An Exploration of Discrepant Recalls Between AI and Human Readers of Malignant Lesions in Digital Mammography Screening
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Retrospective Screening Data
2.3. Ground Truth
2.4. Artificial Intelligence
2.5. Case Categorization
2.6. Online Reader Study
2.6.1. Facilitating Software
2.6.2. Study Readers
2.7. Statistical Analysis
2.7.1. Discriminative Ability of AI
2.7.2. Breast Composition
2.7.3. Lesion Visibility
2.7.4. PoM Scores
2.7.5. Visible Mass Characteristics
2.7.6. Lesion Size
3. Results
3.1. Discriminative Ability of AI
3.2. Selection of Discrepant Cases
3.3. Reader Study Results
Lesion Size
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van der Waal, D.; Verbeek, A.; den Heeten, G.; Ripping, T.; Tjan-Heijnen, V.; Broeders, M. Breast cancer diagnosis and death in The Netherlands: A changing burden. Eur. J. Public Health 2014, 25, 320–324. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, Z.; Yang, Z.; Xu, F.; Lu, H.; Zhu, Z.; Shi, W.; Jiang, J.; Yao, P.P.; Zhu, H.P. Risk factors and preventions of breast cancer. Int. J. Biol. Sci. 2017, 13, 1387–1397. [Google Scholar] [CrossRef]
- Hakema, M.; Coleman, P.; Alexe, D.M.; Auvine, A. Cancer screening: Evidence and practice in Europe 2008. Eur. J. Cancer 2008, 44, 404–413. [Google Scholar] [CrossRef]
- Weerakkody, Y.; Manning, T.; Lemos, P. Breast Imaging-Reporting and Data System (BI-RADS). Radiopaedia. Available online: https://radiopaedia.org/articles/breast-imaging-reporting-and-data-system-bi-rads?lang=us (accessed on 11 June 2025).
- American College of Radiology. ACR BI-RADS® Atlas: Breast Imaging Reporting and Data System, 5th ed.; American College of Radiology: Reston, VA, USA, 2020. [Google Scholar]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, E19. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Ruiz, A.; Lång, K.; Gubern-Merida, A.; Broeders, M.; Gennaro, G.; Clauser, P.; Helbich, T.H.; Chevalier, M.; Tan, T.; Mertelmeier, T.; et al. Stand-Alone Artificial Intelligence for Breast Cancer Detection in Mammography: Comparison with 101 Radiologists. J. Natl. Cancer Inst. 2019, 111, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Phang, J.; Park, J.; Shen, Y.; Huang, Z.; Zorin, M.; Jastrzębski, S.; Févry, T.; Katsnelson, J.; Kim, E.; et al. Deep neural networks improve radiologists’ performance in breast cancer screening. IEEE Trans. Med. Imaging. 2019, 39, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Sechopoulos, I.; Mann, R.M. Stand-alone artificial intelligence—The future of breast cancer screening? Breast 2020, 49, 254–260. [Google Scholar] [CrossRef]
