Metagenomic Next-Generation Sequencing in Infectious Diseases: Clinical Applications, Translational Challenges, and Future Directions
Abstract
1. Introduction
2. Overview of NGS Technologies in Clinical Microbiology
3. Clinical Applications of NGS in Infectious Disease Diagnosis and Management
3.1. Unbiased Pathogen Detection in Undiagnosed Cases
3.2. Detection of Fastidious, Novel, and Polymicrobial Infections
3.3. AMR Profiling and Resistance Gene Surveillance
3.4. NGS for Outbreak Investigation and Epidemiological Typing
3.5. NGS-Guided Personalized Infectious Disease Management
3.6. Real-World Clinical Impact of mNGS Across Diverse Infectious Syndromes
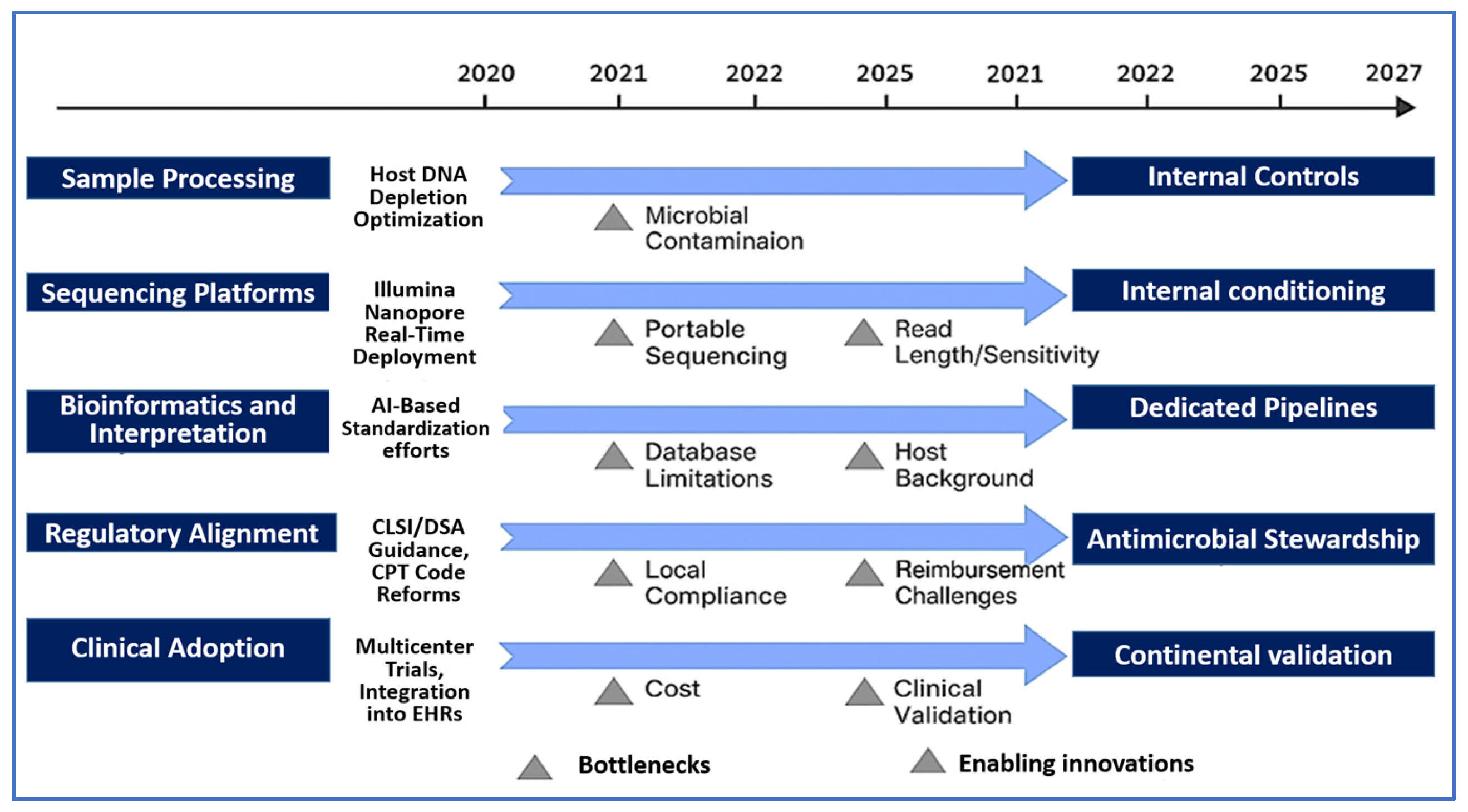
4. Translational Challenges and Future Directions in NGS-Based Infectious Disease Diagnostics
4.1. Technical Barriers and Sample-Processing Complexities
4.2. Bioinformatics Standardization and Interpretive Challenges
4.3. Clinical Integration and Diagnostic Stewardship
4.4. Cost, Turnaround Time, and Health Economics
4.5. Ethical, Legal, and Privacy Considerations
4.6. Regulatory and Reimbursement Landscape
4.7. Future Innovations and Research Priorities
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| mNGS | Metagenomic Next-Generation Sequencing |
| AMR | Antimicrobial Resistance |
| WGS | Whole Genome Sequencing |
| cfDNA | Cell-Free DNA |
| ICU | Intensive Care Unit |
| CAP | Community-Acquired Pneumonia |
| EHRs | Electronic Health Records |
| AI | Artificial Intelligence |
| ML | Machine Learning |
| BALF | Bronchoalveolar Lavage fluid |
| RCT | Randomized Controlled Trial |
| FDA | Food and Drug Administration |
| PCR | Polymerase Chain Reaction |
| ESCMID | European Society of Clinical Microbiology and Infectious Diseases |
| GLASS | Global Antimicrobial Resistance Surveillance System |
| SNP | Single Nucleotide Polymorphism |
| RNA | Ribonucleic Acid |
| DNA | Deoxyribonucleic Acid |
| IDSA | Infectious Diseases Society of America |
| CLSI | Clinical and Laboratory Standards Institute |
| NGS | Next-Generation Sequencing |
| HAP | Hospital-Acquired Pneumonia |
| EU | European Union |
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Flynn, C.E.; Guarner, J. Emerging antimicrobial resistance. Mod. Pathol. 2023, 36, 100249. [Google Scholar] [CrossRef]
- Alara, J.A.; Alara, O.R. An overview of the global alarming increase of multiple drug resistant: A major challenge in clinical diagnosis. Infect. Disord.-Drug Targets 2024, 24, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Miller, S.; Carroll, K.C. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin. Infect. Dis. 2018, 66, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.Y.; Miller, S.A. Clinical metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Salzberg, S.L.; Breitwieser, F.P.; Kumar, A.; Hao, H.; Burger, P.; Rodriguez, F.J.; Lim, M.; Quiñones-Hinojosa, A.; Gallia, G.L.; Tornheim, J.A. Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e251. [Google Scholar] [CrossRef]
- Schlaberg, R.; Chiu, C.Y.; Miller, S.; Procop, G.W.; Weinstock, G.; The Professional Practice Committee; Committee on Laboratory Practices of the American Society for Microbiology; The Microbiology Resource Committee of the College of American Pathologists. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab. Med. 2017, 141, 776–786. [Google Scholar] [CrossRef]
- Wilson, M.R.; Sample, H.A.; Zorn, K.C.; Arevalo, S.; Yu, G.; Neuhaus, J.; Federman, S.; Stryke, D.; Briggs, B.; Langelier, C. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N. Engl. J. Med. 2019, 380, 2327–2340. [Google Scholar] [CrossRef]
- Gu, W.; Deng, X.; Lee, M.; Sucu, Y.D.; Arevalo, S.; Stryke, D.; Federman, S.; Gopez, A.; Reyes, K.; Zorn, K. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 2021, 27, 115–124. [Google Scholar] [CrossRef]
- Miller, S.; Naccache, S.N.; Samayoa, E.; Messacar, K.; Arevalo, S.; Federman, S.; Stryke, D.; Pham, E.; Fung, B.; Bolosky, W.J. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019, 29, 831–842. [Google Scholar] [CrossRef]
- Brown, J.R.; Bharucha, T.; Breuer, J. Encephalitis diagnosis using metagenomics: Application of next generation sequencing for undiagnosed cases. J. Infect. 2018, 76, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Casto, A.M.; Fredricks, D.N.; Hill, J.A. Diagnosis of infectious diseases in immunocompromised hosts using metagenomic next generation sequencing-based diagnostics. Blood Rev. 2022, 53, 100906. [Google Scholar] [CrossRef] [PubMed]
- Quick, J.; Loman, N.J.; Duraffour, S.; Simpson, J.T.; Severi, E.; Cowley, L.; Bore, J.A.; Koundouno, R.; Dudas, G.; Mikhail, A. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016, 530, 228–232. [Google Scholar] [CrossRef]
- Leggett, R.M.; Clark, M.D. A world of opportunities with nanopore sequencing. J. Exp. Bot. 2017, 68, 5419–5429. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Quick, J.; Claro, I.M.; Theze, J.; de Jesus, J.G.; Giovanetti, M.; Kraemer, M.U.; Hill, S.C.; Black, A.; da Costa, A.C. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 2017, 546, 406–410. [Google Scholar] [CrossRef]
- Meredith, L.W.; Hamilton, W.L.; Warne, B.; Houldcroft, C.J.; Hosmillo, M.; Jahun, A.S.; Curran, M.D.; Parmar, S.; Caller, L.G.; Caddy, S.L. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: A prospective genomic surveillance study. Lancet Infect. Dis. 2020, 20, 1263–1271. [Google Scholar] [CrossRef]
