Enhancing YOLOv11 with Large Kernel Attention and Multi-Scale Fusion for Accurate Small and Multi-Lesion Bone Tumor Detection in Radiographs
Abstract
1. Introduction
- We propose an improved YOLOv11-based architecture for bone tumor detection and classification, addressing the clinical gap of small bone tumor detection. Small lesions, which are frequently missed in real-world datasets, exhibit a miss rate of 43% in the BTXRD dataset using YOLOv8. Our method, YOLOv11-MTB, reduces this miss rate to 22%, improving detection precision and increasing the overall mAP from 73.1 to 79.6.
- We perform extensive evaluation on two multi-center datasets, including the publicly available BTXRD dataset, verifying the generalizability and robustness of the proposed method across imaging centers and conditions.
- Our method achieves state-of-the-art results on both detection and classification tasks, particularly improving mAP for small-object and multi-lesion categories.
- We provide detailed ablation studies to illustrate how our architectural improvements contribute to the final performance, with implications for future clinical deployment.
2. Methods
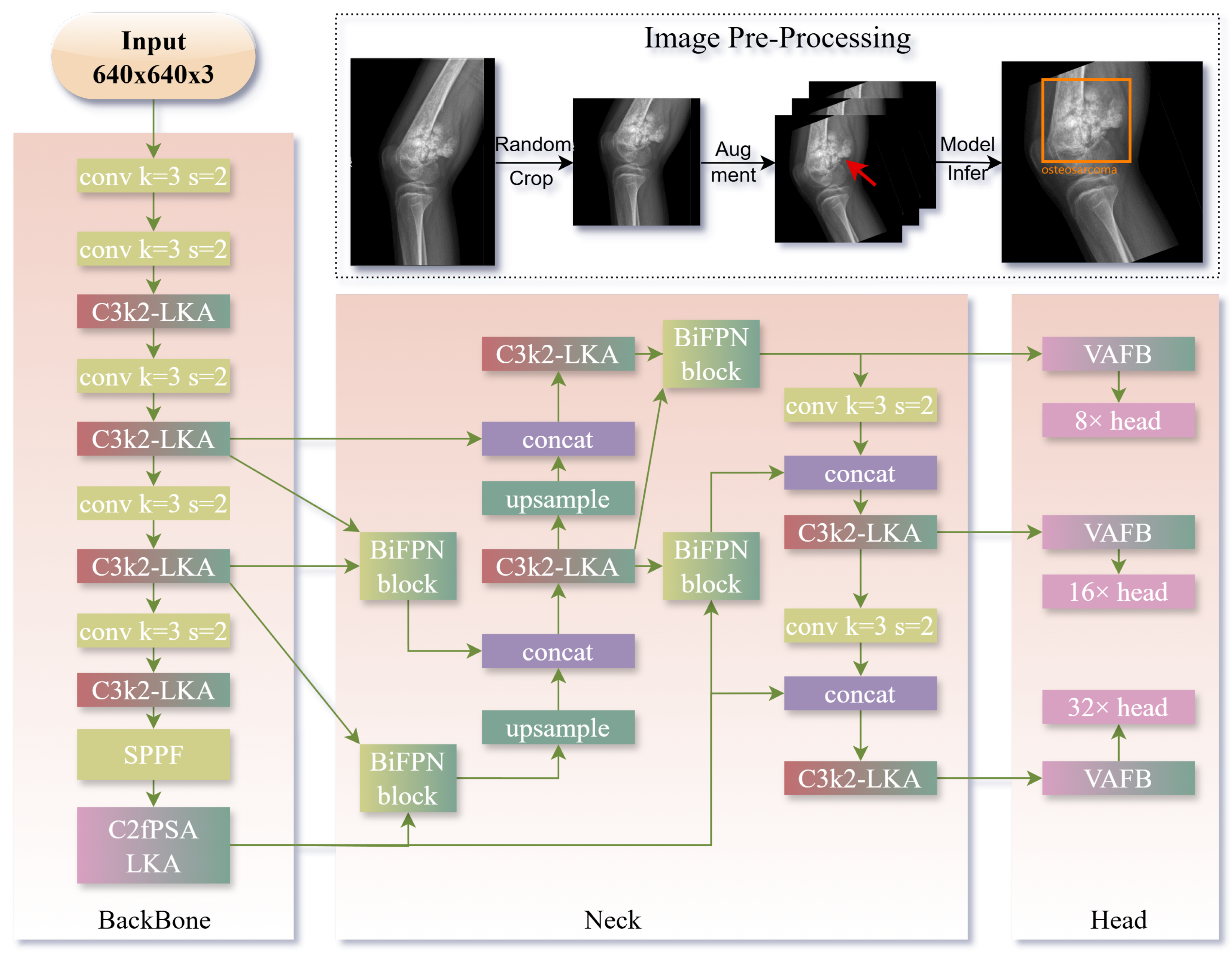
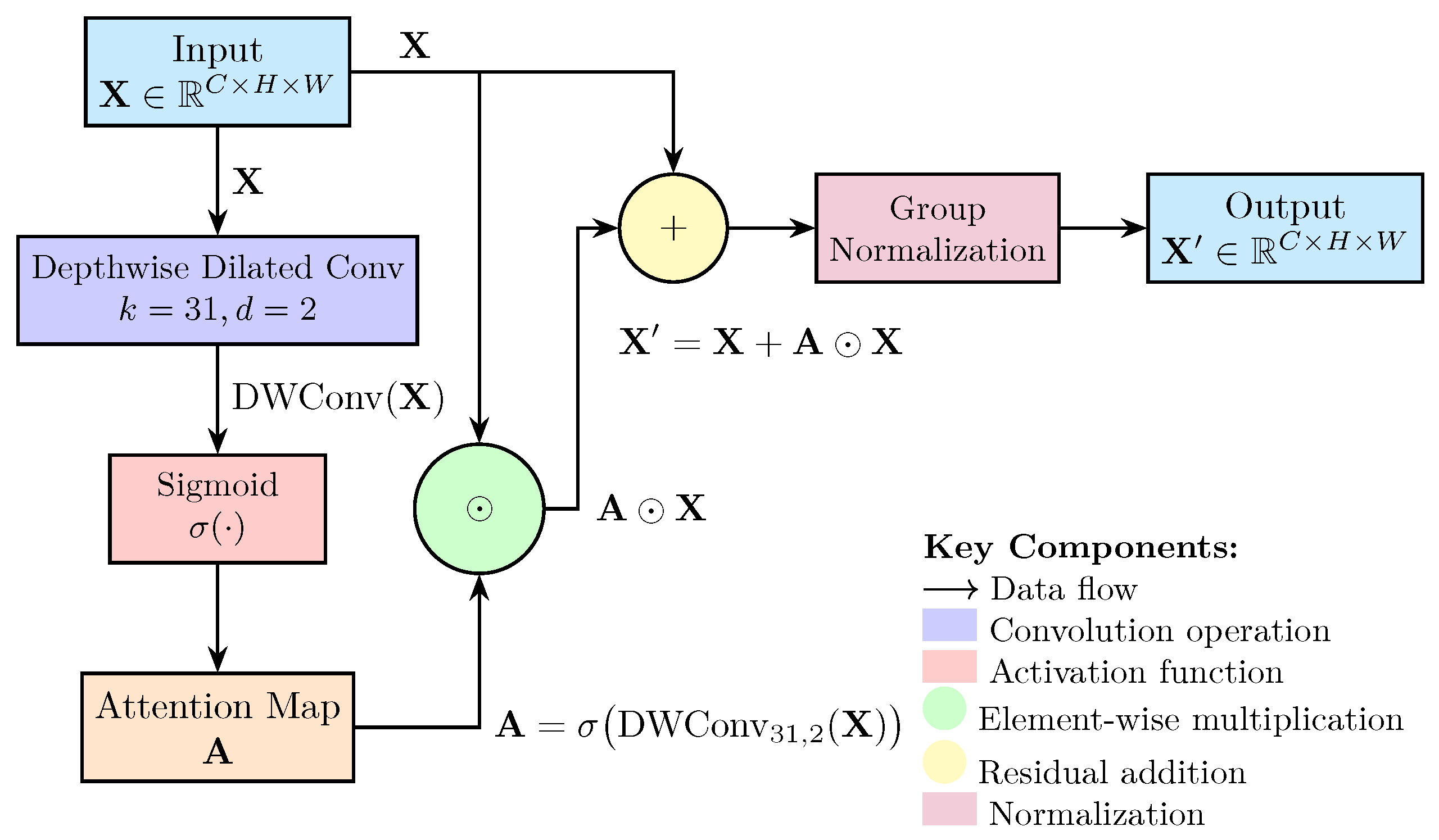
2.1. Model Improvements for Bone Tumor Detection
2.2. Implementation Details
- Initial learning rate: ;
- Weight decay: ;
- Scheduler: Cosine Annealing with warm-up for the first 5 epochs.
- Training duration: 300 maximum epochs;
- Evaluation: Metrics computed every 100 iterations;
- Early stopping: Activated if no improvement for 10 consecutive evaluations;
- Model selection: Final results reported using the best-performing weights.
- Random crop;
- Resize to (640, 640);
- Horizontal flipping (p = 0.5);
- Random rotation in range ;
- Contrast-limited adaptive histogram equalization (CLAHE);
- Random brightness/contrast adjustments.
