | 97.6 |
| # Features | LSVM | QSVM | CSVM | MGSVM | CGSVM | LDA | ESD |
|---|---|---|---|---|---|---|---|
| 10 | 96.1 | 95.9 | 94.7 | 95.8 | 95.8 | 95.5 | 95.1 |
| 20 | 97.4 | 97.1 | 96.4 | 97.1 | 97.4 | 97.0 | 96.8 |
| 30 | 97.6 | 97.3 | 96.9 | 97.4 | 97.5 | 97.0 | 97.0 |
| 40 | 97.7 | 97.4 | 97.2 | 97.4 | 97.6 | 97.0 | 96.9 |
| 50 | 97.7 | 97.5 | 97.3 | 97.4 | 97.6 | 97.0 | 96.9 |
| 60 | 97.6 | 97.4 | 97.2 | 97.3 | 97.6 | 97.1 | 97.0 |
| 70 | 97.8 | 97.5 | 97.2 | 97.4 | 97.5 | 97.0 | 97.0 |
| 80 | 97.6 | 97.5 | 97.2 | 97.4 | 97.6 | 97.2 | 97.2 |
| 90 | 97.8 | 97.7 | 97.3 | 97.5 | 97.7 | 97.2 | 97.2 |
| 100 | 97.7 | 97.6 | 97.5 | 97.4 | 97.6 | 97.1 | 97.3 |
| # Features | LSVM | QSVM | CSVM | MGSVM | CGSVM | LDA | ESD |
|---|---|---|---|---|---|---|---|
| 10 | 96.4 | 95.9 | 95.0 | 95.9 | 95.9 | 95.2 | 94.9 |
| 20 | 98.0 | 97.9 | 97.2 | 97.7 | 97.4 | 97.7 | 97.5 |
| 30 | 98.2 | 98.1 | 97.8 | 97.8 | 98.0 | 98.3 | 98.1 |
| 40 | 98.2 | 98.3 | 97.9 | 97.9 | 98.1 | 98.3 | 98.2 |
| 50 | 98.3 | 98.1 | 98.0 | 97.9 | 98.0 | 98.2 | 98.1 |
| 60 | 98.2 | 98.1 | 97.9 | 97.8 | 98.0 | 98.2 | 98.2 |
| 70 | 98.3 | 98.2 | 98.1 | 97.8 | 98.0 | 98.1 | 98.1 |
| 80 | 98.2 | 98.2 | 98.0 | 97.6 | 97.9 | 98.0 | 98.1 |
| 90 | 98.2 | 98.2 | 97.9 | 97.7 | 98.0 | 98.0 | 98.0 |
| 100 | 98.4 | 98.1 | 98.0 | 97.7 | 98.0 | 98.1 | 98.2 |
| Classifier | Sensitivity | Specificity | Precision | F1-Score | MCC |
|---|---|---|---|---|---|
| Kvasir v2 | |||||
| LSVM | 0.9779 | 0.9969 | 0.9781 | 0.9779 | 0.9749 |
| QSVM | 0.9766 | 0.9965 | 0.9768 | 0.9766 | 0.9732 |
| CSVM | 0.9745 | 0.9960 | 0.9747 | 0.9745 | 0.9710 |
| MGSVM | 0.9749 | 0.9961 | 0.9750 | 0.9749 | 0.9714 |
| CGSVM | 0.9765 | 0.9964 | 0.9765 | 0.9765 | 0.9729 |
| LDA | 0.9718 | 0.9956 | 0.9719 | 0.9718 | 0.9678 |
| ESD | 0.9729 | 0.9958 | 0.9729 | 0.9729 | 0.9689 |
| HyperKvasir Dataset | |||||
| LSVM | 0.983 | 0.998 | 0.984 | 0.983 | 0.981 |
| QSVM | 0.982 | 0.998 | 0.983 | 0.982 | 0.980 |
| CSVM | 0.982 | 0.998 | 0.983 | 0.982 | 0.980 |
| MGSVM | 0.980 | 0.998 | 0.981 | 0.980 | 0.978 |
| CGSVM | 0.979 | 0.998 | 0.980 | 0.979 | 0.977 |
| LDA | 0.981 | 0.998 | 0.982 | 0.981 | 0.979 |
| ESD | 0.980 | 0.998 | 0.981 | 0.980 | 0.978 |
| Study | Method(s) | Accuracy |
|---|---|---|
| [30] | 5-fold cross-validation with 3 new CNN models, followed by stacking ensemble (ML classifier at the second level) and McNemar statistics test | Kvasir v2: 98.42% HyperKvasir: %98.53 |
| [31] | Combining three pre-trained models (DenseNet201, Inception V3, ResNet50) with ensemble method (model average and weighted average) | Kvasir v2: Model Average: %92.96, Weighted Average: %95.00 |