- Schwab, T.C.; Joseph, L.; Moono, A.; Göller, P.C.; Motsei, M.; Muula, G.; Evans, D.; Neuenschwander, S.; Günther, G.; Bolton, C. Field evaluation of nanopore targeted next-generation sequencing to predict drug-resistant tuberculosis from native sputum in South Africa and Zambia. J. Clin. Microbiol. 2025, 63, e0139024. [Google Scholar] [CrossRef]
- Tyson, J.R.; James, P.; Stoddart, D.; Sparks, N.; Wickenhagen, A.; Hall, G.; Choi, J.H.; Lapointe, H.; Kamelian, K.; Smith, A.D. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv 2020. [Google Scholar] [CrossRef]
- Köser, C.U.; Holden, M.T.; Ellington, M.J.; Cartwright, E.J.; Brown, N.M.; Ogilvy-Stuart, A.L.; Hsu, L.Y.; Chewapreecha, C.; Croucher, N.J.; Harris, S.R. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 2012, 366, 2267–2275. [Google Scholar] [CrossRef]
- Gordon, N.; Price, J.; Cole, K.; Everitt, R.; Morgan, M.; Finney, J.; Kearns, A.; Pichon, B.; Young, B.; Wilson, D. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J. Clin. Microbiol. 2014, 52, 1182–1191. [Google Scholar] [CrossRef]
- Yue, Y.; Shen, M.; Liu, X.; Hao, Q.; Kang, Y.; Che, Y.; Li, F.; Chen, S.; Xu, S.; Jing, H. Whole-genome sequencing-based prediction and analysis of antimicrobial resistance in Yersinia enterocolitica from Ningxia, China. Front. Microbiol. 2022, 13, 936425. [Google Scholar] [CrossRef]
- Allard, M.W.; Strain, E.; Melka, D.; Bunning, K.; Musser, S.M.; Brown, E.W.; Timme, R. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 2016, 54, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.; Merker, M.; Kohl, T.; Crook, D.; Niemann, S.; Peto, T. Whole genome sequencing for M/XDR tuberculosis surveillance and for resistance testing. Clin. Microbiol. Infect. 2017, 23, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Jajou, R.; van der Laan, T.; de Zwaan, R.; Kamst, M.; Mulder, A.; de Neeling, A.; Anthony, R.; van Soolingen, D. WGS more accurately predicts susceptibility of Mycobacterium tuberculosis to first-line drugs than phenotypic testing. J. Antimicrob. Chemother. 2019, 74, 2605–2616. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tan, G.; Sha, W.; Liu, H.; Yang, J.; Guo, Y.; Shen, X.; Wu, Z.; Shen, H.; Yu, F. Use of whole-genome sequencing to predict Mycobacterium tuberculosis complex drug resistance from early positive liquid cultures. Microbiol. Spectr. 2022, 10, e02516–e02521. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Stoesser, N.; Sheppard, A.E.; Peirano, G.; Anson, L.W.; Pankhurst, L.; Sebra, R.; Phan, H.T.; Kasarskis, A.; Mathers, A.J.; Peto, T.E. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci. Rep. 2017, 7, 5917. [Google Scholar] [CrossRef]
- Charalampous, T.; Kay, G.L.; OeGrady, J. Applying clinical metagenomics for the detection and characterisation of respiratory infections. In Lung Microbiome (ERS Monogr.); European Respiratory Society: Sheffield, UK, 2019; pp. 35–49. [Google Scholar] [CrossRef]
- Kalantar, K.L.; Carvalho, T.; de Bourcy, C.F.; Dimitrov, B.; Dingle, G.; Egger, R.; Han, J.; Holmes, O.B.; Juan, Y.-F.; King, R. IDseq—An open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Gigascce 2020, 9, giaa111. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Tsalik, E.L.; Henao, R.; Nichols, M.; Burke, T.; Ko, E.R.; McClain, M.T.; Hudson, L.L.; Mazur, A.; Freeman, D.H.; Veldman, T. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 2016, 8, 322ra311. [Google Scholar] [CrossRef]
- Herberg, J.A.; Kaforou, M.; Wright, V.J.; Shailes, H.; Eleftherohorinou, H.; Hoggart, C.J.; Cebey-López, M.; Carter, M.J.; Janes, V.A.; Gormley, S. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA 2016, 316, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.E.; Wong, H.R.; Khatri, P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci. Transl. Med. 2016, 8, 346ra391. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Tarumoto, N.; Misawa, K.; Runtuwene, L.R.; Sakai, J.; Hayashida, K.; Eshita, Y.; Maeda, R.; Tuda, J.; Murakami, T. A novel diagnostic method for malaria using loop-mediated isothermal amplification (LAMP) and MinION™ nanopore sequencer. BMC Infect. Dis. 2017, 17, 621. [Google Scholar] [CrossRef] [PubMed]
- Votintseva, A.A.; Bradley, P.; Pankhurst, L.; del Ojo Elias, C.; Loose, M.; Nilgiriwala, K.; Chatterjee, A.; Smith, E.G.; Sanderson, N.; Walker, T.M. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J. Clin. Microbiol. 2017, 55, 1285–1298. [Google Scholar] [CrossRef]
- Van El, C.G.; Cornel, M.C.; Borry, P.; Hastings, R.J.; Fellmann, F.; Hodgson, S.V.; Howard, H.C.; Cambon-Thomsen, A.; Knoppers, B.M.; Meijers-Heijboer, H. Whole-genome sequencing in health care. Eur. J. Hum. Genet. 2013, 21, 580–584. [Google Scholar] [CrossRef]
- Dove, E.S.; Townend, D.; Meslin, E.M.; Bobrow, M.; Littler, K.; Nicol, D.; de Vries, J.; Junker, A.; Garattini, C.; Bovenberg, J. Ethics review for international data-intensive research. Science 2016, 351, 1399–1400. [Google Scholar] [CrossRef]
- Roberts, M.C.; Holt, K.E.; Del Fiol, G.; Baccarelli, A.A.; Allen, C.G. Precision public health in the era of genomics and big data. Nat. Med. 2024, 30, 1865–1873. [Google Scholar] [CrossRef]
- Wang, M. Next-generation sequencing (NGS). In Clinical Molecular Diagnostics; Springer: Singapore, 2021; pp. 305–327. [Google Scholar] [CrossRef]
- Kumar, K.R.; Cowley, M.J.; Davis, R.L. Next-generation sequencing and emerging technologies. In Seminars in Thrombosis and Hemostasis; Thieme Medical Publishers: New York, NY, USA, 2024; pp. 1026–1038. [Google Scholar]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-generation sequencing technologies: An overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Mardis, E.R. DNA sequencing technologies: 2006–2016. Nat. Protoc. 2017, 12, 213–218. [Google Scholar] [CrossRef]
