Machine and Deep Learning for the Diagnosis, Prognosis, and Treatment of Cervical Cancer: A Scoping Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Study Selection
2.3. Eligibility Criteria
2.4. Study Design
2.5. Language
2.6. Publication Date
2.7. Exclusion Criteria
2.8. Data Extraction
2.9. Outcome Measurement
2.10. Quality Assessment
2.11. Synthesis of Results
2.12. Statistical Analysis
3. Results
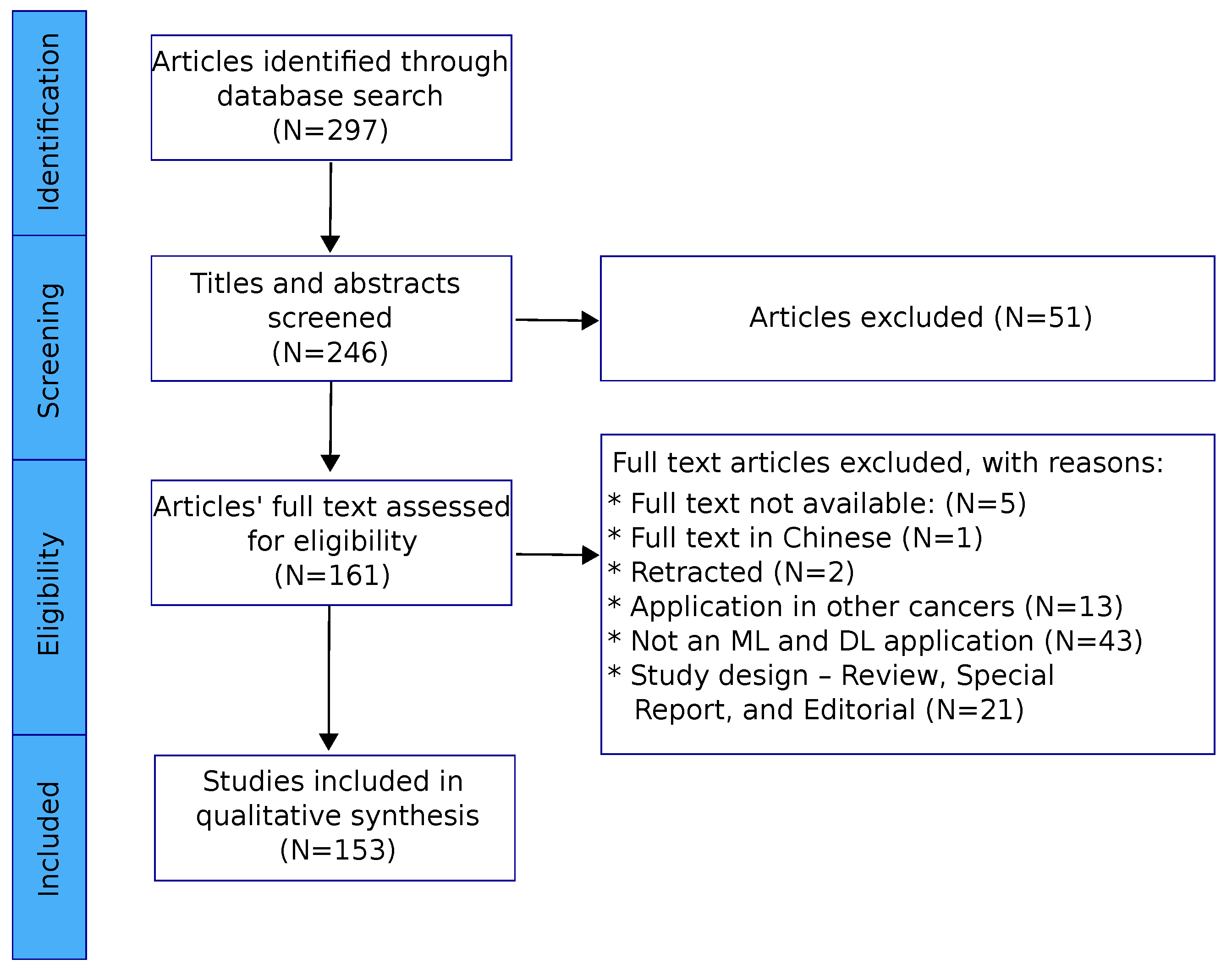