2.3. Evaluation Metrics
3. Results
3.1. Dataset
3.1.1. BTXRD
3.1.2. Munich Dataset
3.1.3. Dataset Statistics
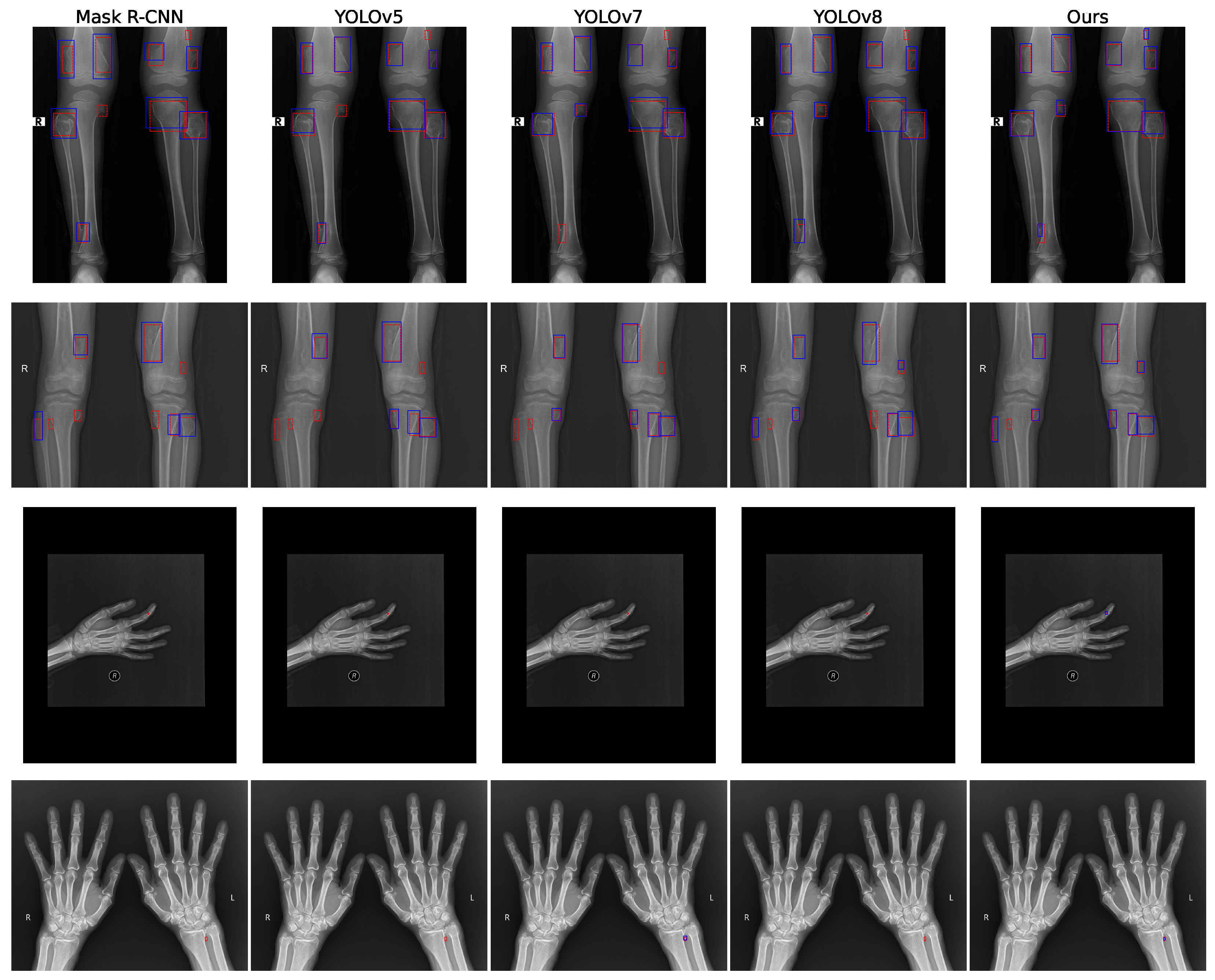
3.2. Comparison with SOTA Methods

3.3. Ablation Study
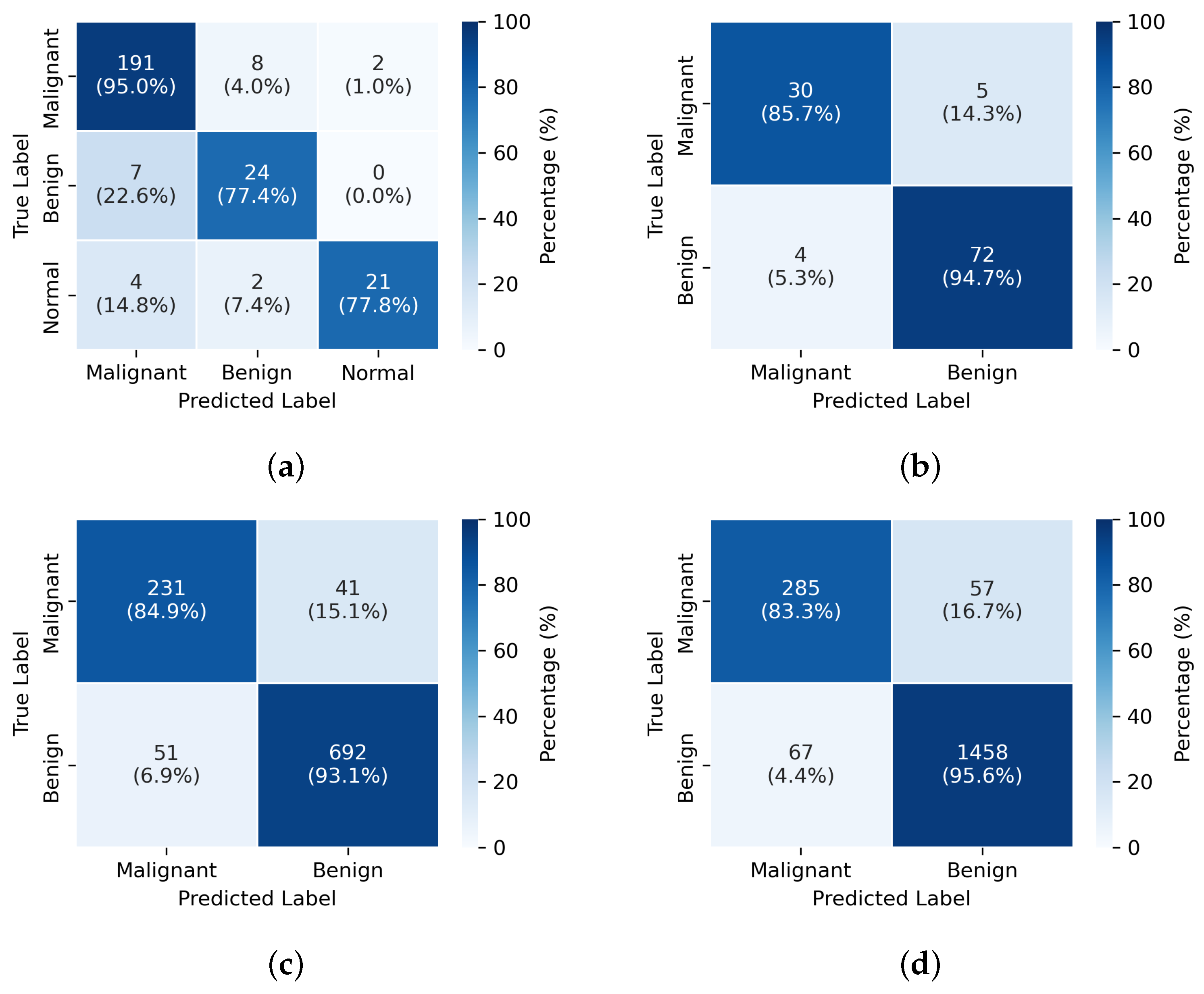
3.4. Visualization
4. Discussion
4.1. Clinical Significance and Performance Analysis
4.2. Architectural Innovations and Technical Contributions
4.3. Generalization and Robustness Analysis
4.4. Computational Efficiency and Clinical Deployment Considerations
4.5. Limitations and Future Directions
4.6. Broader Implications for Medical AI
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ong, W.; Zhu, L.; Tan, Y.L.; Teo, E.C.; Tan, J.H.; Kumar, N.; Vellayappan, B.; Ooi, B.; Quek, S.; Makmur, A.; et al. Application of Machine Learning for Differentiating Bone Malignancy on Imaging: A Systematic Review. Cancers 2023, 15, 1837. [Google Scholar] [CrossRef] [PubMed]
- Do, B.H.; Langlotz, C.P.; Beaulieu, C.F. Bone Tumor Diagnosis Using a Naïve Bayesian Model of Demographic and Radiographic Features. J. Digit. Imaging 2017, 30, 640–647. [Google Scholar] [CrossRef]
- Song, L.; Li, C.; Lilian, T.; Wang, M.; Chen, X.; Ye, Q.; Li, S.; Zhang, R.; Zeng, Q.; Xie, Z.; et al. A deep learning model to enhance the classification of primary bone tumors based on incomplete multimodal images in X-ray, CT, and MRI. Cancer Imaging 2024, 24, 135. [Google Scholar] [CrossRef] [PubMed]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.X.; Huang, Y.; Wang, X.Y.; Zhang, Y.; Paixao, I.C.; Wang, Z.; Chai, C.L.; Wang, H.; Lu, D.; Webb, G.I.; et al. A Radiograph Dataset for the Classification, Localization, and Segmentation of Primary Bone Tumors. Sci. Data 2025, 12, 88. [Google Scholar] [CrossRef]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef]
- Hassan, C.; Misawa, M.; Rizkala, T.; Mori, Y.; Sultan, S.; Facciorusso, A.; Antonelli, G.; Spadaccini, M.; Houwen, B.B.; Rondonotti, E.; et al. Computer-aided diagnosis for leaving colorectal polyps in situ: A systematic review and meta-analysis. Ann. Intern. Med. 2024, 177, 919–928. [Google Scholar] [CrossRef]
- Teodorescu, B.; Gilberg, L.; Melton, P.W.; Hehr, R.M.; Guzel, H.E.; Koc, A.M.; Baumgart, A.; Maerkisch, L.; Ataide, E.J.G. A systematic review of deep learning-based spinal bone lesion detection in medical images. Acta Radiol. 2024, 65, 1115–1125. [Google Scholar] [CrossRef]
