Interpretable Deep Learning Models for Arrhythmia Classification Based on ECG Signals Using PTB-X Dataset
Abstract
1. Introduction
- A custom convolutional neural network-based deep learning model is proposed, specifically tailored for ECG arrhythmia classification. The model supports various diagnostic tasks, including binary classification (e.g., normal vs. specific conditions), and multiclass classification into 5, 10, or 15 clinically meaningful categories based on the PTB-XL dataset, allowing for both broad and detailed arrhythmia detection.
- To enhance contextual accuracy, the model integrates demographic attributes, namely age and gender, which contribute to a deeper understanding of arrhythmia patterns and lead to improved detection, particularly for less-represented classes.
- This research utilizes the PTB-XL dataset, a large and heterogeneous collection of ECG recordings, to ensure comprehensive training and validation. This choice supports the model’s generalizability across a broad range of cardiac conditions.
- An extensive experimental evaluation is conducted to assess the influence of incorporating demographic data. This study compares classification performance across binary and multiclass tasks, both with and without demographic features, demonstrating the added value of contextual inputs.
- In addition, the model offers interpretability by examining the influence of individual ECG leads in arrhythmia detection. This lead-specific analysis provides clinical insight into which signal channels most significantly contribute to the diagnostic process.
2. Literature Review
3. Methodology
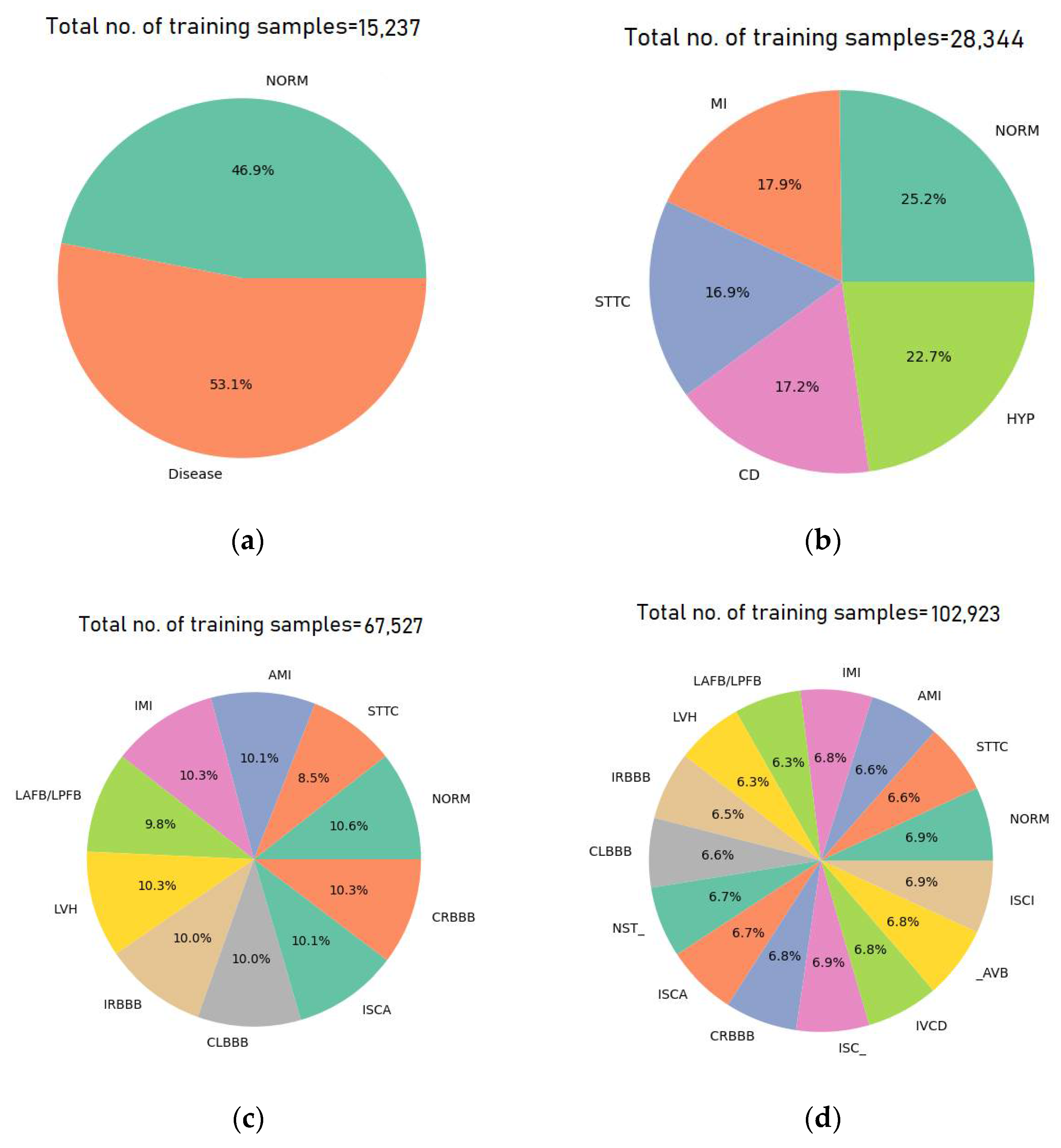
3.1. Dataset Description
- Folds 1–8 were used for training.
- Fold 9 was used for validation.
- Fold 10 was used for testing.
3.2. Experimental Setup
3.3. Diagnostic Classes
- Five-Class Supercategory Classification: In this multiclass classification task, the model categorizes ECG signals into one of five broad superclasses: NORM, MI, CD, STTC, and HYP.
- Ten-Class Subcategory Classification: In this configuration, the model classifies ECG signals into one of ten specific subclasses: NORM, STTC, AMI, IMI, LAFB/LPFB, LVH, IRBBB, CLBBB, ISCA, and CRBBB. These ten subclasses represent the most frequently occurring categories in the dataset, with all other less-represented subclasses being excluded.
- Fifteen-Class Subcategory Classification: This scenario expands the classification to fifteen subclasses, including: NORM, STTC, AMI, IMI, LAFB/LPFB, LVH, IRBBB, CLBBB, NST_, ISCA, CRBBB, IVCD, ISC_, _AVB, and ISCI. As with the ten-class configuration, only the top fifteen subclasses based on the number of records are considered.