- Kristina Lång, K.; Joseffson, V.; Larsson, A.M.; Larsson, S.; Högberg, C.; Sartor, H.; Hofvind, S.; Andersson, I.; Rosso, A. Artificial intelligence-supported screen reading versus standard double reading in the Mammography Screening with Artificial Intelligence trial (MASAI). Lancet Oncol. 2023, 24, P936–P944. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, A.; Lång, K.; Gubern-Merida, A.; Teuwen, J.; Broeders, M.; Gennaro, G.; Clauser, P.; Helbich, T.H.; Chevalier, M.; Mertelmeier, T.; et al. Can we reduce the workload of mammographic screening by automatic identification of normal exams with artificial intelligence? A feasibility study. Eur. Radiol. 2019, 29, 4825–4832. [Google Scholar] [CrossRef]
- Yala, A.; Schuster, T.; Miles, R.; Barzilay, R.; Lehman, C. A deep learning model to triage screening mammograms: A simulation study. Radiology 2019, 1, 38–46. [Google Scholar] [CrossRef]
- Dembrower, K.; Wåhlin, E.; Liu, Y.; Salim, M.; Smith, K.; Lindholm, P.; Eklund, M.; Strand, F. Effect of artificial intelligence-based triaging of breast cancer screening mammograms on cancer detection and radiologist workload: A retrospective simulation study. Lancet Digit. Health 2020, 2, e468–e474. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, A.; Krupinski, E.; Mordang, J.J.; Schilling, K.; Heywang-Köbrunner, S.H.; Sechopoulos, I.; Mann, R.M. Detection of Breast Cancer with Mammography: Effect of an Artificial Intelligence Support System. Radiology 2019, 290, 305–314. [Google Scholar] [CrossRef] [PubMed]
- van Winkel, S.L.; Rodríguez-Ruiz, A.; Appelman, L.; Gubern-Mérida, A.; Karssemeijer, N.; Teuwen, J.; Wanders, A.J.; Sechopoulos, I.; Mann, R.M. Impact of artificial intelligence support on accuracy and reading time in breast tomosynthesis image interpretation: A multi-reader multi-case study. Eur. Radiol. 2021, 31, 8682–8691. [Google Scholar] [CrossRef]
- Sechopoulos, I.; Teuwen, J.; Mann, R. Artificial intelligence for breast cancer detection in mammography and digital breast tomosynthesis: State of the art. Semin. Cancer Biol. 2021, 72, 214–225. [Google Scholar] [CrossRef]
- Hupse, R.; Samulsky, M.; Lobbes, M.; Heeten Ad Imhoff-Tas, N.K.M.W.; Beijerick, D.; Pijnappel, R.; Boetes, C.; Karssemeijer, N. Stand-alone computer-aided detection compared to radiologists’ performance for the detection of mamographic masses. Eur. Radiol. 2013, 23, 93–100. [Google Scholar] [CrossRef]
- Gennaro, G.; Hendrick, R.; Ruppel, P.; Chersevani, R.; di Maggio, C.; La Grassa, M.; Pescarini, L.; Polico, I.; Proietti, A.; Baldan, E.; et al. Performance comparison of single view digital breast thomosynthesis plus single view digital mammography with tweo-view digital mammography. Eur. Radiol. 2013, 23, 664–672. [Google Scholar] [CrossRef]