3.1. Included Studies
3.2. Quality Assessment of Included Studies
3.3. Clinical Applications of ML and DL in Cervical Cancer
3.4. Prediction Tasks in CC
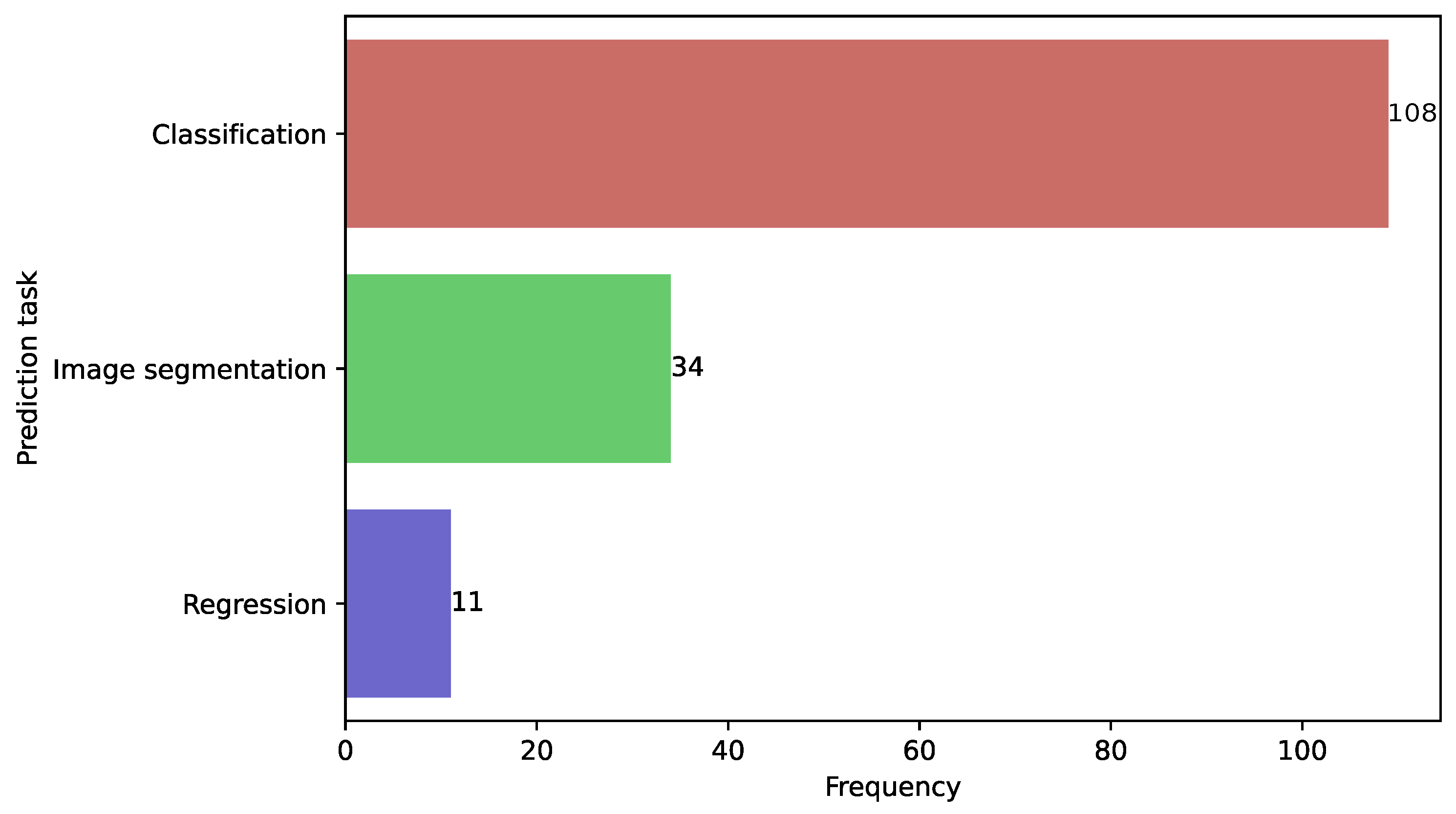
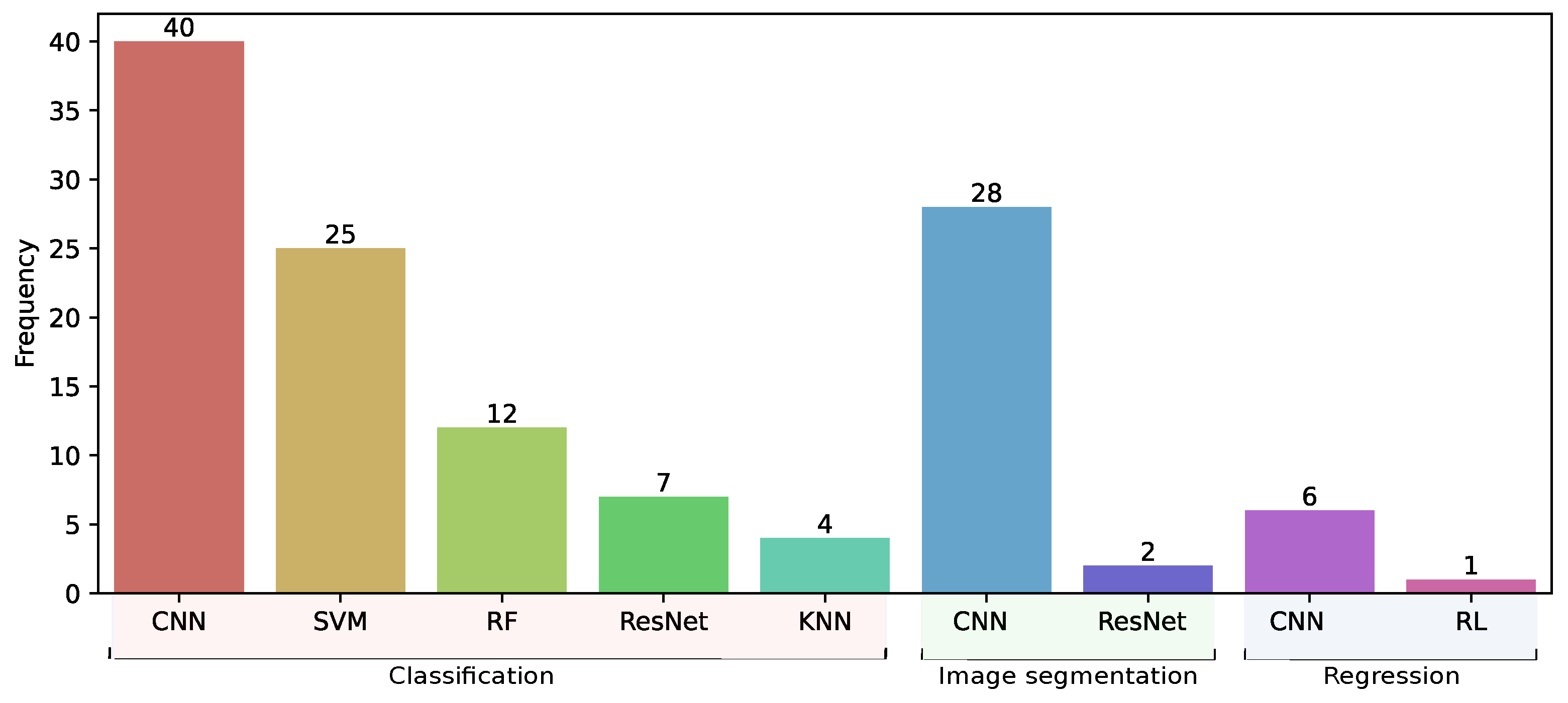