3.4. Data Augmentation
3.5. The Proposed Models
3.5.1. The Proposed CNN Model Architecture
3.5.2. VGG Model Architecture
3.6. Model Interpretability Using the SHAP Method
3.7. Performance Evaluation Metrics
4. Experimental Results and Discussion
4.1. Experimental Results Using the CNN Model
4.1.1. Binary Classification Test Results
4.1.2. Multiclass Classification Results
4.2. Experimental Results Using the VGG16 Model
4.3. Performance Evaluation of CNN Against VGG16
4.4. Performance Comparison with Recently Published Works
4.5. Augmentation-Based Model Results
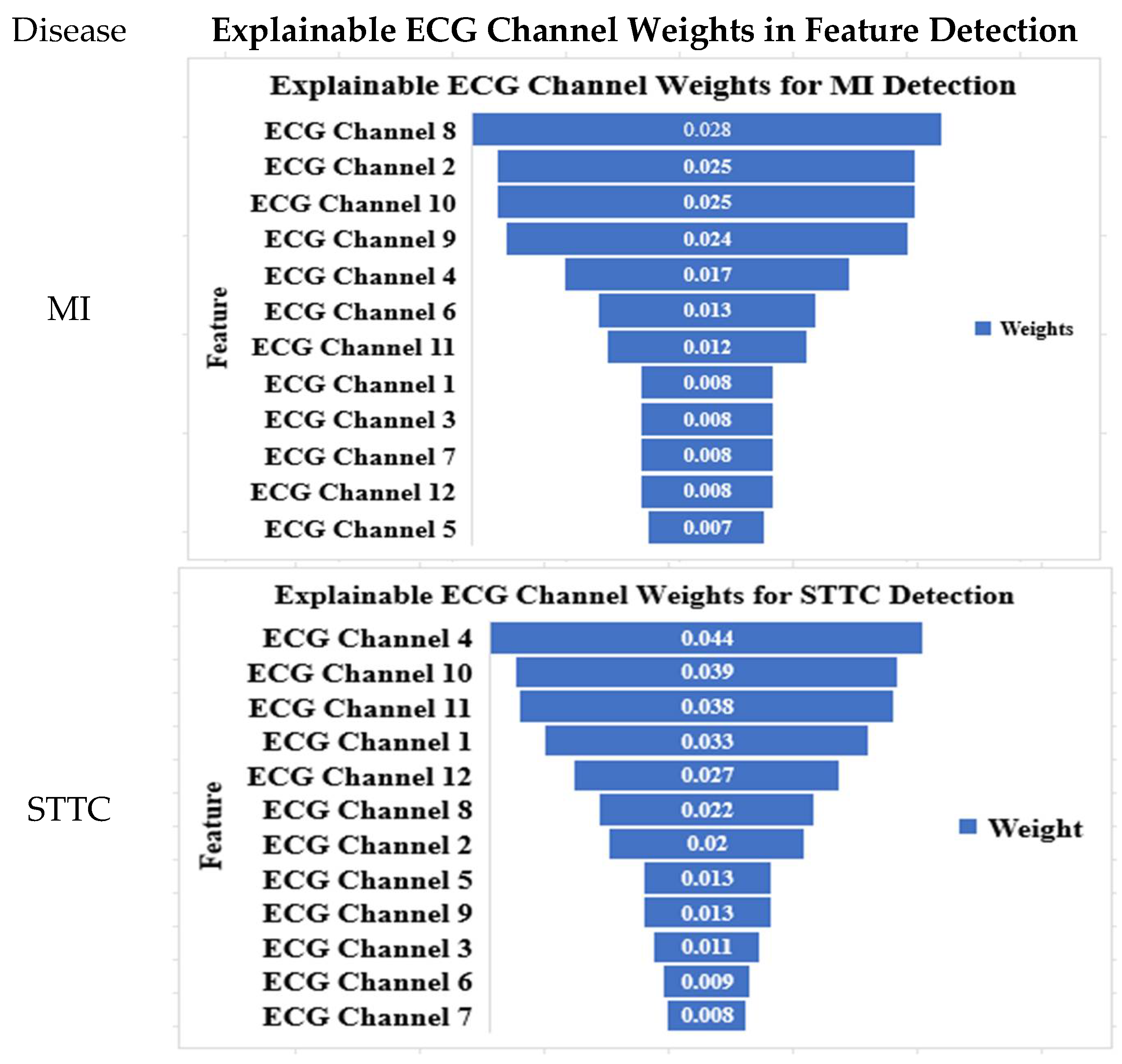
4.6. Explainable ECG Channel Contributions for Arrhythmia Detection
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Surawicz, B.; Knilans, T.K. Chou’s Electrocardiography in Clinical Practice; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar] [CrossRef]
- Strodthoff, N.; Wagner, P.; Schaeffter, T.; Samek, W. Deep Learning for ECG Analysis: Benchmarks and Insights from PTB-XL. IEEE J. Biomed. Health Inform. 2021, 25, 1519–1528. [Google Scholar] [CrossRef]
- Clifford, G.D.; Liu, C.; Moody, B.; Lehman, L.H.; Silva, I.; Li, Q.; Johnson, A.E.; Mark, R.G. AF Classification from a Short Single Lead ECG Recording: The Physionet Computing in Cardiology Challenge 2017. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017. [Google Scholar] [CrossRef]
- Warrick, P.A.; Lostanlen, V.; Eickenberg, M.; Homsi, M.N.; Rodraguez, A.C.; Anden, J. Arrhythmia Classification of Reduced-Lead Electrocardiograms by Scattering-Recurrent Networks. In Proceedings of the 2021 Computing in Cardiology (CinC), Brno, Czech Republic, 13–15 September 2021; IEEE: New York, NY, USA; pp. 1–4. [Google Scholar] [CrossRef]
- Jin, Y.; Li, Z.; Wang, M.; Liu, J.; Tian, Y.; Liu, Y.; Wei, X.; Zhao, L.; Liu, C. Cardiologist-level interpretable knowledge-fused deep neural network for automatic arrhythmia diagnosis. Commun. Med. 2024, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Weimann, K.; Conrad, T.O.F. Transfer learning for ECG classification. Sci. Rep. 2021, 11, 5251. [Google Scholar] [CrossRef] [PubMed]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Chen, J.; Zhou, L. A transformer-based deep neural network for arrhythmia detection using continuous ECG signals. Comput. Biol. Med. 2022, 144, 105325. [Google Scholar] [CrossRef]
- Aziz, S.; Ahmed, S.; Alouini, M.-S. ECG-based machine-learning algorithms for heartbeat classification. Sci. Rep. 2021, 11, 18738. [Google Scholar] [CrossRef]
- Figueroa-Gil, L.E.; López-Cons, I.V.; Orjuela-Cañón, A.D. Machine Learning Techniques for Classifying Cardiac Arrhythmias. In Proceedings of the XLVII Mexican Conference on Biomedical Engineering, Hermosillo, Mexico, 7–9 November 2025; Springer: Berlin/Heidelberg, Germany, 2025; pp. 27–39. [Google Scholar] [CrossRef]
- Pandey, S.K.; Janghel, R.R.; Vani, V. Patient Specific Machine Learning Models for ECG Signal Classification. Procedia Comput. Sci. 2020, 167, 2181–2190. [Google Scholar] [CrossRef]
- Zabihi, F.; Safara, F.; Ahadzadeh, B. An electrocardiogram signal classification using a hybrid machine learning and deep learning approach. Healthc. Anal. 2024, 6, 100366. [Google Scholar] [CrossRef]
- Jambukia, S.H.; Dabhi, V.K.; Prajapati, H.B. Classification of ECG signals using machine learning techniques: A survey. In Proceedings of the 2015 International Conference on Advances in Computer Engineering and Applications, Ghaziabad, India, 19–20 March 2015; IEEE: New York, NY, USA; pp. 714–721. [Google Scholar] [CrossRef]
- Baghdadi, N.A.; Abdelaliem, S.M.F.; Malki, A.; Gad, I.; Ewis, A.; Atlam, E. Advanced machine learning techniques for cardiovascular disease early detection and diagnosis. J. Big Data 2023, 10, 144. [Google Scholar] [CrossRef]
- Sraitih, M.; Jabrane, Y.; El Hassani, A.H. An Automated System for ECG Arrhythmia Detection Using Machine Learning Techniques. J. Clin. Med. 2021, 10, 5450. [Google Scholar] [CrossRef]