- Tagini, F.; Greub, G. Bacterial genome sequencing in clinical microbiology: A pathogen-oriented review. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2007–2020. [Google Scholar] [CrossRef]
- Zhou, Y.; Shi, W.; Wen, Y.; Mao, E.; Ni, T. Comparison of pathogen detection consistency between metagenomic next-generation sequencing and blood culture in patients with suspected bloodstream infection. Sci. Rep. 2023, 13, 9460. [Google Scholar] [CrossRef]
- Wen, L.; Yang, L.; Chen, C.; Li, J.; Fu, J.; Liu, G.; Kan, Q.; Ho, C.-T.; Huang, Q.; Lan, Y. Applications of multi-omics techniques to unravel the fermentation process and the flavor formation mechanism in fermented foods. Crit. Rev. Food Sci. Nutr. 2024, 64, 8367–8383. [Google Scholar] [CrossRef]
- Blauwkamp, T.A.; Thair, S.; Rosen, M.J.; Blair, L.; Lindner, M.S.; Vilfan, I.D.; Kawli, T.; Christians, F.C.; Venkatasubrahmanyam, S.; Wall, G.D. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 2019, 4, 663–674. [Google Scholar] [CrossRef]
- Chen, J.; Qin, Z.; Jia, Z. The application status of sequencing technology in global respiratory infectious disease diagnosis. Infection 2024, 52, 2169–2181. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, W.; Zhang, X. Application of metagenomic next-generation sequencing in the diagnosis of infectious diseases. Front. Cell. Infect. Microbiol. 2024, 14, 1458316. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, V.; Vashisht, A.; Mondal, A.K.; Farmaha, J.; Alptekin, A.; Singh, H.; Ahluwalia, P.; Srinivas, A.; Kolhe, R. Genomics for emerging pathogen identification and monitoring: Prospects and obstacles. BioMedInformatics 2023, 3, 1145–1177. [Google Scholar] [CrossRef]
- Shetty, M. Genomics in Infectious Diseases. Pediatr. Infect. Dis. 2021, 3, 57–64. [Google Scholar] [CrossRef]
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 319–338. [Google Scholar] [CrossRef]
- Benoit, P.; Brazer, N.; de Lorenzi-Tognon, M.; Kelly, E.; Servellita, V.; Oseguera, M.; Nguyen, J.; Tang, J.; Omura, C.; Streithorst, J. Seven-year performance of a clinical metagenomic next-generation sequencing test for diagnosis of central nervous system infections. Nat. Med. 2024, 30, 3522–3533. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, C.; Chen, W.; Gai, W.; Zheng, Y.; Guo, Y.; Wang, Z.; Chen, Y.; Cai, Z. Clinical application of metagenomic next-generation sequencing in sepsis patients with early antibiotic treatment. Infect. Drug Resist. 2024, 17, 4695–4706. [Google Scholar] [CrossRef]
- Zuo, Y.-H.; Wu, Y.-X.; Hu, W.-P.; Chen, Y.; Li, Y.-P.; Song, Z.-J.; Luo, Z.; Ju, M.-J.; Shi, M.-H.; Xu, S.-Y. The clinical impact of metagenomic next-generation sequencing (mNGS) test in hospitalized patients with suspected sepsis: A multicenter prospective study. Diagnostics 2023, 13, 323. [Google Scholar] [CrossRef]
- Lai, L.M.; Chen, Q.-G.; Liu, Y.; Zhao, R.; Cao, M.L.; Yuan, L. The value of metagenomic next-generation sequencing in the diagnosis of fever of unknown origin. Sci. Rep. 2025, 15, 1963. [Google Scholar] [CrossRef]
- Zhao, J.; Zhuge, R.; Hu, B.; Wang, Y.; Wang, X.; Zhang, Y.; Yuan, L.; Qiu, C.; Yan, Y.; Zhang, X. Clinical impact of bronchoalveolar lavage fluid metagenomic next-generation sequencing in immunocompromised patients with severe community-acquired pneumonia in ICU: A multicenter retrospective study. Infection 2025, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhang, S.; Zhao, Y.; Ding, X.; Yang, F.; Zhao, Y. Diagnostic value of metagenomic next-generation sequencing in sepsis and bloodstream infection. Front. Cell. Infect. Microbiol. 2023, 13, 1117987. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, O.; Liu, W.; Gan, L.; Li, X.; Ma, Q.; Hu, X.; Jian, X. Coxiella burnetii and Bartonella endocarditis diagnosed by metagenomic next-generation sequencing. J. Clin. Med. 2022, 11, 7150. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Wang, K.; Qiu, L.; Liang, Y.; Tu, C.; Chen, M.; Wang, Z.; Wu, J.; Huang, Y.; Tan, C. Tropheryma whipplei detection by metagenomic next-generation sequencing in bronchoalveolar lavage fluid: A cross-sectional study. Front. Cell. Infect. Microbiol. 2022, 12, 961297. [Google Scholar] [CrossRef] [PubMed]
- Briese, T.; Paweska, J.T.; McMullan, L.K.; Hutchison, S.K.; Street, C.; Palacios, G.; Khristova, M.L.; Weyer, J.; Swanepoel, R.; Egholm, M. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever–associated arenavirus from Southern Africa. PLoS Pathog. 2009, 5, e1000455. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Ladner, J.T.; Kraemer, M.U.; Dudas, G.; Tan, A.L.; Gangavarapu, K.; Wiley, M.R.; White, S.; Thézé, J.; Magnani, D.M. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 2017, 546, 401–405. [Google Scholar] [CrossRef]
- Langelier, C.; Kalantar, K.L.; Moazed, F.; Wilson, M.R.; Crawford, E.D.; Deiss, T.; Belzer, A.; Bolourchi, S.; Caldera, S.; Fung, M. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl. Acad. Sci. USA 2018, 115, E12353–E12362. [Google Scholar] [CrossRef]
- Köser, C.U.; Ellington, M.J.; Cartwright, E.J.; Gillespie, S.H.; Brown, N.M.; Farrington, M.; Holden, M.T.; Dougan, G.; Bentley, S.D.; Parkhill, J. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 2012, 8, e1002824. [Google Scholar] [CrossRef]
- Ellington, M.; Ekelund, O.; Aarestrup, F.M.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.; Hopkins, K.L. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef]