3.5. Models Trained in Classification, Segmentation, and Regression Tasks
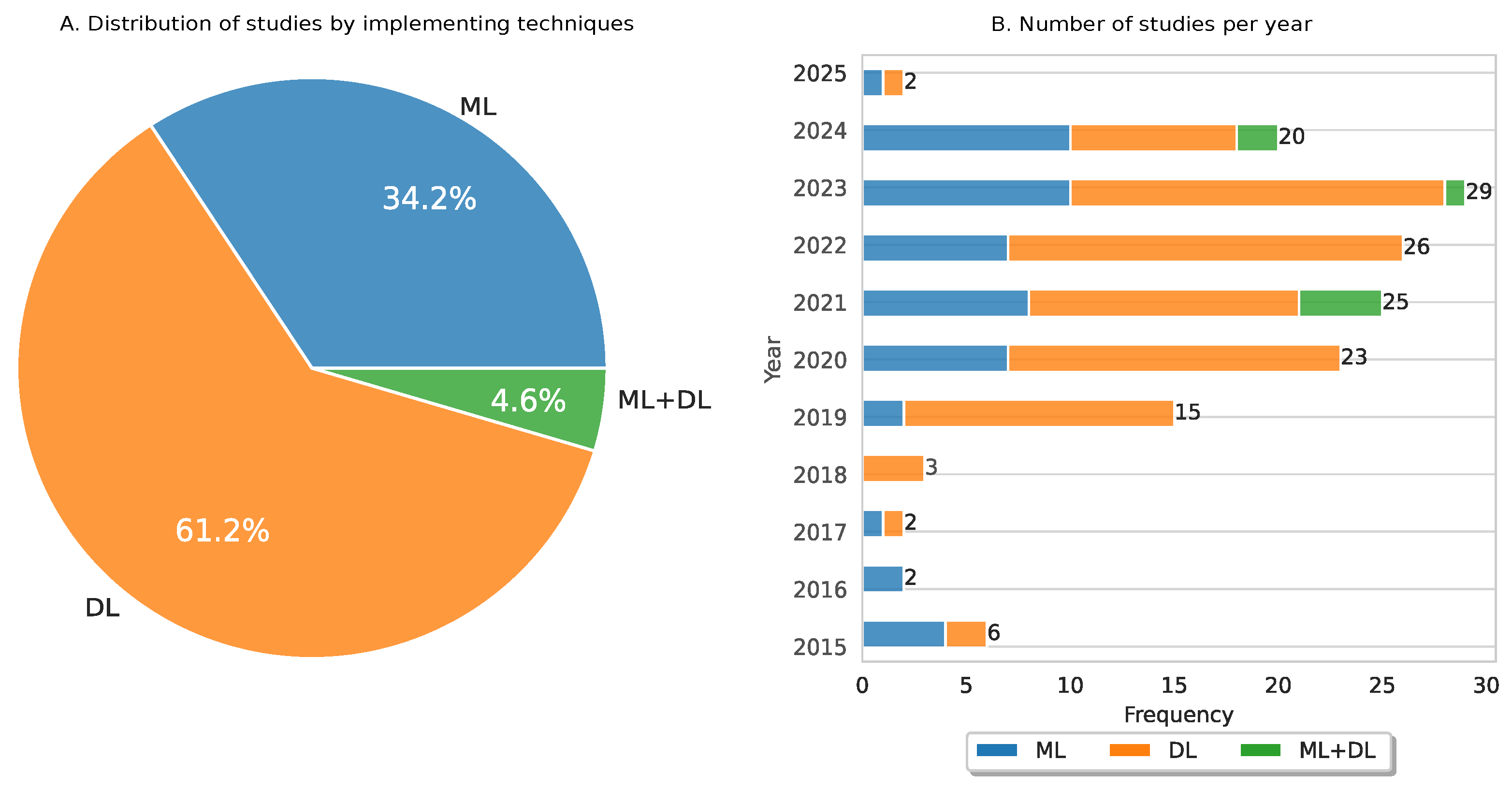
3.6. Temporal Analysis of ML and DL Implemented in CC
3.7. Analysis of ML-Based Models Implemented in CC