- Malik, M.; Dua, T.; Snigdha. Biomedical Signal Processing: ECG Signal Analysis Using Machine Learning in MATLAB. In Recent Advances in Metrology; Springer: Berlin/Heidelberg, Germany, 2023; pp. 121–127. [Google Scholar] [CrossRef]
- Hassaballah, M.; Wazery, Y.M.; Ibrahim, I.E.; Farag, A. ECG Heartbeat Classification Using Machine Learning and Metaheuristic Optimization for Smart Healthcare Systems. Bioengineering 2023, 10, 429. [Google Scholar] [CrossRef]
- Ben-Moshe, N.; Brimer, S.B.; Tsutsui, K.; Suleiman, M.; Sörnmo, L.; Behar, J.A. Machine learning for ranking f-wave extraction methods in single-lead ECGs. Biomed. Signal Process. Control 2025, 99, 106817. [Google Scholar] [CrossRef]
- Alfaras, M.; Soriano, M.C.; Ortín, S. A Fast Machine Learning Model for ECG-Based Heartbeat Classification and Arrhythmia Detection. Front. Phys. 2019, 7, 103. [Google Scholar] [CrossRef]
- Hemakom, A.; Atiwiwat, D.; Israsena, P. ECG and EEG based machine learning models for the classification of mental workload and stress levels for women in different menstrual phases, men, and mixed sexes. Biomed. Signal Process. Control 2024, 95, 106379. [Google Scholar] [CrossRef]
- Pal, A.; Srivastva, R.; Singh, Y.N. CardioNet: An Efficient ECG Arrhythmia Classification System Using Transfer Learning. Big Data Res. 2021, 26, 100271. [Google Scholar] [CrossRef]
- Chorney, W.; Wang, H. Towards federated transfer learning in electrocardiogram signal analysis. Comput. Biol. Med. 2024, 170, 107984. [Google Scholar] [CrossRef]
- Nguyen, C.V.; Do, C.D. Transfer Learning in ECG Diagnosis: Is It Effective? Available online: http://arxiv.org/abs/2402.02021 (accessed on 21 June 2025).
- Ahmad, M.; Ahmed, A.; Hashim, H.; Farsi, M.; Mahmoud, N. Enhancing Heart Disease Diagnosis Using ECG Signal Reconstruction and Deep Transfer Learning Classification with Optional SVM Integration. Diagnostics 2025, 15, 1501. [Google Scholar] [CrossRef]
- Ao, R.; He, G. Image Based Deep Learning in 12-Lead ECG Diagnosis. medRxiv 2022. [Google Scholar] [CrossRef]
- Bontinck, L.; Fonteyn, K.; Dhaene, T.; Deschrijver, D. ECGencode: Compact and computationally efficient deep learning feature encoder for ECG signals. Expert. Syst. Appl. 2024, 255, 124775. [Google Scholar] [CrossRef]
- Butt, F.S.; Wagner, M.F.; Schäfer, J.; Ullate, D.G. Toward Automated Feature Extraction for Deep Learning Classification of Electrocardiogram Signals. IEEE Access 2022, 10, 118601–118616. [Google Scholar] [CrossRef]
- Aarthy, S.T.; Iqbal, J.L.M. A novel deep learning approach for early detection of cardiovascular diseases from ECG signals. Med. Eng. Phys. 2024, 125, 104111. [Google Scholar] [CrossRef]
- Akalın, F.; Çavdaroğlu, P.D.; Orhan, M.F. Arrhythmia detection with transfer learning architecture integrating the developed optimization algorithm and regularization method. BMC Biomed Eng. 2025, 7, 8. [Google Scholar] [CrossRef]
- Ansari, Y.; Mourad, O.; Qaraqe, K.; Serpedin, E. Deep learning for ECG Arrhythmia detection and classification: An overview of progress for period 2017–2023. Front. Physiol. 2023, 14, 1246746. [Google Scholar] [CrossRef]
- Narotamo, H.; Dias, M.; Santos, R.; Carreiro, A.V.; Gamboa, H.; Silveira, M. Deep learning for ECG classification: A comparative study of 1D and 2D representations and multimodal fusion approaches. Biomed. Signal Process. Control 2024, 93, 106141. [Google Scholar] [CrossRef]
- Śmigiel, S.; Pałczyński, K.; Ledziński, D. Deep learning techniques in the classification of ecg signals using r-peak detection based on the ptb-xl dataset. Sensors 2021, 21, 8174. [Google Scholar] [CrossRef] [PubMed]
- Kuila, S.; Dhanda, N.; Joardar, S. ECG signal classification to detect heart arrhythmia using ELM and CNN. Multimed. Tools Appl. 2023, 82, 29857–29881. [Google Scholar] [CrossRef]
- Alamatsaz, N.; Tabatabaei, L.S.; Yazdchi, M.; Payan, H.; Alamatsaz, N.; Nasimi, F. A Lightweight Hybrid CNN-LSTM Model for ECG-Based Arrhythmia Detection. Available online: http://arxiv.org/abs/2209.00988 (accessed on 21 June 2025).