- Kersh, E.N.; Pham, C.D.; Papp, J.R.; Myers, R.; Steece, R.; Kubin, G.; Gautom, R.; Nash, E.E.; Sharpe, S.; Gernert, K.M. Expanding US Laboratory capacity for Neisseria gonorrhoeae antimicrobial susceptibility testing and whole-genome sequencing through the CDC’s antibiotic resistance laboratory network. J. Clin. Microbiol. 2020, 58, e01461-19. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.; Dessai, U.; McGarry, S.; Gerner-Smidt, P. Use of whole-genome sequencing for food safety and public health in the United States. Foodborne Pathog. Dis. 2019, 16, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Saputra, D.; Sicheritz-Ponten, T.; Lund, O.; Svendsen, C.A.; Frimodt-Møller, N.; Aarestrup, F.M. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 2014, 52, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Kaas, R.S.; Seyfarth, A.M.; Agersø, Y.; Lund, O.; Larsen, M.V.; Aarestrup, F.M. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 771–777. [Google Scholar] [CrossRef]
- Su, M.; Satola, S.W.; Read, T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2019, 57, e01405-18. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Lakin, S.M.; Dean, C.; Noyes, N.R.; Dettenwanger, A.; Ross, A.S.; Doster, E.; Rovira, P.; Abdo, Z.; Jones, K.L.; Ruiz, J. MEGARes: An antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017, 45, D574–D580. [Google Scholar] [CrossRef]
- Gorrie, C.L.; Mirčeta, M.; Wick, R.R.; Edwards, D.J.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.F.; Garlick, J.S.; Watson, K.M.; Pilcher, D.V. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 2017, 65, 208–215. [Google Scholar] [CrossRef]
- Eyre, D.W.; Cule, M.L.; Wilson, D.J.; Griffiths, D.; Vaughan, A.; O’Connor, L.; Ip, C.L.; Golubchik, T.; Batty, E.M.; Finney, J.M. Diverse sources of C. difficile infection identified on whole-genome sequencing. N. Engl. J. Med. 2013, 369, 1195–1205. [Google Scholar] [CrossRef]
- Sundermann, A.J.; Kumar, P.; Griffith, M.P.; Waggle, K.D.; Rangachar Srinivasa, V.; Raabe, N.; Mills, E.G.; Coyle, H.; Ereifej, D.; Creager, H.M. Real-Time Genomic Surveillance for Enhanced Healthcare Outbreak Detection and Control: Clinical and Economic Impact. Clin. Infect. Dis. 2025, ciaf216. [Google Scholar] [CrossRef] [PubMed]
- Silvotti, M.G.; Scaltriti, E.; Bolzoni, L.; Zerbi, B.; Tocci, G.; Zappavigna, A.; Lamberti, G.; Donati, F.; Federici, F.; Pongolini, S. Outbreak of carbapenem resistant Klebsiella pneumoniae in a neurorehabilitation unit: Genomic epidemiology reveals complex transmission pattern in a tertiary care hospital. J. Glob. Antimicrob. Resist. 2025, 41, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Gire, S.K.; Goba, A.; Andersen, K.G.; Sealfon, R.S.; Park, D.J.; Kanneh, L.; Jalloh, S.; Momoh, M.; Fullah, M.; Dudas, G. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 2014, 345, 1369–1372. [Google Scholar] [CrossRef] [PubMed]
- Gudbjartsson, D.F.; Helgason, A.; Jonsson, H.; Magnusson, O.T.; Melsted, P.; Norddahl, G.L.; Saemundsdottir, J.; Sigurdsson, A.; Sulem, P.; Agustsdottir, A.B. Spread of SARS-CoV-2 in the Icelandic population. N. Engl. J. Med. 2020, 382, 2302–2315. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Y.; Li, Y.; Song, C.; Pan, P. Next-generation sequencing guides the treatment of severe community-acquired pneumonia with empiric antimicrobial therapy failure: A propensity-score-matched study. PLOS Neglected Trop. Dis. 2024, 18, e0012701. [Google Scholar] [CrossRef]
- Lai, L.M.; Dai, Q.-B.; Cao, M.L.; Liu, Y.; Zhao, R.; Yuan, L. Clinical utility of metagenomic next-generation sequencing in pathogen detection for lower respiratory tract infections. Sci. Rep. 2025, 15, 19039. [Google Scholar] [CrossRef]
- Zheng, H.; Peng, P.; Wang, S.; Zhang, B.; Yang, L.; Wang, Y.; Li, L.; Pang, G. Microbiological Diagnostic Performance and Clinical Effect of Metagenomic Next-Generation Sequencing for the Detection of Immunocompromised Patients with Community-Acquired Pneumonia. Infect. Drug Resist. 2025, 18, 1223–1236. [Google Scholar] [CrossRef]
- Cai, Y.; Fang, X.; Chen, Y.; Huang, Z.; Zhang, C.; Li, W.; Yang, B.; Zhang, W. Metagenomic next generation sequencing improves diagnosis of prosthetic joint infection by detecting the presence of bacteria in periprosthetic tissues. Int. J. Infect. Dis. 2020, 96, 573–578. [Google Scholar] [CrossRef]
- Xiang, C.; Wu, X.; Li, T.; Tang, X.; Zhang, Y.; Zeng, F.; Xiang, H.; Chen, T.; Kuang, Z.; Liu, F. Effect of metagenomic next-generation sequencing on clinical outcomes in adults with severe pneumonia post-cardiac surgery: A single-center retrospective study. Sci. Rep. 2024, 14, 28907. [Google Scholar] [CrossRef]
- Shi, T.; Lin, Y.; Zheng, X.; Ruan, H.; Zhang, R.; Liu, Y.; Xu, S.; Wang, H. Metagenomic next-generation sequencing for the clinical identification of spinal infection-associated pathogens. Front. Cell. Infect. Microbiol. 2025, 14, 1437665. [Google Scholar] [CrossRef]
- Huang, H.; Shi, J.; Zheng, M.; Su, S.; Chen, W.; Ming, J.; Ren, T.; Qu, D. Pathogen detection in suspected spinal infection: Metagenomic next-generation sequencing versus culture. Eur. Spine J. 2023, 32, 4220–4228. [Google Scholar] [CrossRef]
- Shi, T.; Chen, H.; Liu, Y.; Wu, Y.; Lin, F. Clinical applications of metagenomic next-generation sequencing in the identification of pathogens in periprosthetic joint infections: A retrospective study. J. Orthop. Surg. Res. 2024, 19, 301. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, R.; Zhu, Y.; Hu, L.; Xia, H.; Li, J.; Ye, Y. Metagenomic next-generation sequencing of plasma cell-free DNA improves the early diagnosis of suspected infections. BMC Infect. Dis. 2024, 24, 187. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, Y.; Zhang, X.; Liu, S.; Yin, Y.; Guo, Y.; Wang, X.; Zhang, Y.; Zhao, C.; Gai, W. Clinical evaluation of cell-free and cellular metagenomic next-generation sequencing of infected body fluids. J. Adv. Res. 2024, 55, 119–129. [Google Scholar] [CrossRef]