- Afnouch, M.; Gaddour, O.; Hentati, Y.; Bougourzi, F.; Abid, M.; Alouani, I.; Ahmed, A.T. BM-Seg: A new bone metastases segmentation dataset and ensemble of CNN-based segmentation approach. Expert Syst. Appl. 2023, 228, 120376. [Google Scholar] [CrossRef]
- Karako, K.; Mihara, Y.; Arita, J.; Ichida, A.; Bae, S.K.; Kawaguchi, Y.; Ishizawa, T.; Akamatsu, N.; Kaneko, J.; Hasegawa, K.; et al. Automated liver tumor detection in abdominal ultrasonography with a modified faster region-based convolutional neural networks (Faster R-CNN) architecture. Hepatobiliary Surg. Nutr. 2022, 11, 675. [Google Scholar] [CrossRef]
- Weikert, T.; Jaeger, P.; Yang, S.; Baumgartner, M.; Breit, H.; Winkel, D.; Sommer, G.; Stieltjes, B.; Thaiss, W.; Bremerich, J.; et al. Automated lung cancer assessment on 18F-PET/CT using Retina U-Net and anatomical region segmentation. Eur. Radiol. 2023, 33, 4270–4279. [Google Scholar] [CrossRef] [PubMed]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med. Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef] [PubMed]
- Materka, A.; Jurek, J. Using deep learning and b-splines to model blood vessel lumen from 3d images. Sensors 2024, 24, 846. [Google Scholar] [CrossRef] [PubMed]
- von Schacky, C.V.; Wilhelm, N.J.; Schäfer, V.S.; Leonhardt, Y.; Gassert, F.; Foreman, S.; Gassert, F.; Jung, M.; Jungmann, P.; Russe, M.; et al. Multitask Deep Learning for Segmentation and Classification of Primary Bone Tumors on Radiographs. Radiology 2021, 301, 398–406. [Google Scholar] [CrossRef]
- Liu, R.; Pan, D.; Xu, Y.; Zeng, H.; He, Z.; Lin, J.; Zeng, W.; Wu, Z.; Luo, Z.; Qin, G.; et al. A deep learning–machine learning fusion approach for the classification of benign, malignant, and intermediate bone tumors. Eur. Radiol. 2022, 32, 1371–1383. [Google Scholar] [CrossRef]
- Xie, Z.; Zhao, H.; Song, L.; Ye, Q.; Zhong, L.; Li, S.; Zhang, R.; Wang, M.; Chen, X.; Lu, Z.; et al. A radiograph-based deep learning model improves radiologists’ performance for classification of histological types of primary bone tumors: A multicenter study. Eur. J. Radiol. 2024, 176, 111496. [Google Scholar] [CrossRef]