- Swaroop, P.; Badolia, N.; Ranjan, R.; Kumar, M. Arrhythmia Classification Using Hybrid CNN-LSTM Model. In Proceedings of the 2024 First International Conference on Electronics, Communication and Signal Processing (ICECSP), New Delhi, India, 8–10 August 2024; IEEE: New York, NY, USA, 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Selvam, I.J.; Madhavan, M.; Kumarasamy, S.K. Detection and classification of electrocardiography using hybrid deep learning models. Hell. J. Cardiol. 2024, 81, 75–84. [Google Scholar] [CrossRef]
- Varalakshmi, P.; Sankaran, A.P. An improved hybrid AI model for prediction of arrhythmia using ECG signals. Biomed. Signal Process. Control 2023, 80, 104248. [Google Scholar] [CrossRef]
- Shah, H.A.; Saeed, F.; Diyan, M.; Almujally, N.A.; Kang, J. ECG-TransCovNet: A hybrid transformer model for accurate arrhythmia detection using Electrocardiogram signals. In CAAI Transactions on Intelligence Technology; Wiley: Hoboken, NJ, USA, 2024; pp. 1–14. [Google Scholar] [CrossRef]
- Talukder, M.A.; Khalid, M.; Kazi, M.; Jahan Muna, N.; Nur-e-Alam, M.; Halder, S.; Sultana, N. A hybrid cardiovascular arrhythmia disease detection using ConvNeXt-X models on electrocardiogram signals. Sci. Rep. 2024, 14, 30366. [Google Scholar] [CrossRef]
- Bai, X.; Dong, X.; Li, Y.; Liu, R.; Zhang, H. A hybrid deep learning network for automatic diagnosis of cardiac arrhythmia based on 12-lead ECG. Sci. Rep. 2024, 14, 24441. [Google Scholar] [CrossRef]
- Jing, J.; Zhang, J.; Liu, A.; Gao, M.; Qian, R.; Chen, X. ECG-Based Multiclass Arrhythmia Classification Using Beat-Level Fusion Network. J. Healthc. Eng 2023, 2023, 1755121. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, Y.; Zhang, Z.; Liu, J.; Yuan, Y.; Wang, K.; He, R. Learning with incomplete labels of multisource datasets for ECG classification. Pattern Recognit. 2024, 150, 110321. [Google Scholar] [CrossRef]
- Nguyen, C.V.; Duong, H.M.; Do, C.D. MELEP: A Novel Predictive Measure of Transferability in Multi-Label ECG Diagnosis. J. Heal. Inform. Res. 2024, 8, 506–522. [Google Scholar] [CrossRef]
- Pałczyński, K.; Śmigiel, S.; Ledziński, D.; Bujnowski, S. Study of the Few-Shot Learning for ECG Classification Based on the PTB-XL Dataset. Sensors 2022, 22, 904. [Google Scholar] [CrossRef] [PubMed]
- Bhanja, N. Design and Comparison of Deep Learning Model for ECG Classification Using PTB-XL Dataset. Available online: https://www.researchgate.net/publication/374061560 (accessed on 21 June 2025).