- Marotz, C.; Sanders, J.; Zuniga, C.; Zaramela, L.; Knight, R.; Zengler, K. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome 2018, 6, 42. [Google Scholar] [CrossRef]
- He, Y.; Fang, K.; Shi, X.; Yang, D.; Zhao, L.; Yu, W.; Zheng, Y.; Xu, Y.; Ma, X.; Chen, L. Enhanced DNA and RNA pathogen detection via metagenomic sequencing in patients with pneumonia. J. Transl. Med. 2022, 20, 195. [Google Scholar] [CrossRef]
- Gu, W.; Crawford, E.D.; O’Donovan, B.; Wilson, M.R.; Chow, E.D.; Retallack, H.; DeRisi, J.L. Depletion of Abundant Sequences by Hybridization (DASH): Using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016, 17, 41. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Diao, Z.; Lai, H.; Han, Y.; Xie, J.; Zhang, R.; Li, J. Multilaboratory assessment of metagenomic next-generation sequencing for unbiased microbe detection. J. Adv. Res. 2022, 38, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, R.; Minich, J.J.; Marotz, C.; Cooper, A.; Knight, R.; Weyrich, L.S. Contamination in low microbial biomass microbiome studies: Issues and recommendations. Trends Microbiol. 2019, 27, 105–117. [Google Scholar] [CrossRef]
- Minich, J.J.; Sanders, J.G.; Amir, A.; Humphrey, G.; Gilbert, J.A.; Knight, R. Quantifying and understanding well-to-well contamination in microbiome research. mSystems 2019, 4, e00186-19. [Google Scholar] [CrossRef]
- Karstens, L.; Asquith, M.; Davin, S.; Fair, D.; Gregory, W.T.; Wolfe, A.J.; Braun, J.; McWeeney, S. Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments. mSystems 2019, 4, e00290-19. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Almeida, A.; Mitchell, A.L.; Boland, M.; Forster, S.C.; Gloor, G.B.; Tarkowska, A.; Lawley, T.D.; Finn, R.D. A new genomic blueprint of the human gut microbiota. Nature 2019, 568, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Zepeda, A.; Godoy-Lozano, E.E.; Raggi, L.; Segovia, L.; Merino, E.; Gutiérrez-Rios, R.M.; Juarez, K.; Licea-Navarro, A.F.; Pardo-Lopez, L.; Sanchez-Flores, A. Analysis of sequencing strategies and tools for taxonomic annotation: Defining standards for progressive metagenomics. Sci. Rep. 2018, 8, 12034. [Google Scholar] [CrossRef] [PubMed]
- Minot, S.S.; Krumm, N.; Greenfield, N.B. One codex: A sensitive and accurate data platform for genomic microbial identification. bioRxiv 2015, 027607. [Google Scholar] [CrossRef]
- Besser, J.; Carleton, H.A.; Gerner-Smidt, P.; Lindsey, R.L.; Trees, E. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin. Microbiol. Infect. 2018, 24, 335–341. [Google Scholar] [CrossRef]
- Junier, T.; Huber, M.; Schmutz, S.; Kufner, V.; Zagordi, O.; Neuenschwander, S.; Ramette, A.; Kubacki, J.; Bachofen, C.; Qi, W. Viral metagenomics in the clinical realm: Lessons learned from a Swiss-wide ring trial. Genes 2019, 10, 655. [Google Scholar] [CrossRef]
- Shean, R.C.; Garrett, E.; Malleis, J.; Lieberman, J.A.; Bradley, B.T. A retrospective observational study of mNGS test utilization to examine the role of diagnostic stewardship at two academic medical centers. J. Clin. Microbiol. 2024, 62, e00605–e00624. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Guo, Y.; Xue, X.-H.; Pang, F. Application of metagenomic next-generation sequencing in the diagnosis of infectious diseases of the central nervous system after empirical treatment. World J. Clin. Cases 2022, 10, 7760. [Google Scholar] [CrossRef]
- Charalampous, T.; Kay, G.L.; Richardson, H.; Aydin, A.; Baldan, R.; Jeanes, C.; Rae, D.; Grundy, S.; Turner, D.J.; Wain, J. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 2019, 37, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Charalampous, T.; Alcolea-Medina, A.; Snell, L.B.; Alder, C.; Tan, M.; Williams, T.G.; Al-Yaakoubi, N.; Humayun, G.; Meadows, C.I.; Wyncoll, D.L. Routine metagenomics service for ICU patients with respiratory infection. Am. J. Respir. Crit. Care Med. 2024, 209, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Sutton, A.J.; Lupu, D.S.; Bergin, S.P.; Holland, T.L.; McAdams, S.A.; Dadwal, S.S.; Nguyen, K.; Nolte, F.S.; Tremblay, G.; Perkins, B.A. Cost-Effectiveness of Plasma Microbial Cell-Free DNA Sequencing When Added to Usual Care Diagnostic Testing for Immunocompromised Host Pneumonia. PharmacoEconomics 2024, 42, 1029–1045. [Google Scholar] [CrossRef] [PubMed]
- Jelsig, A.M.; Qvist, N.; Brusgaard, K.; Ousager, L.B. Research participants in NGS studies want to know about incidental findings. Eur. J. Hum. Genet. 2015, 23, 1423–1426. [Google Scholar] [CrossRef]
- Azencott, C.-A. Machine learning and genomics: Precision medicine versus patient privacy. Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 2018, 376, 20170350. [Google Scholar] [CrossRef]
- Chiang, C.W. The opportunities and challenges of integrating population histories into genetic studies for diverse populations: A motivating example from Native Hawaiians. Front. Genet. 2021, 12, 643883. [Google Scholar] [CrossRef]
- Johnson, S.B.; Slade, I.; Giubilini, A.; Graham, M. Rethinking the ethical principles of genomic medicine services. Eur. J. Hum. Genet. 2020, 28, 147–154. [Google Scholar] [CrossRef]
- Chiu, C.Y.; López-Labrador, F.X.; Wilson, M.R.; de Vries, J.J.; The Regulatory Landscape for Clinical Metagenomic Testing. JAMA Neurology. Available online: https://jamanetwork.com/journals/jamaneurology/article-abstract/2831973 (accessed on 12 July 2025).