- Wallis, M.G.; Moa, E.; Zanca, F.; Leifland, K.; Danielsson, M. Two-view and single-view tomosynthesis versus full-filed digital mammography: High- resolotion x-ray imaging observer study. Eur. Radiol. 2012, 262, 788–796. [Google Scholar] [CrossRef]
- Visser, R.; Veldkamp, W.J.; Beijerinck, D.; Bun, P.A.; Deurenberg, J.J.; Imhof-Tas, M.W.; Schuur, K.H.; Snoeren, M.M.; den Heeten, G.J.; Karssemeijer, N.; et al. Increase in perceived case suspiciousness due to local contrast optimisation in digital screening mammography. Eur. Radiol. 2012, 22, 908–914. [Google Scholar] [CrossRef]
- Garayoa, J.; Chevalier, M.; Castillio, M.; Mahillo-Fernandez, I.; Ouahabi, N.A.E.; Estrada, C.; Tejerina, A.; Benitez, O.; Valverde, J. Diagnostic value of the stand-alone synthetic image in digital breast tomosynthesis examinations. Eur. Radiol. 2018, 28, 565–572. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, A.; Gubern-Merida, A.; Imhof-Tas, M.; Lardenoije, S.; Wanders, A.J.T.; Andersson, I.; Zackrisson, S.; Lång, K.; Dustler, M.; Karssemeijer, N.; et al. One-view digital breast tomosynthesis as a atand-alone modality for breast cancer detection: Do we need more? Eur. Radiol. 2018, 28, 1938–1948. [Google Scholar] [CrossRef]
- Clauser, P.; Baltzer, P.; Kapetas, P.; Woitek, R.; Weber, M.; Leon, F.; Bernathova, M.; Helbich, T.H. Synthetic 2d mammography can replace digital mammography as an adjuct to wide-angle digital breast tomosynthesis. Investig. Radiol. 2019, 54, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Perry, N.; Broeders, M.; de Wolf, C.; Törnberg, S.; Holland, R.; von Karsa, L. European Guidelines for quality assurance in breast cancer screening and diagnosis. Ann. Oncol. 2008, 19, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Nikitin, V.; Filatov, A.; Bagotskaya, N.; Kil, I.; Lossev, I.; Losseva, N. Improvement in ROC curves of readers with next generation of mammography CAD. In Proceedings of the European Congress of Radiology (ECR) 2014, Vienna, Austria, 6–10 March 2014. [Google Scholar] [CrossRef]
- Mansour, S.; Soliman, S.; Kansakar, A.; Marey, A.; Hunold, C.; Hanafy, M.M. Strengths and challenges of the artificial intelligence in the assessment of dense breasts. BJR Open 2022, 4, 20220018. [Google Scholar] [CrossRef]
- Meakin, J.; Gerke, P.K.; Kerkstra, S.; Groeneveld, M.; van Leeuwen, K.; Mickan, A.; Overkamp, M.; van Run, C.; van Zeeland, H.; van Ginneken, B. Grand-Challenge.org [Software], Version v2023.05; Zenodo: Geneva, Switzerland, 2023. [CrossRef]