- Wagner, P.; Strodthoff, N.; Bousseljot, R.-D.; Kreiseler, D.; Lunze, F.I.; Samek, W.; Schaeffter, T. PTB-XL, a large publicly available electrocardiography dataset. Sci Data 2020, 7, 154. [Google Scholar] [CrossRef]
- Hambarde, S.; Paithane, A.; Lambhate, P.; Hambarde, A.S.; Kalyankar, P.A. Smart Arrhythmia Detection Using Single Lead ECG Signal and Hybridized Deep Neural Network Model. Web Intell. 2025, 23, 155–171. [Google Scholar] [CrossRef]
- Strodthoff, N.; Mehari, T.; Nagel, C.; Aston, P.J.; Sundar, A.; Graff, C.; Kanters, J.K.; Haverkamp, W.; Dössel, O.; Loewe, A.; et al. PTB-XL+, a comprehensive electrocardiographic feature dataset. Sci. Data 2023, 10, 279. [Google Scholar] [CrossRef]
- Krasteva, V.; Stoyanov, T.; Naydenov, S.; Schmid, R.; Jekova, I. Detection of Atrial Fibrillation in Holter ECG Recordings by ECHOView Images: A Deep Transfer Learning Study. Diagnostics 2025, 15, 865. [Google Scholar] [CrossRef]
- Wickramasinghe, N.L.; Athif, M. Multi-label classification of reduced-lead ECGs using an interpretable deep convolutional neural network. Physiol. Meas. 2022, 43, 064002. [Google Scholar] [CrossRef]
- Gour, A.; Gupta, M.; Wadhvani, R.; Shukla, S. ECG Based Heart Disease Classification: Advancement and Review of Techniques. In Procedia Computer Science; Elsevier: Amsterdam, The Netherlands, 2024; pp. 1634–1648. [Google Scholar] [CrossRef]
- Safdar, M.F.; Nowak, R.M.; Pałka, P. Pre-Processing Techniques and Artificial Intelligence Algorithms for Electrocardiogram (ECG) Signals Analysis: A Comprehensive Review; Elsevier: Amsterdam, The Netherlands, 2024; Volume 1. [Google Scholar] [CrossRef]
- Feyisa, D.W.; Debelee, T.G.; Ayano, Y.M.; Kebede, S.R.; Assore, T.F. Lightweight Multireceptive Field CNN for 12-Lead ECG Signal Classification. Comput. Intell. Neurosci. 2022, 2022, 8413294. [Google Scholar] [CrossRef]
- Śmigiel, S.; Pałczyński, K.; Ledziński, D. ECG Signal Classification Using Deep Learning Techniques Based on the PTB-XL Dataset. Entropy 2021, 23, 1121. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, F.M.; Schoenfeld, M.H.; Barrett, C.; Edgerton, J.R.; Ellenbogen, K.A.; Gold, M.R.; Goldschlager, N.F.; Hamilton, R.M.; Joglar, J.A.; Kim, R.J.; et al. 2018 ACC/AHA/HRS Guideline on the Evaluation and Management of Patients with Bradycardia and Cardiac Conduction Delay: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Circulation 2019, 140, E382–E482. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.H.; Timothy, K.W.; Frankovich, D.; Fromm, B.S.; Keating, M.; Locati, E.H.; Taggart, R.; A Towbin, J.; Moss, A.J.; Schwartz, P.J.; et al. Age-Gender Influence on the Rate-Corrected QT Interval and the QT-Heart Rate Relation in Families with Genotypically Characterized Long QT Syndrome. J. Am. Coll. Cardiol. 1997, 29, 93–99. [Google Scholar] [CrossRef]
- Okin, P.M.; Roman, M.J.; Devereux, R.B.; Kligfield, P. Gender Differences and the Electrocardiogram in Left Ventricular Hypertrophy. Hypertension 1995, 25, 242–249. [Google Scholar] [CrossRef]
- Antzelevitch, C.; Patocskai, B. Brugada Syndrome: Clinical, Genetic, Molecular, Cellular, and Ionic Aspects. Curr. Probl. Cardiol. 2016, 41, 7–57. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From precision, recall, and F-measure to ROC, informedness, markedness, and correlation. arXiv 2011, arXiv:2010.16061. [Google Scholar]
- Rashed, A.E.E.; Bahgat, W.M.; Ahmed, A.; Farrag, T.A.; Atwa, A.E.M. Efficient machine learning models across multiple datasets for autism spectrum disorder diagnoses. Biomed. Signal Process Control 2025, 100, 106949. [Google Scholar] [CrossRef]
- Misumida, N.; Kobayashi, A.; Fox, J.T.; Hanon, S.; Schweitzer, P.; Kanei, Y. Predictive Value of ST-Segment Elevation in Lead aVR for Left Main and/or Three-Vessel Disease in Non-ST-Segment Elevation Myocardial Infarction. Ann. Noninvasive Electrocardiol. 2016, 21, 91–97. [Google Scholar] [CrossRef]
- Razin, V.; Krasnov, A.; Karchkov, D.; Moskalenko, V.; Rodionov, D.; Zolotykh, N.; Smirnov, L.; Osipov, G. Solving the Problem of Diagnosing a Disease by ECG on the PTB-XL Dataset Using Deep Learning. In Advances in Neural Computation, Machine Learning, and Cognitive Research VII; Springer: Berlin/Heidelberg, Germany, 2023; pp. 13–21. [Google Scholar] [CrossRef]
- Surawicz, B.; Childers, R.; Deal, B.J.; Gettes, L.S. AHA/ACCF/HRS Recommendations for the Standardization and Interpretation of the Electrocardiogram. Circulation 2009, 119, e235–e240. [Google Scholar] [CrossRef]
- Hancock, E.W.; Deal, B.J.; Mirvis, D.M.; Okin, P.; Kligfield, P.; Gettes, L.S. AHA/ACCF/HRS Recommendations for the Standardization and Interpretation of the Electrocardiogram. Circulation 2009, 119, e251–e261. [Google Scholar] [CrossRef]