- American Medical Association. CPT® Overview and Code Approval. Available online: https://www.ama-assn.org/practice-management/cpt/cpt-overview-and-code-approval (accessed on 29 July 2025).
- Phillips, K.A.; Deverka, P.A.; Marshall, D.A.; Wordsworth, S.; Regier, D.A.; Christensen, K.D.; Buchanan, J. Methodological issues in assessing the economic value of next-generation sequencing tests: Many challenges and not enough solutions. Value Health 2018, 21, 1033–1042. [Google Scholar] [CrossRef]
- Kang, S.-Y.; Odouard, I.; Gresenz, C.R. Claim Denials for Cancer-Related Next-Generation Sequencing in Medicare. JAMA Netw. Open 2025, 8, e255785. [Google Scholar] [CrossRef]
- Aamot, H.V.; Claas, E.C.; Couto, N.; Westh, H.; Greub, G.; Rossen, J.W. Next-generation sequencing in routine clinical microbiology and infectious diseases: An ESGMD-ESGEM ESCMID postgraduate course. New Microbes New Infect. 2022, 49, 101046. [Google Scholar] [CrossRef]
- Olatunji, I.; Bardaji, D.K.R.; Miranda, R.R.; Savka, M.A.; Hudson, A.O. Artificial intelligence tools for the identification of antibiotic resistance genes. Front. Microbiol. 2024, 15, 1437602. [Google Scholar] [CrossRef]
- Fan, S.; Si, M.; Xu, N.; Yan, M.; Pang, M.; Liu, G.; Gong, J.; Wang, H. Metagenomic next-generation sequencing-guided antimicrobial treatment versus conventional antimicrobial treatment in early severe community-acquired pneumonia among immunocompromised patients (MATESHIP): A study protocol. Front. Microbiol. 2022, 13, 927842. [Google Scholar] [CrossRef]
- Schulz, E.; Grumaz, S.; Hatzl, S.; Gornicec, M.; Valentin, T.; Huber-Kraßnitzer, B.; Kriegl, L.; Uhl, B.; Deutsch, A.; Greinix, H. Pathogen detection by metagenomic next-generation sequencing during neutropenic fever in patients with hematological malignancies. Open Forum Infect. Dis. 2022, 9, ofac393. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.-M.; Du, B.; Qin, H.-Y.; Wang, Q.; Shi, Y. Metagenomic next-generation sequencing for the diagnosis of suspected pneumonia in immunocompromised patients. J. Infect. 2021, 82, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Sun, T.; He, H.; Xing, L.; Cheng, Z.; Geng, S.; Xu, D.; Luo, H.; Chen, C.; Jiang, M. Effect of metagenomic next-generation sequencing on clinical outcomes of patients with severe community-acquired pneumonia in the ICU: A multicenter, randomized controlled trial. Chest 2025, 167, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, L.; Zhu, C.; Jin, J.; Song, C.; Dong, H.; Li, Z.; Wang, Z.; Chen, Y.; Yang, Z. Clinical value of metagenomic next-generation sequencing by Illumina and Nanopore for the detection of pathogens in bronchoalveolar lavage fluid in suspected community-acquired pneumonia patients. Front. Cell. Infect. Microbiol. 2022, 12, 1021320. [Google Scholar] [CrossRef]
- Xie, Y.; Dai, B.; Zhou, X.; Liu, H.; Wu, W.; Yu, F.; Zhu, B. Diagnostic value of metagenomic next-generation sequencing for multi-pathogenic pneumonia in HIV-infected patients. Infect. Drug Resist. 2023, 16, 607–618. [Google Scholar] [CrossRef]
| NGS Modality | Sequencing Scope | Advantages | Limitations | Clinical Use Cases |
|---|---|---|---|---|
| WGS [22] | Complete genome (from cultured isolate) | High resolution; AMR/virulence detection; outbreak tracing | Requires culture; slower turnaround | Bacterial typing; resistance surveillance |
| mNGS [8,9] | All DNA/RNA in sample (unbiased) | Detects unknown/rare pathogens; no culture needed | High host DNA background; expensive; complex analysis | Meningitis; sepsis of unknown origin; rare pathogens |