- Li, J.; Li, S.; Li, X.l.; Miao, S.; Dong, C.; Gao, C.P.; Liu, X.; Hao, D.; Xu, W.; Huang, M.; et al. Primary bone tumor detection and classification in full-field bone radiographs via YOLO deep learning model. Eur. Radiol. 2022, 33, 4237–4248. [Google Scholar] [CrossRef]
- Schacky, C.E.V.; Wilhelm, N.J.; Schäfer, V.S.; Leonhardt, Y.; Jung, M.; Jungmann, P.M.; Russe, M.F.; Foreman, S.C.; Gassert, F.G.; Gassert, F.T.; et al. Development and evaluation of machine learning models based on X-ray radiomics for the classification and differentiation of malignant and benign bone tumors. Eur. Radiol. 2022, 32, 6247–6257. [Google Scholar] [CrossRef]
- He, Y.; Pan, I.; Bao, B.; Halsey, K.; Chang, M.; Liu, H.; Peng, S.; Sebro, R.; Guan, J.; Yi, T.; et al. Deep learning-based classification of primary bone tumors on radiographs: A preliminary study. EBioMedicine 2020, 62, 103121. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Wang, Z.; Jin, L.; Wang, S.; Xu, H. Apple stem/calyx real-time recognition using YOLO-v5 algorithm for fruit automatic loading system. Postharvest Biol. Technol. 2022, 185, 111808. [Google Scholar] [CrossRef]
- Sapkota, R.; Flores-Calero, M.; Qureshi, R.; Badgujar, C.; Nepal, U.; Poulose, A.; Zeno, P.; Vaddevolu, U.B.P.; Khan, S.; Shoman, M.; et al. YOLO advances to its genesis: A decadal and comprehensive review of the You Only Look Once (YOLO) series. Artif. Intell. Rev. 2025, 58, 1–83. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7464–7475. [Google Scholar]
- Sohan, M.; Sai Ram, T.; Rami Reddy, C.V. A review on yolov8 and its advancements. In Proceedings of the International Conference on Data Intelligence and Cognitive Informatics, Tirunelveli, India, 18–20 November 2024; pp. 529–545. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning transferable visual models from natural language supervision. In Proceedings of the International Conference on Machine Learning, Online, 18–24 July 2021; pp. 8748–8763. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Li, J.; Li, D.; Xiong, C.; Hoi, S. Blip: Bootstrapping language-image pre-training for unified vision-language understanding and generation. In Proceedings of the International Conference on Machine Learning, Baltimore, MD, USA, 17–23 July 2022; pp. 12888–12900. [Google Scholar]