- Weintraub, R.G.; Alexander, P.M.A. Outcomes in Pediatric Dilated Cardiomyopathy. J. Am. Coll. Cardiol. 2017, 70, 2674–2676. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Su, H.; Di Lallo, A.; Murphy, R.R.; Taylor, R.H.; Garibaldi, B.T.; Krieger, A. Physical human–robot interaction for clinical care in infectious environments. Nat. Mach. Intell. 2021, 3, 184–186. [Google Scholar] [CrossRef]



















| Ref. | Dataset | Models Applied | Key Results | DOI/Link |
|---|---|---|---|---|
| [2] | PTB-XL, ICBEB2018 | ResNet, Inception, Transfer Learning, xresnet1d101 | Demonstrated PTB-XL as a benchmark for ECG analysis; transfer learning showed promising results for small datasets. | https://doi.org/10.1109/JBHI.2020.3022989 |
| [5] | Chinese ECG Benchmark | Knowledge-Fused DNN | Achieved higher performance than cardiologists on arrhythmia classification in remote settings. | https://doi.org/10.1038/s43856-024-00464-4 |
| [23] | PTB-XL, CPSC2018, Georgia, Ribeiro | Transfer Learning (Various CNNs, RNNs) | Transfer learning was effective on small datasets; fine-tuning did not consistently outperform training from scratch on larger dataset. | https://doi.org/10.48550/arXiv.2402.02021 |
| [25] | PTB-XL, CPSC2018, Shaoxing, Tongji | Convolutional Neural Networks (CNNs) | Excellent AUROC, AUPRC on test data from PTB-XL; lower performance on unseen datasets]. | https://doi.org/10.1101/2022.11.21.22282586 |
| [27] | PTB-XL | CNN-LSTM, Attention Transformer | Achieved 91.07% accuracy (MI detection) | https://doi.org/10.1109/ACCESS.2022.3220670 |
| [31] | PTB-XL | 1D CNN, LSTM, GRU, Multimodal Fusion, Attention-based models | GRU achieved 79.67% sensitivity and 81.04% specificity. The 1D representation outperformed 2D representations | https://doi.org/10.1016/j.bspc.2024.106141 |
| [32] | PTB-XL | CNN, QRS complex extraction, Entropy-Based Features | Improved performance by adding entropy-based features to raw signals. | https://doi.org/10.3390/s21248174 |
| [41] | PTB-XL | Beat-Level Fusion Network (BLF-Net) | Outperformed state-of-the-art methods in multiclass arrhythmia classification. | https://doi.org/10.1155/2023/1755121 |
| [43] | PTB-XL, various smaller datasets | MELEP, CNNs, RNNs | MELEP effectively predicted transferability; strong correlation with fine-tuning performance (0.6+ correlation) | https://doi.org/10.1007/s41666-024-00168-3 |
| [44] | PTB-XL | Few-Shot Learning (FSL), CNN | FSL achieved 93.2% accuracy for 2-class classification, outperforming softmax-based models. | https://doi.org/10.3390/s22030904 |
| [45] | PTB-XL | AlexNet, LeNet | AlexNet performed better with high classification accuracy for cardiac conditions | https://www.researchgate.net/publication/374061560 accessed on 21 June 2025 |
| [48] | PTB-XL | ECGDeli, Marquette 12SL | Introduced ECGDeli, Marquette 12SL feature sets for enhanced ECG interpretation. | https://doi.org/10.1038/s41597-023-02153-8 |
| [53] | PTB-XL | Multi-Receptive Field CNN (MRF-CNN) | Achieved 0.72 F1 score, 0.93 AUC for 5 superclasses on PTB-XL dataset | https://doi.org/10.1155/2022/8413294 |
| [54] | PTB-XL | CNN, SincNet, Entropy-Based Features | Best performance with convolutional network + entropy features. | https://doi.org/10.3390/e23091121 |
| Subset | Folds | Size |
|---|---|---|
| Train | 1–8 | 15,237 |
| Validation | 9 | 1886 |
| Test | 10 | 1903 |
| Superclass | Description | Subclass | Description |
|---|---|---|---|
| NORM. | Normal ECG | NORM | Normal ECG |
| CD | Conduction Disturbance | LAFB/LPFB | Left anterior/Left posterior fascicular block |
| IRBBB | Incomplete right bundle branch block | ||
| ILBBB | Incomplete left bundle branch block | ||
| CLBBB | Complete left bundle branch block | ||
| CRBBB | Complete right bundle branch block | ||
| _AVB | AV block | ||
| IVCB | Non-specific intraventricular conduction disturbance (block) | ||
| WPW | Wolff–Parkinson–White syndrome | ||
| HYP | Hypertrophy | LVH | Left ventricular hypertrophy |
| RHV | Right ventricular hypertrophy | ||
| LAO/LAE | Left atrial overload/enlargement | ||
| RAO/RAE | Right atrial overload/enlargement | ||
| SEHYP | Septal hypertrophy | ||
| MI | Myocardial Infarction | AMI | Anterior myocardial infarction |
| IMI | Inferior myocardial infarction | ||
| LMI | Lateral myocardial infarction | ||
| PMI | Posterior myocardial infarction | ||
| STTC | ST/T change | ISCA | Ischemic in anterior leads |
| ISCI | Ischemic in inferior leads | ||
| ISC_ | Non-specific ischemic | ||
| STTC | ST-T changes | ||
| NST_ | Non-specific ST changes |
| Technique | Description |
|---|---|
| Adding noise | Adding noise of Gaussian distribution with a mean of 0 and a standard deviation of 0.01. |
| Amplification | Multiply by a gain k = 1 + x, x ∈ [0.001, 0.01]. |
| Attenuation | Multiply by a gain k = 1 − x, x ∈ [0.001, 0.01]. |
| Parameter | Value |
|---|---|
| Convolutional layers for Branch 1 (ECG signals) | 32, 64, 128, 256, 512 |
| Convolutional layer size for Branch 1 | 3 × 1 |