| Targeted NGS Panels [28] | Predefined microbial/resistance genes | Faster; lower cost; easier interpretation | Limited to panel design; misses unexpected targets | Syndromic panels (respiratory, GI, sepsis) |
| Long-read Sequencing (ONT/PacBio) [13,34] | Long fragments (up to >10 kb) | Portable; real-time sequencing; resolves complex genomic structures | Higher error rates; infrastructure variable | Point-of-care outbreak diagnostics; TB genomics |
| Transcriptomics/Single-cell RNA-seq [31,33] | Host RNA expression | Host response profiling; immune status insights | Emerging; complex interpretation; mostly research stage | Disease severity prediction; host–pathogen studies |
| Automated Platforms [46] | Integrated sample-to-answer systems | Same-day results; minimal manual input | Initial setup cost; platform-dependent limitations | Rapid diagnostics in hospital labs |
| Cloud-based Pipelines [29,30] | Data analysis only | Removes need for in-house bioinformatics; scalable | Dependent on internet; privacy/security concerns | Clinical metagenomics (low-resource labs) |
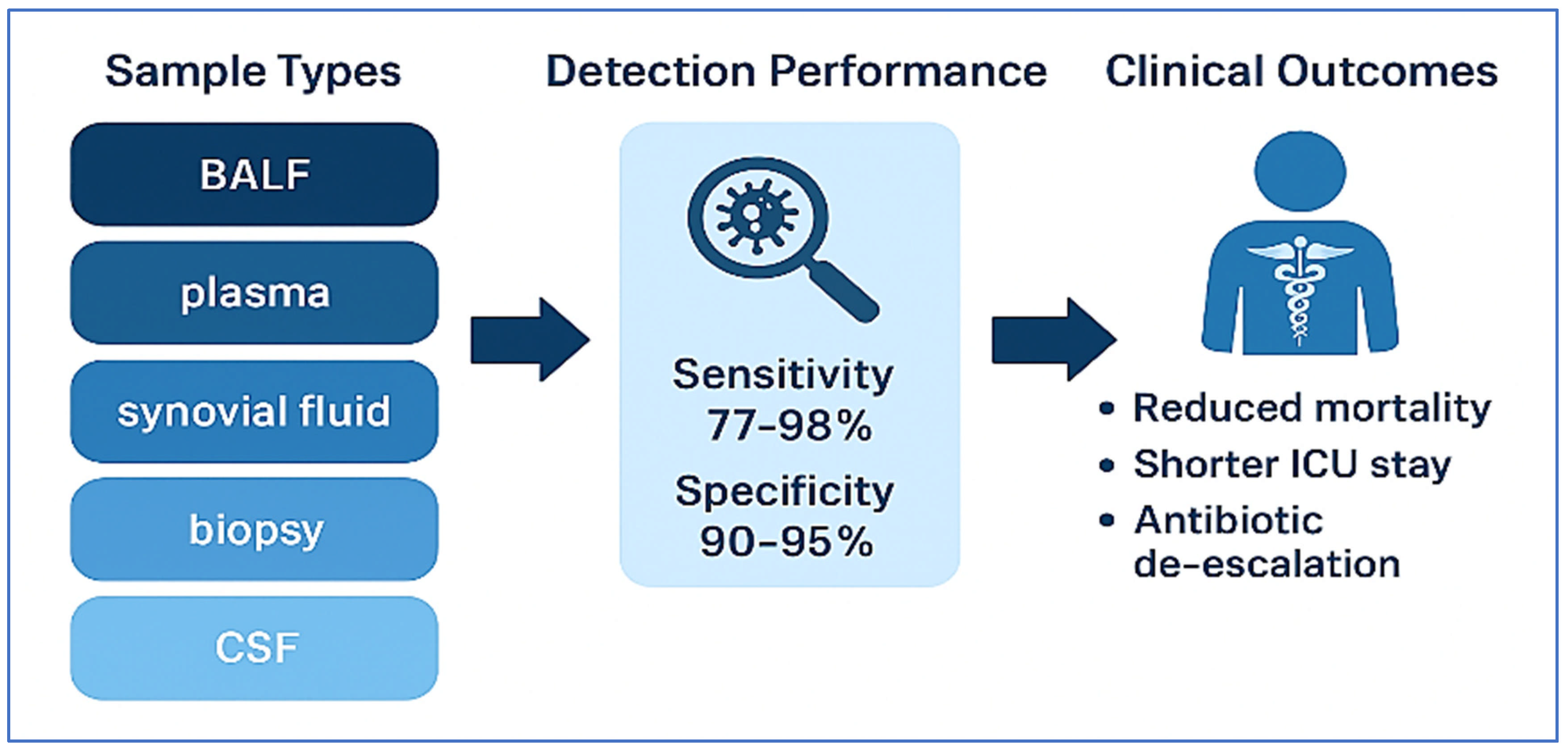
| Study | Clinical Syndrome | Sample Type | Key Findings | Clinical Impact |
|---|---|---|---|---|
| Xiang et al. [82] | Post-cardiac surgery pneumonia | BALF | 98.2% diagnostic yield vs. 58.4% by culture | Reduced mechanical ventilation duration, improved SOFA scores, shorter ICU stays |
| Shi et al. [83] | Vertebral osteomyelitis | Biopsy | 77.8% detection by mNGS vs. 27.2% by culture; identified anaerobes and polymicrobial infections | High specificity (90.3%); effective despite prior antibiotics |
| Huang et al. [84] | PJI | Synovial Fluid | 89% sensitivity, 95% specificity | Enabled antibiotic de-escalation without compromising efficacy |
| Shi et al. [85] | PJI | Synovial Fluid | 89.1% sensitivity, 94.7% specificity; detected fastidious organisms | Supported tailored therapy and improved diagnostic confidence |
| Zhang et al. [86] | Systemic infections/sepsis | Plasma (cfDNA) | 74.4% mNGS detection vs. 12.1% by culture | 70.3% had antimicrobial therapy adjusted; earlier sampling linked to shorter hospital stay |
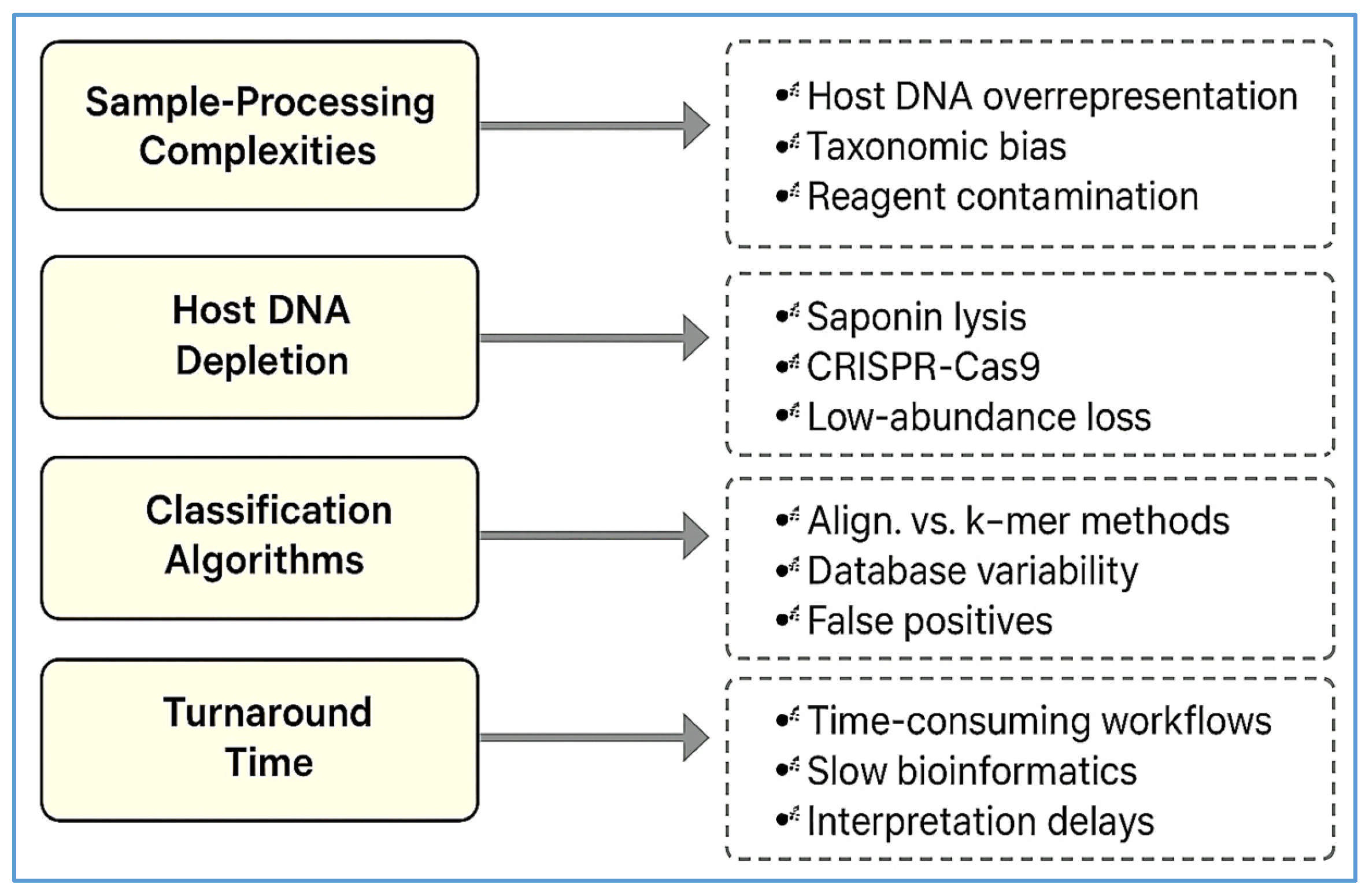
| Challenge | Description | Potential Solutions | References |
|---|---|---|---|
| High host DNA background | Over 90% of reads in plasma/CSF may be host-derived, masking microbial signals | Host depletion via saponin lysis, DNase digestion, or CRISPR-Cas9 (DASH) | [89,90] |
| Environmental contamination (“kitome”) | Reagent-based or lab-introduced contaminants skew microbial profiles | Inclusion of no-template/extraction blanks; contamination-aware bioinformatics | [94,95] |
| Bioinformatics inconsistency | Non-standardized taxonomic classification and AMR annotation lead to variable results | Harmonized pipelines, curated databases, confidence thresholds, consensus guidelines | [4,70,71] |
| Clinical interpretation uncertainty | Difficulty distinguishing colonization, contamination, or infection in low-abundance reads | Expert stewardship teams, integrated EHR tools, multidisciplinary training | [5,78,103] |
| Cost and turnaround time | mNGS costs USD 100–USD 300/sample; results take 24–72 h; delays care in some settings | Streamlined nanopore workflows, automation, targeted panels | [105,106] |