- Zhang, X.; Xu, M.; Qiu, D.; Yan, R.; Lang, N.; Zhou, X. Mediclip: Adapting clip for few-shot medical image anomaly detection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; pp. 458–468. [Google Scholar]
- Manzari, O.N.; Ahmadabadi, H.; Kashiani, H.; Shokouhi, S.B.; Ayatollahi, A. MedViT: A robust vision transformer for generalized medical image classification. Comput. Biol. Med. 2023, 157, 106791. [Google Scholar] [CrossRef]
- Chen, Q.; Hong, Y. Medblip: Bootstrapping language-image pre-training from 3d medical images and texts. In Proceedings of the Asian Conference on Computer Vision, Hanoi, Vietnam, 8–12 December 2024; pp. 2404–2420. [Google Scholar]
- Gutiérrez-Jimeno, M.; Panizo-Morgado, E.; Tamayo, I.; San Julián, M.; Catalán-Lambán, A.; Alonso, M.M.; Patiño-García, A. Somatic and germline analysis of a familial Rothmund–Thomson syndrome in two siblings with osteosarcoma. NPJ Genom. Med. 2020, 5, 51. [Google Scholar] [CrossRef]
| Dataset | Normal | Benign Tumor | Malignant Tumor | Total |
|---|---|---|---|---|
| BTXRD (Center 1) | 1593 | 1110 | 235 | 2938 |
| BTXRD (Center 2) | 259 | 214 | 76 | 549 |
| BTXRD (Center 3) | 27 | 201 | 31 | 259 |
| BTXRD Total | 1879 | 1525 | 342 | 3746 |
| Munich (Internal) | – | 667 | 267 | 934 |
| Munich (External) | – | 76 | 35 | 111 |
| Experiment | Model | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|---|
| Exp1: BTXRD Center1/2 → Center3 | CLIP [31] | 83.21 ± 0.83 | 81.35 ± 0.75 | 79.42 ± 0.93 | 80.36 ± 0.85 | 88.15 ± 0.68 |
| ViT [32] | 85.73 ± 0.61 | 83.97 ± 0.53 | 82.15 ± 0.75 | 83.04 ± 0.65 | 89.67 ± 0.58 | |
| Swin [29] | 87.92 ± 0.54 | 85.63 ± 0.45 | 84.27 ± 0.63 | 84.93 ± 0.58 | 91.24 ± 0.40 | |
| BLIP [33] | 86.45 ± 0.72 | 84.12 ± 0.69 | 83.56 ± 0.81 | 83.83 ± 0.73 | 90.17 ± 0.51 | |
| Ours | 91.38 ± 0.49 | 89.76 ± 0.33 | 88.93 ± 0.51 | 89.34 ± 0.42 | 94.62 ± 0.39 | |
| Exp2: Munich Internal → External | CLIP | 82.16 ± 1.21 | 78.34 ± 1.36 | 80.25 ± 1.13 | 79.28 ± 1.29 | 87.43 ± 0.84 |
| ViT | 84.95 ± 0.96 | 81.67 ± 1.02 | 83.14 ± 0.85 | 82.39 ± 0.92 | 89.26 ± 0.73 | |
| Swin | 86.32 ± 0.82 | 83.45 ± 0.90 | 84.97 ± 0.79 | 84.20 ± 0.84 | 90.85 ± 0.64 | |
| BLIP | 85.14 ± 1.02 | 82.78 ± 1.14 | 83.56 ± 0.95 | 83.16 ± 1.02 | 89.73 ± 0.73 | |