| Max pooling layer size | 2 × 1 |
| Dense layers for Branch 1 | 100, 32 |
| Dense layers for Branch 2 (demographic data) | 100, 64, 32, 16 |
| Drop out size for Branch 1 | 0.4 |
| Drop out size for Branch 2 | 0.4 |
| Dense layers for concatenated branch | 10, 10, num_classes (5, 10, 15) |
| Drop out size for concatenated branch | 0.2 |
| Convolutional layer activation function | ReLU |
| Output activation function | Softmax |
| Loss function | Crossentropy |
| Optimizer | Adam (Learning rate = 0.001) |
| Callbacks | EarlyStopping, Reduce LearningRate, Save CheckPoint |
| Max_Epochs | 60 |
| Batch size | 16 |
| Parameter | Value |
|---|---|
| Convolutional layers for Branch 1 (ECG signals) | 64, 64, 128, 128, 256, 256, 512, 512 |
| Convolutional layer size for Branch 1 | 3 × 1 |
| Max pooling layer size | 2 × 1 |
| Dense layers for Branch 1 | 512, 512 |
| Dense layers for Branch 2 (demographic data) | 100, 64, 32, 16 |
| Drop out size for Branch 1 | 0.5 |
| Drop out size for Branch 2 | 0.4 |
| Dense layers for concatenated branch | 10, 10, num_classes (5, 10, 15) |
| Drop out size for concatenated branch | 0.2 |
| Convolutional layer activation function | ReLU |
| Output activation function | Softmax |
| Loss function | Crossentropy |
| Optimizer | Adam (Learning rate = 0.001) |
| Callbacks | EarlyStopping, Reduce LearningRate, Save CheckPoint |
| Max_Epochs | 60 |
| Batch size | 16 |
| Actual/Predicted | Predicted Positive | Predicted Negative |
|---|---|---|
| Actual Positive | True Positive (TP) | False Negative (FN) |
| Actual Negative | False Positive (FP) | True Negative (TN) |
| Without Patient Data | With Patient Data | |||||||
|---|---|---|---|---|---|---|---|---|
| NORM and MI | NORM and STTC | NORM and CD | NORM and HYP | NORM and MI | NORM and STTC | NORM and CD | NORM and HYP | |
| Precision | 0.9652 | 0.9397 | 0.9195 | 0.9752 | 0.9745 | 0.9410 | 0.9244 | 0.9770 |
| Recall | 0.9652 | 0.9397 | 0.9195 | 0.9752 | 0.9745 | 0.9410 | 0.9244 | 0.9770 |
| F1_score | 0.9279 | 0.9114 | 0.8690 | 0.8765 | 0.9321 | 0.9132 | 0.8794 | 0.8995 |
| Accuracy | 0.9652 | 0.9397 | 0.9195 | 0.9752 | 0.9745 | 0.9410 | 0.9244 | 0.9770 |
| Training time (s) | 260 | 252 | 223 | 179 | 289 | 338 | 309 | 306 |
| Average accuracy | 0.9499 | 0.9542 | ||||||
| NORM and STTC | NORM and AMI | NORM and IMI | NORM and LAFB/ LPFB | NORM and LVH | NORM and IRBBB | NORM and CLBBB | NORM and ISCA | NORM and CRBBB | |
|---|---|---|---|---|---|---|---|---|---|
| Precision | 0.9243 | 0.9838 | 0.9678 | 0.9687 | 0.9855 | 0.9627 | 0.9965 | 0.9925 | 0.9987 |
| Recall | 0.9243 | 0.9838 | 0.9678 | 0.9687 | 0.9855 | 0.9627 | 0.9965 | 0.9925 | 0.9987 |
| F1_score | 0.8456 | 0.9641 | 0.8832 | 0.8990 | 0.9440 | 0.8435 | 0.9538 | 0.9050 | 0.9907 |
| Accuracy | 0.9243 | 0.9838 | 0.9678 | 0.9687 | 0.9855 | 0.9627 | 0.9965 | 0.9925 | 0.9987 |
| Training time (s) | 242 | 227 | 197 | 198 | 176 | 181 | 206 | 184 | 241 |
| Average accuracy | 0.9756 | ||||||||
| NORM and STTC | NORM and AMI | NORM and IMI | NORM and LAFB/LPFB | NORM and LVH | NORM and IRBBB | NORM and CLBBB | NORM and ISCA | NORM and CRBBB | |
|---|---|---|---|---|---|---|---|---|---|
| Precision | 0.9255 | 0.9850 | 0.9747 | 0.9744 | 0.9797 | 0.9708 | 0.9986 | 0.9932 | 0.9986 |
| Recall | 0.9255 | 0.9850 | 0.9747 | 0.9744 | 0.9797 | 0.9708 | 0.9986 | 0.9932 | 0.9986 |
| F1_score | 0.8609 | 0.9647 | 0.9092 | 0.9168 | 0.9227 | 0.8621 | 0.9739 | 0.9122 | 0.9739 |
| Accuracy | 0.9255 | 0.9850 | 0.9747 | 0.9744 | 0.9797 | 0.9708 | 0.9986 | 0.9932 | 0.9986 |
| Training time (s) | 311 | 283 | 224 | 289 | 219 | 276 | 312 | 236 | 310 |
| Average accuracy | 0.9778 | ||||||||
| Without Patient Data | With Patient Data | |
|---|---|---|
| Precision | 0.8821 | 0.8934 |
| Recall | 0.8821 | 0.8934 |
| F1_score | 0.8808 | 0.8929 |
| Accuracy | 0.8821 | 0.8934 |
| Training time (s) | 325 | 495 |
| Metric | Without Patient Data | With Patient Data | ||||
|---|---|---|---|---|---|---|
| 5 Superclasses | 10 Subclasses | 15 Subclasses | 5 Superclasses | 10 Subclasses | 15 Subclasses | |
| Precision | 0.7986 | 0.8145 | 0.7913 | 0.8270 | 0.8341 | 0.8131 |
| Recall | 0.7895 | 0.7613 | 0.7141 | 0.7659 | 0.7523 | 0.6613 |
| F1_score | 0.6804 | 0.6469 | 0.4272 | 0.6757 | 0.6261 | 0.3889 |
| Accuracy | 0.7949 | 0.7835 | 0.7370 | 0.7955 | 0.7960 | 0.7261 |
| Training time (s) | 340 | 304 | 338 | 666 | 626 | 571 |
| Metric | With Patient Data | |||
|---|---|---|---|---|
| NORM and MI | NORM and STTC | NORM and CD | NORM and HYP | |
| Precision | 0.9564 | 0.9202 | 0.9297 | 0.9752 |
| Recall | 0.9564 | 0.9202 | 0.9297 | 0.9752 |
| F1_score | 0.9306 | 0.8771 | 0.8916 | 0.8823 |
| Accuracy | 0.9564 | 0.9202 | 0.9297 | 0.9752 |
| Training time (s) | 398 | 459 | 460 | 446 |
| Average accuracy | 0.9454 | |||