- Landis, J.R.; Koch, G.C. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Lamb, L.; Mohallem, F.; Verma, R.; Seely, J. Missed breast cancer: Effects of subconscious bias and lesion characteristics. Radiographics 2020, 40, 941–960. [Google Scholar] [CrossRef]
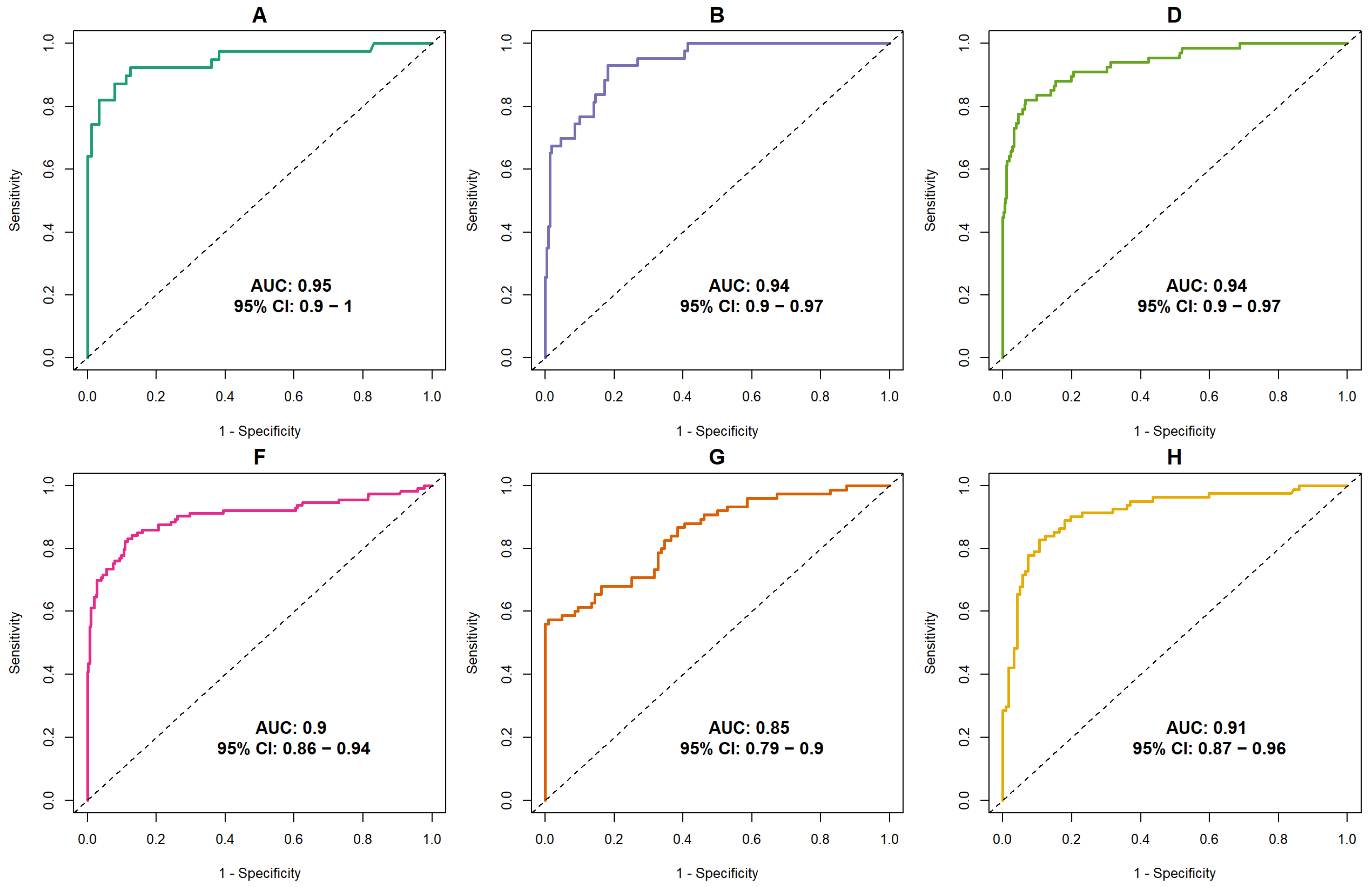
| Dataset (Year) | Vendor (s) | Population | Exam Type | Total Exams | Cancer, n (%) | Benign Lesion | Normal | Readers | Experience (Years) | Mean Recall Rate (%) Human Readers | Scoring Scale |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A (2011) | GE Sectra | 40–80 (mean of 56) years, Screening (86), Clinical (43) | Bilateral no priors | 129 | 40 (31) | 23 | 66 | 14 | 3–25 (mean of 10) | 60 | BI-RADS |
| B (2012) | GE | 51–86 (mean of 60) years, Screening | Bilateral + priors | 263 | 43 (16) | 110 | 110 | 6 | 1–34 | 38 | PoM |
| D (2013) | GE | >40 years, 50–74 years, Screening + Clinical | Unilateral + priors | 469 | 68 (15) | 200 | 201 | 6 | 5–30 | 43 | BI-RADS |
| F (2017) | Hologic | 34–92 (mean of 55) y, Screening + Clinical | Bilateral + priors | 585 | 113 (19) | 160 | 313 | 3 | 10–20 | 41 | BI-RADS |
| G (2017) | Siemens | 30–88 (mean of 52) years, Screening + Clinical | Unilateral + priors | 179 | 75 (42) | 49 | 55 | 6 | 3–44 (mean of 22) | 47 | PoM BI-RADS |
| H (2018) | Siemens | 36–84 (mean of 56) years, Screening + Clinical | Bilateral + priors | 204 | 82 (40) | 43 | 80 | 4 | >5 | 49 | BI-RADS |
| Lesion Type | AI-Missed (n = 15) | Human-Missed (n = 11) | p-Value | ||
|---|---|---|---|---|---|
| Breast Density | AI-Scored Observations, n (%) | Human-Scored Observations, n (%) | AI-Scored Observations, n (%) | Human-Scored Observations, n (%) | |
| A | 2 (13) | 24/210 (11) | 0 (-) | 0/154 (-) | |
| B | 8 (53) | 98/210 (47) | 3 (27) | 43/154 (28) | |
| C | 3 (20) | 67/210 (32) | 4 (36) | 62/154 (40) | |
| D | 2 (13) | 21/210 (10) | 4 (36) | 49/154 (32) | |
| Fleiss’ Kappa (human-scored breast density) | 0.40 (95%CI: 0.36–0.44; p < 0.0001) | 0.47 (95%CI: 0.43–0.52; p < 0.0001) | |||
| Lesion Type | Observations | Observations | |||
| Architectural distortion | 26/210 (12.4) | 24/154 (15.6) | ≈1 Adj. p ≈ 1 | ||
| Calcifications | 38/210 (18.1) | 42/154 (27.3) | 0.396 Adj. p ≈ 1 | ||
| Mass | 85/210 (40.5) | 64/154 (41.6) | 0.211 Adj. p = 0.845 | ||
| No visible lesion | 61/210 (29.0) | 24/154 (15.6) | 0.001 Adj. p = 0.003 | ||
| Kappa (lesion type) | 0.27 (95%CI: 0.24–0.29; p < 0.0001) | 0.26 (95%CI: 0.24–0.28; p < 0.0001) | |||
| Lesion Visibility | Observations | Observations | |||
| Slight | 56/210 (26.7) | 42/154 (27.3) | 0.185 Adj. p = 0.738 | ||