| Ours | 89.87 ± 0.72 | 87.62 ± 0.89 | 88.34 ± 0.66 | 87.98 ± 0.78 | 93.45 ± 0.53 | |
| Exp3: BTXRD → Munich | CLIP | 70.54 ± 1.87 | 68.12 ± 1.92 | 69.23 ± 1.85 | 68.67 ± 1.89 | 84.12 ± 1.76 |
| ViT | 61.67 ± 7.15 | 59.12 ± 6.89 | 60.45 ± 7.32 | 59.78 ± 7.10 | 74.12 ± 6.45 | |
| Swin | 75.12 ± 2.32 | 73.45 ± 2.12 | 74.12 ± 2.45 | 73.78 ± 2.32 | 82.45 ± 2.12 | |
| BLIP | 80.26 ± 0.84 | 78.14 ± 0.91 | 79.27 ± 0.70 | 78.70 ± 0.83 | 86.87 ± 0.61 | |
| Ours | 87.62 ± 0.81 | 85.93 ± 0.60 | 86.45 ± 0.65 | 86.19 ± 0.58 | 92.78 ± 0.47 | |
| Exp4: Munich → BTXRD | CLIP | 75.93 ± 1.12 | 73.45 ± 1.25 | 74.12 ± 1.15 | 73.78 ± 1.18 | 82.45 ± 1.05 |
| ViT | 57.45 ± 6.78 | 55.12 ± 6.45 | 56.34 ± 6.89 | 55.78 ± 6.67 | 70.12 ± 5.98 | |
| Swin | 85.93 ± 0.57 | 83.25 ± 0.64 | 84.76 ± 0.42 | 84.00 ± 0.50 | 90.62 ± 0.33 | |
| BLIP | 82.47 ± 0.61 | 79.83 ± 0.78 | 81.25 ± 0.54 | 80.53 ± 0.69 | 87.95 ± 0.42 | |
| Ours | 88.74 ± 0.41 | 86.82 ± 0.59 | 87.63 ± 0.31 | 87.22 ± 0.49 | 93.87 ± 0.31 |
| Configuration | mAP | Small mAP | Multi-Lesion mAP | F1-Score | FPS |
|---|---|---|---|---|---|
| Baseline YOLOv11 | 72.3 | 45.2 | 51.2 | 0.751 | 125.10 |
| + Large Kernel Attention | 75.1 | 50.3 | 56.8 | 0.769 | 100.47 |
| + Multi-scale Fusion | 75.8 | 52.4 | 58.9 | 0.782 | 99.81 |
| + Angle-aware Encoding | 78.2 | 53.1 | 60.7 | 0.791 | 97.85 |
| + Adaptive Focal Loss | 79.6 | 55.8 | 63.2 | 0.804 | 97.85 |
| Experiment | Model | mAP | Small mAP | Multi-Lesion mAP | F1-Score | FPS | Params (M) |
|---|---|---|---|---|---|---|---|
| Exp1: BTXRD Center1/2 → Center3 | Mask R-CNN | 65.8 ± 1.12 | 33.2 ± 1.25 | 38.5 ± 1.15 | 0.682 ± 1.08 | 95.4 ± 1.3 | 42.3 |
| YOLOv5 | 69.6 ± 0.92 | 38.5 ± 1.05 | 42.8 ± 0.95 | 0.718 ± 0.88 | 120.3 ± 1.2 | 7.2 | |
| YOLOv7 | 71.2 ± 0.83 | 40.8 ± 0.95 | 45.2 ± 0.88 | 0.735 ± 0.82 | 115.6 ± 1.1 | 9.6 | |
| YOLOv8 | 73.1 ± 0.75 | 43.2 ± 0.88 | 48.5 ± 0.82 | 0.752 ± 0.78 | 110.4 ± 1.3 | 10.9 | |
| Ours | 79.6 ± 0.62 | 55.8 ± 0.71 | 63.2 ± 0.65 | 0.804 ± 0.68 | 80.9 ± 1.5 | 15.4 | |
| Exp2: Munich Internal → External | Mask R-CNN | 67.3 ± 1.22 | 35.5 ± 1.35 | 40.8 ± 1.22 | 0.692 ± 1.25 | 92.8 ± 1.4 | 42.3 |
| YOLOv5 | 67.6 ± 1.18 | 36.2 ± 1.32 | 41.5 ± 1.15 | 0.698 ± 1.25 | 118.7 ± 1.4 | 7.2 | |
| YOLOv7 | 69.0 ± 1.12 | 37.8 ± 1.25 | 42.7 ± 1.05 | 0.712 ± 1.18 | 116.5 ± 1.3 | 9.6 | |