| Metric | NORM and STTC | NORM and AMI | NORM and IMI | NORM and LAFB/ LPFB | NORM and LVH | NORM and IRBBB | NORM and CLBBB | NORM and ISCA | NORM and CRBBB |
|---|---|---|---|---|---|---|---|---|---|
| Precision | 0.9390 | 0.9805 | 0.9731 | 0.9695 | 0.9787 | 0.9634 | 0.9974 | 0.9631 | 0.9994 |
| Recall | 0.9390 | 0.9805 | 0.9731 | 0.9695 | 0.9787 | 0.9634 | 0.9974 | 0.9631 | 0.9994 |
| F1_score | 0.8724 | 0.9533 | 0.9014 | 0.8952 | 0.9136 | 0.8677 | 0.9650 | 0.8952 | 0.9964 |
| Accuracy | 0.9390 | 0.9805 | 0.9731 | 0.9695 | 0.9787 | 0.9634 | 0.9974 | 0.9631 | 0.9994 |
| Training time (s) | 408 | 394 | 608 | 371 | 484 | 486 | 314 | 517 | 785 |
| Average accuracy | 0.9738 | ||||||||
| Precision | Recall | F1_score | Accuracy | Training time (s) |
|---|---|---|---|---|
| 0.8729 | 0.8729 | 0.8701 | 0.8729 | 557 |
| Metric | 5 Superclasses | 10 Subclasses | 15 Subclasses |
|---|---|---|---|
| Precision | 0.7685 | 0.7884 | 0.7179 |
| Recall | 0.7628 | 0.7515 | 0.6855 |
| F1_score | 0.7656 | 0.7695 | 0.7013 |
| Accuracy | 0.7653 | 0.7685 | 0.6929 |
| Training time (s) | 1024 | 1005 | 1001 |
| Ref. | Year | Model | Average Accuracy | |||||
|---|---|---|---|---|---|---|---|---|
| Binary | Multiclass | |||||||
| Normal and Abnormal | Normal vs. One Superclass | Normal vs. One Subclass | 5 Classes | 10 Classes | 15 Classes | |||
| [27] | 2022 | CNN-LSTM | 90.94% | 74.33% | ||||
| [31] | 2024 | GRU | 80.69% | |||||
| [32] | 2021 | QRS entropy+ Raw signal | 89.8% | 75.8% | ||||
| [44] | 2022 | FSL + XGBoost | 88.9% | 75.2% | ||||
| [54] | 2021 | CNN + Entropy features. | 89.2% | 76.5% | ||||
| [62] | 2023 | CNN | 70.11% | |||||
| Proposed | 2025 | VGG (Incorporating patient data) | 87.29% | 94.54% | 97.38% | 76.53% | 76.85% | 69.29% |
| Proposed | 2025 | CNN (Incorporating patient data) | 89.34% | 95.42% | 97.78% | 79.55% | 79.60% | 72.61% |
| Metric | NORM and MI | NORM and STTC | NORM and CD | NORM and HYP |
|---|---|---|---|---|
| Precision | 0.9499 | 0.9246 | 0.9251 | 0.9714 |
| Recall | 0.9499 | 0.9246 | 0.9251 | 0.9714 |
| F1_score | 0.9175 | 0.8977 | 0.8874 | 0.8846 |
| Accuracy | 0.9499 | 0.9246 | 0.9251 | 0.9714 |
| Training time (s) | 294 | 235 | 257 | 274 |
| Average accuracy | 0.9428 | |||
| Metric | NORM and STTC | NORM and AMI | NORM and IMI | NORM and LAFB/LPFB | NORM and LVH | NORM and IRBBB | NORM and CLBBB | NORM and ISCA | NORM and CRBBB |
|---|---|---|---|---|---|---|---|---|---|
| Precision | 0.9267 | 0.9883 | 0.9725 | 0.9711 | 0.9763 | 0.9641 | 1.0000 | 0.9926 | 1.0000 |
| Recall | 0.9267 | 0.9883 | 0.9725 | 0.9711 | 0.9763 | 0.9641 | 1.0000 | 0.9926 | 1.0000 |
| F1_score | 0.8669 | 0.9728 | 0.9035 | 0.8972 | 0.9159 | 0.8693 | 0.9839 | 0.9060 | 1.0000 |
| Accuracy | 0.9267 | 0.9883 | 0.9725 | 0.9711 | 0.9763 | 0.9641 | 1.0000 | 0.9926 | 1.0000 |
| Training time (s) | 277 | 273 | 278 | 294 | 266 | 297 | 381 | 276 | 288 |
| Average accuracy | 0.9768 | ||||||||
| Precision | Recall | F1_score | Accuracy | Training time (s) |
|---|---|---|---|---|
| 0.8893 | 0.8893 | 0.8820 | 0.8893 | 322 |
| Metric | 5 Superclasses | 10 Subclasses | 15 Subclasses |
|---|---|---|---|
| Precision | 0.7732 | 0.7906 | 0.7220 |
| Recall | 0.7666 | 0.7867 | 0.7144 |
| F1_score | 0.6460 | 0.6499 | 0.4370 |
| Accuracy | 0.7689 | 0.7889 | 0.7147 |
| Training time (s) | 631 | 1294 | 1868 |
| Dataset | Binary Classification | Multiclass Classification | ||||
|---|---|---|---|---|---|---|
| Binary (Normal vs. Abnormal) | Normal vs. One of 5-Super Classes | Normal vs. One of 10-Subclasses | 5 Classes | 10 Classes | 15 Classes | |
| PTB- XL dataset without augmentation | 0.8934 | 0.9542 | 0.9778 | 0.7955 | 0.7960 | 0.7261 |
| PTB- XL dataset with augmentation | 0.8893 | 0.9428 | 0.9768 | 0.7689 | 0.7889 | 0.7147 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atwa, A.E.M.; Atlam, E.-S.; Ahmed, A.; Atwa, M.A.; Abdelrahim, E.M.; Siam, A.I. Interpretable Deep Learning Models for Arrhythmia Classification Based on ECG Signals Using PTB-X Dataset. Diagnostics 2025, 15, 1950. https://doi.org/10.3390/diagnostics15151950
Atwa AEM, Atlam E-S, Ahmed A, Atwa MA, Abdelrahim EM, Siam AI. Interpretable Deep Learning Models for Arrhythmia Classification Based on ECG Signals Using PTB-X Dataset. Diagnostics. 2025; 15(15):1950. https://doi.org/10.3390/diagnostics15151950
Chicago/Turabian StyleAtwa, Ahmed E. Mansour, El-Sayed Atlam, Ali Ahmed, Mohamed Ahmed Atwa, Elsaid Md. Abdelrahim, and Ali I. Siam. 2025. "Interpretable Deep Learning Models for Arrhythmia Classification Based on ECG Signals Using PTB-X Dataset" Diagnostics 15, no. 15: 1950. https://doi.org/10.3390/diagnostics15151950
APA StyleAtwa, A. E. M., Atlam, E.-S., Ahmed, A., Atwa, M. A., Abdelrahim, E. M., & Siam, A. I. (2025). Interpretable Deep Learning Models for Arrhythmia Classification Based on ECG Signals Using PTB-X Dataset. Diagnostics, 15(15), 1950. https://doi.org/10.3390/diagnostics15151950