| Moderate | 51/210 (24.3) | 45/154 (29.2) | 0.598 Adj. p ≈ 1 | ||
| Very | 36/210 (17.1) | 40/154 (26.0) | ≈1 Adj. p ≈ 1 | ||
| Not Applicable | 67/210 (31.9) | 27/154 (17.5) | <0.001 Adj. p < 0.001 | ||
| Kappa (visibility) | 0.12 (95%CI: 0.107–0.129; p < 0.0001) | 0.15 (0.95%CI: 0.14–0.16; p < 0.0001) | |||
| Probability of Malignancy (PoM) | Observations | Observations | |||
| Average | 25.8% | 33.8% | |||
| SD | 27.0% | 25.8% | |||
| Median | 15.0% | 30.0% | 0.002 | ||
| IQR | 35.0% | 40.0% | |||
| Min | 0.0% | 0.0% | |||
| Max | 100.0% | 96.0% | |||
| ICC | 0.40 (95%CI: 0.24–0.64; p < 0.0001) | 0.15 (95%CI: 0.05–0.41; p < 0.001) | |||
| Lesion Shape | Observations | Observations | |||
| Round | 27/210 (12.8) | 17/154 (11.0) | 0.522 Adj. p ≈ 1 | ||
| Oval | 16/210 (7.6) | 16/154 (10.4) | 1.000 Adj. p ≈ 1 | ||
| Irregular | 49/210 (23.3) | 40/154 (26.0) | 0.456 Adj. p ≈ 1 | ||
| Not Applicable | 118/210 (56.2) | 81/154 (52.6) | 0.002 Adj. p ≈ 0.006 | ||
| Kappa (shape) | 0.27 (95%CI: 0.25-0.30; p < 0.0001) | 0.18 (95%CI: 0.16–0.19; p < 0.0001) | |||
| Lesion Density | Observations | Observations | |||
| Fat | 1/210 (0.5) | 1/154 (0.6) | 1.000 Adj. p ≈ 1 | ||
| Low | 10/210 (4.8) | 5/154 (3.2) | 0.423 Adj. p ≈ 1 | ||
| Equal | 68/210 (32.4) | 32/154 (20.8) | 0.002 Adj. p = 0.009 | ||
| High | 31/210 (14.8) | 44/154 (28.6) | 0.166 Adj. p = 0.829 | ||
| Not Applicable | 100/210 (47.6) | 72/154 (46.8) | 0.007 Adj. p = 0.037 | ||
| Kappa (lesion density) | 0.20 (0.95%CI: 0.18, 0.21; p < 0.0001) | 0.26 (0.95%CI: 0.23–0.28; p < 0.0001) | |||
| Margins of a Visible Mass | Observations | Observations | |||
| Circumscribed | 19/210 (9.0) | 12/154 (7.8) | 0.845 Adj. p ≈ 1 | ||
| Obscured | 14/210 (6.7) | 14/154 (9.1) | 1.000 Adj. p ≈ 1 | ||
| Microlobulated | 13/210 (6.2) | 5/154 (3.2) | 0.099 Adj. p = 0.594 | ||
| Indistinct | 26/210 (12.4) | 29/154 (18.8) | 0.784 Adj. p ≈ 1 | ||
| Spiculated | 26/210 (12.4) | 16/154 (10.4) | 0.165 Adj. p = 0.989 | ||
| Not Applicable | 112/210 (53.3) | 78/154 (50.6) | 0.003 Adj. p ≈ 1 | ||
| Kappa (margins) | 0.29 (95%CI: 0.26–0.31; p < 0.0001) | 0.20 (0.95%CI: 0.18–0.21; p < 0.0001) | |||
| Presence and Type of Calcifications | Observations | Observations | |||
| None | 87/210 (41.4) | 38/154 (25.0) | <0.001 Adj. p < 0.001 | ||
| Typically benign | 94/210 (44.8) | 72/154 (46.8) | 0.091 Adj. p = 0.272 | ||
| Typically suspected of malignancy | 29/210 (13.8) | 44/154 (28.6) | 0.382 Adj. p ≈ 1 | ||
| Kappa (calcifications) | 0.30 (95%CI: 0.27–0.32; p < 0.0001) | 0.22 (0.95%CI: 0.20–0.25; p < 0.0001) | |||
| Likelihood of Malignancy | Observations | Observations | |||