| [33] | Image enhancement (contrast enhancement), segmentation with deep saliency maps, MobileNet-V2-based transfer learning, hyperparameter adjustment with Bayesian optimization, attribute extraction with average pooling, attribute selection with hybrid whale optimization algorithm, classification with Extreme Learning Machine | Kvasir v2: 98.02% |
| [35] | EfficientNet-B0, ResNet101v2, InceptionV3, InceptionResNetV2 with Intelligent Learning Rate Controller (ILRC); transfer learning, layer freezing, fine-tuning, residual learning, regularization techniques | Kvasir v2: 98.06% |
| [36] | Integrates case-specific dynamic weighting with Grad-CAM, leveraging three CNNs: DenseNet201, InceptionV3, and VGG19. | Kvasir v2: 91.00% |
| [37] | Improved StarGAN + Perspective Transformation Module + Viewpoint Attention Module + EfficientNetB7 | Kvasir v2: 95.25% |
| [38] | -SC-DSAN(SparseConvolutionalDenseNet201+Self-Attention) -CNN-GRU -Network-levelfusion -Featureselection:EMPA -Hyperparameteroptimization:BayesianOptimization -Classification: Shallow Wide Neural Network (SWNN) | Kvasir v2: 95.10% |
| [39] | EfficientNet B3 + explainable AI | Kvasir v2: 94.25% |
| [40] | Discrete Wavelet Transform + Deep CNN (2-stage) | Kvasir v2: 97.25% |
| [43] | Automatic and modular pre-processing (edge removal, contrast enhancement, filtering, color mapping, scaling, gamma correction); NASNet DL model | Kvasir v2: 97.35% |
| [48] | ResNet-50 | HyperKvasir: 94.75% |
| [54] | Teacher–student Framework | HyperKvasir: 89.3% |
| [55] | Inception + Class Imbalance Loss | HyperKvasir: 91.55% |
| Proposed EndoNet | Combining two deep features from two layers of three CNNs and reducing their dimensions using NNMF and mRMR methods | Kvasir v2: 97.8% HyperKvasir: 98.4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attallah, O.; Aslan, M.F.; Sabanci, K. EndoNet: A Multiscale Deep Learning Framework for Multiple Gastrointestinal Disease Classification via Endoscopic Images. Diagnostics 2025, 15, 2009. https://doi.org/10.3390/diagnostics15162009
Attallah O, Aslan MF, Sabanci K. EndoNet: A Multiscale Deep Learning Framework for Multiple Gastrointestinal Disease Classification via Endoscopic Images. Diagnostics. 2025; 15(16):2009. https://doi.org/10.3390/diagnostics15162009
Chicago/Turabian StyleAttallah, Omneya, Muhammet Fatih Aslan, and Kadir Sabanci. 2025. "EndoNet: A Multiscale Deep Learning Framework for Multiple Gastrointestinal Disease Classification via Endoscopic Images" Diagnostics 15, no. 16: 2009. https://doi.org/10.3390/diagnostics15162009
APA StyleAttallah, O., Aslan, M. F., & Sabanci, K. (2025). EndoNet: A Multiscale Deep Learning Framework for Multiple Gastrointestinal Disease Classification via Endoscopic Images. Diagnostics, 15(16), 2009. https://doi.org/10.3390/diagnostics15162009