| YOLOv8 | 71.2 ± 0.98 | 40.5 ± 1.12 | 45.8 ± 0.95 | 0.735 ± 1.05 | 113.2 ± 1.1 | 10.9 | |
| Ours | 78.3 ± 0.81 | 50.2 ± 0.92 | 58.1 ± 0.76 | 0.792 ± 0.85 | 81.4 ± 1.4 | 15.4 | |
| Exp3: BTXRD → Munich | Mask R-CNN | 62.5 ± 1.42 | 31.8 ± 1.52 | 35.4 ± 1.35 | 0.658 ± 1.35 | 92.8 ± 1.6 | 42.3 |
| YOLOv5 | 65.8 ± 1.12 | 34.8 ± 1.25 | 39.2 ± 1.05 | 0.685 ± 1.18 | 117.9 ± 1.4 | 7.2 | |
| YOLOv7 | 67.9 ± 0.98 | 37.5 ± 1.12 | 42.0 ± 0.95 | 0.705 ± 1.05 | 114.8 ± 1.3 | 9.6 | |
| YOLOv8 | 70.1 ± 0.88 | 40.2 ± 0.98 | 45.5 ± 0.83 | 0.725 ± 0.92 | 111.5 ± 1.2 | 10.9 | |
| Ours | 77.4 ± 0.72 | 51.3 ± 0.81 | 59.4 ± 0.65 | 0.780 ± 0.78 | 79.6 ± 1.5 | 15.4 | |
| Exp4: Munich → BTXRD | Mask R-CNN | 63.8 ± 1.32 | 32.5 ± 1.42 | 36.5 ± 1.25 | 0.670 ± 1.28 | 91.9 ± 1.5 | 42.3 |
| YOLOv5 | 66.3 ± 1.15 | 35.2 ± 1.28 | 39.7 ± 1.12 | 0.690 ± 1.22 | 116.2 ± 1.3 | 7.2 | |
| YOLOv7 | 68.5 ± 1.02 | 37.8 ± 1.15 | 42.3 ± 0.98 | 0.710 ± 1.08 | 113.4 ± 1.2 | 9.6 | |
| YOLOv8 | 70.8 ± 0.91 | 40.3 ± 1.05 | 45.8 ± 0.88 | 0.730 ± 1.02 | 110.1 ± 1.4 | 10.9 | |
| Ours | 78.0 ± 0.75 | 52.4 ± 0.85 | 60.8 ± 0.68 | 0.790 ± 0.82 | 80.3 ± 1.6 | 15.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Peng, Y.; Liu, Y.; Wang, P.; Liu, T. Enhancing YOLOv11 with Large Kernel Attention and Multi-Scale Fusion for Accurate Small and Multi-Lesion Bone Tumor Detection in Radiographs. Diagnostics 2025, 15, 1988. https://doi.org/10.3390/diagnostics15161988
Chen S, Peng Y, Liu Y, Wang P, Liu T. Enhancing YOLOv11 with Large Kernel Attention and Multi-Scale Fusion for Accurate Small and Multi-Lesion Bone Tumor Detection in Radiographs. Diagnostics. 2025; 15(16):1988. https://doi.org/10.3390/diagnostics15161988
Chicago/Turabian StyleChen, Sihan, Youcheng Peng, Yingxuan Liu, Pengyu Wang, and Tao Liu. 2025. "Enhancing YOLOv11 with Large Kernel Attention and Multi-Scale Fusion for Accurate Small and Multi-Lesion Bone Tumor Detection in Radiographs" Diagnostics 15, no. 16: 1988. https://doi.org/10.3390/diagnostics15161988
APA StyleChen, S., Peng, Y., Liu, Y., Wang, P., & Liu, T. (2025). Enhancing YOLOv11 with Large Kernel Attention and Multi-Scale Fusion for Accurate Small and Multi-Lesion Bone Tumor Detection in Radiographs. Diagnostics, 15(16), 1988. https://doi.org/10.3390/diagnostics15161988