| Unlikely | 61/210 (29.0) | 29/154 (18.8) | 0.003 Adj. p = 0.012 | ||
| Uncertain | 87/210 (41.4) | 62/154 (40.3) | 0.068 Adj. p = 0.271 | ||
| Very likely | 41/210 (19.5) | 50/154 (32.5) | 0.828 Adj. p ≈ 1 | ||
| Not Applicable | 21/210 (10.0) | 13/154 (8.4) | 0.137 Adj. p = 0.550 | ||
| Kappa (likelihood of malignancy) | 0.16 (95%CI: 0.15–0.18; p < 0.0001) | 0.05 (95%CI: 0.04–0.05; p = 0.016) | |||
| Subset | Lesion Size [mm] | |||||
|---|---|---|---|---|---|---|
| Median | Mean | SD | Min. | Max. | IQR | |
| All CC views | 12.0 | 16.9 | 14.6 | 3.0 | 53.0 | 8.0–19.0 |
| All MLO views | 11.0 | 16.5 | 15.7 | 2.4 | 64.0 | 7.0–16.0 |
| All views combined | 11.5 | 16.7 | 15.0 | 2.4 | 64.0 | 8.0–18.8 |
| AI-missed CC views | 9.5 | 9.7 | 4.9 | 3.0 | 19.0 | 6.5–12.0 |
| AI-missed MLO views | 9.0 | 9.8 | 5.0 | 2.4 | 21.0 | 7.5–12.0 |
| All AI-missed views combined | 9.0 | 9.7 | 4.9 | 2.4 | 21.0 | 6.5–12.0 |
| Human-missed CC views | 21.0 | 25.9 | 17.9 | 4.0 | 53.0 | 12.0–39.5 |
| Human-missed MLO views | 22.0 | 26.5 | 20.8 | 4.0 | 64.0 | 9.0–43.0 |
| All human-missed views combined | 21.0 | 26.2 | 19.3 | 4.0 | 64.0 | 10.5–41.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Winkel, S.L.; Sechopoulos, I.; Rodríguez-Ruiz, A.; Veldkamp, W.J.H.; Gennaro, G.; Chevalier, M.; Helbich, T.H.; Zhang, T.; Wallis, M.G.; Mann, R.M. An Exploration of Discrepant Recalls Between AI and Human Readers of Malignant Lesions in Digital Mammography Screening. Diagnostics 2025, 15, 1566. https://doi.org/10.3390/diagnostics15121566
van Winkel SL, Sechopoulos I, Rodríguez-Ruiz A, Veldkamp WJH, Gennaro G, Chevalier M, Helbich TH, Zhang T, Wallis MG, Mann RM. An Exploration of Discrepant Recalls Between AI and Human Readers of Malignant Lesions in Digital Mammography Screening. Diagnostics. 2025; 15(12):1566. https://doi.org/10.3390/diagnostics15121566
Chicago/Turabian Stylevan Winkel, Suzanne L., Ioannis Sechopoulos, Alejandro Rodríguez-Ruiz, Wouter J. H. Veldkamp, Gisella Gennaro, Margarita Chevalier, Thomas H. Helbich, Tianyu Zhang, Matthew G. Wallis, and Ritse M. Mann. 2025. "An Exploration of Discrepant Recalls Between AI and Human Readers of Malignant Lesions in Digital Mammography Screening" Diagnostics 15, no. 12: 1566. https://doi.org/10.3390/diagnostics15121566
APA Stylevan Winkel, S. L., Sechopoulos, I., Rodríguez-Ruiz, A., Veldkamp, W. J. H., Gennaro, G., Chevalier, M., Helbich, T. H., Zhang, T., Wallis, M. G., & Mann, R. M. (2025). An Exploration of Discrepant Recalls Between AI and Human Readers of Malignant Lesions in Digital Mammography Screening. Diagnostics, 15(12), 1566. https://doi.org/10.3390/diagnostics15121566