| Regulatory and reimbursement barriers | Lack of FDA approval and CPT codes; insurance denials common | Prospective validation trials; CPT reform; CLSI/IDSA performance guidelines | [4,115] |
| Ethical, legal, and privacy issues | Incidental human findings, re-identification risk, genomic discrimination | Transparent consent, data encryption, privacy laws, governance frameworks | [108,110] |
| Global equity and access | High-income countries dominate infrastructure and data; LMICs underrepresented | Capacity-building, portable devices, federated data sharing | [13,115] |
| Trial Name | Study Design | Population | Infection Type | Intervention Strategy | Primary Outcome(s) | Reference |
|---|---|---|---|---|---|---|
| MATESHIP Trial | Randomized Controlled Trial (RCT) | Immunocompromised ICU patients | Severe Community-Acquired Pneumonia | mNGS-guided therapy vs. standard culture | Antibiotic duration, ICU LOS, mortality | [117] |
| DISQVER Trial | Prospective Observational Study | Septic patients with suspected infection | Bloodstream and respiratory infections | cfDNA mNGS vs. conventional blood cultures | Pathogen ID rate, time to targeted therapy | [118] |
| REMEDID Study | Multicenter Observational Study | Immunosuppressed hematology patients | Hematologic febrile neutropenia | BAL fluid mNGS for pathogen detection | Diagnostic yield, antifungal escalation | [119] |
| GRAIDS Trial | Multicenter Randomized Trial | Critically ill ICU patients | Pulmonary and systemic infections | mNGS plus EHR decision support | Antibiotic use, time to diagnosis | [120] |
| Karius Prospective Study | Prospective Multicenter Cohort | Hospitalized adults with suspected infection | Broad-spectrum infections | cfDNA plasma NGS (Karius test) | Diagnostic yield, time to intervention | [46] |
| NGS-CAP (China) | Prospective Diagnostic Evaluation | Severe CAP patients in ICU | CAP or HAP (non-responders) | Illumina mNGS-based workflow | Detection rate, concordance with cultures | [121] |
| Illumina IDbyDNA Evaluation | Comparative Clinical Validation | Patients with suspected CNS infections | CNS infections (encephalitis, meningitis) | IDbyDNA PathoScope NGS platform | Sensitivity, specificity, turnaround time | [8] |
| NIDP-Fungal mNGS Trial | Prospective Observational Trial | Patients with suspected fungal pneumonia | Invasive fungal infections (IFIs) | Fungal mNGS assay vs. conventional tests | Positive detection rate, clinical impact | [122] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbehiry, A.; Abalkhail, A. Metagenomic Next-Generation Sequencing in Infectious Diseases: Clinical Applications, Translational Challenges, and Future Directions. Diagnostics 2025, 15, 1991. https://doi.org/10.3390/diagnostics15161991
Elbehiry A, Abalkhail A. Metagenomic Next-Generation Sequencing in Infectious Diseases: Clinical Applications, Translational Challenges, and Future Directions. Diagnostics. 2025; 15(16):1991. https://doi.org/10.3390/diagnostics15161991
Chicago/Turabian StyleElbehiry, Ayman, and Adil Abalkhail. 2025. "Metagenomic Next-Generation Sequencing in Infectious Diseases: Clinical Applications, Translational Challenges, and Future Directions" Diagnostics 15, no. 16: 1991. https://doi.org/10.3390/diagnostics15161991
APA StyleElbehiry, A., & Abalkhail, A. (2025). Metagenomic Next-Generation Sequencing in Infectious Diseases: Clinical Applications, Translational Challenges, and Future Directions. Diagnostics, 15(16), 1991. https://doi.org/10.3390/diagnostics15161991