3.8. Analysis of DL-Based Models Implemented in CC
3.9. Analysis of Fusion of ML and DL Models Implemented in CC
3.10. Evaluation Metrics for ML and DL in CC
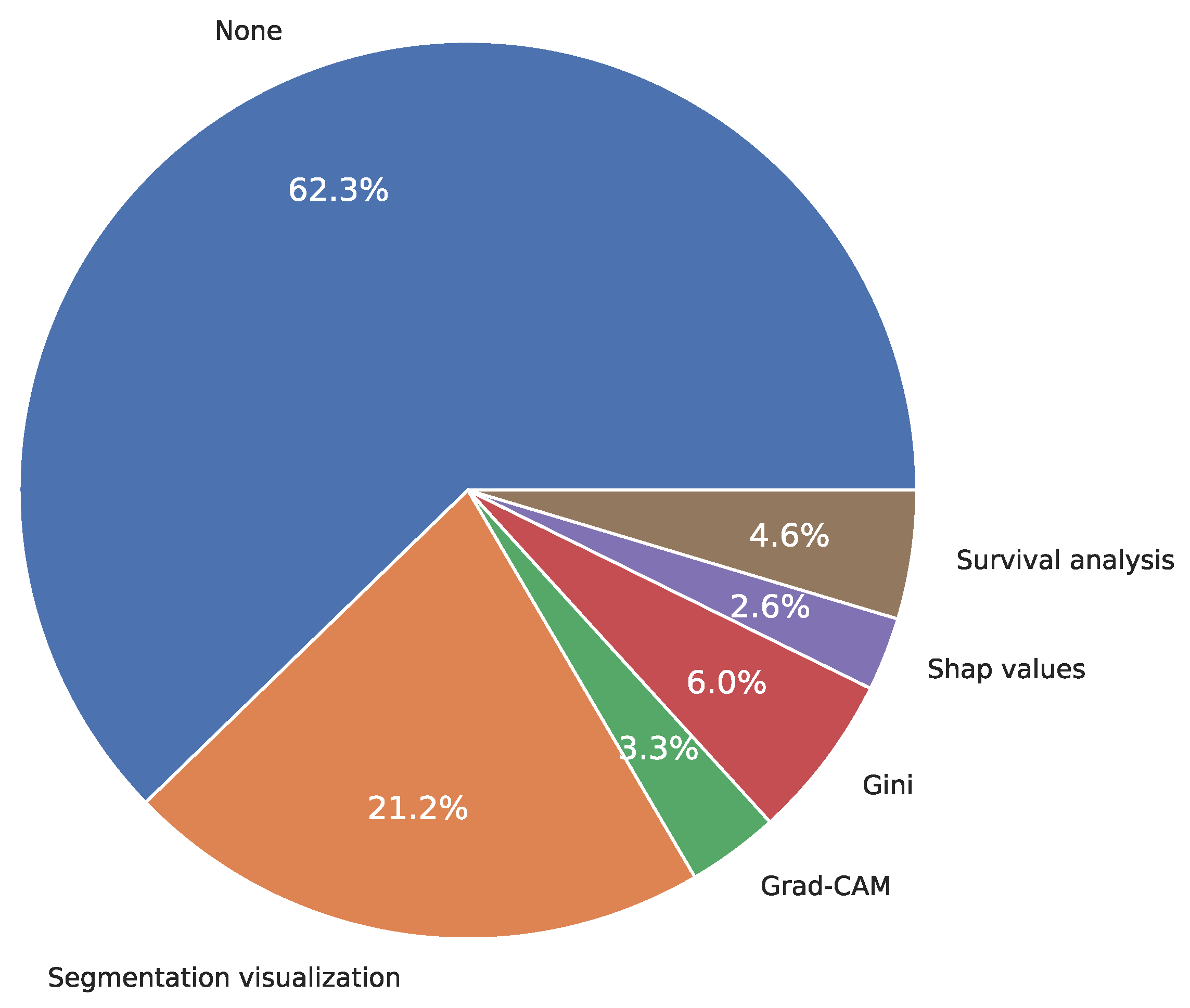
3.11. Explainability
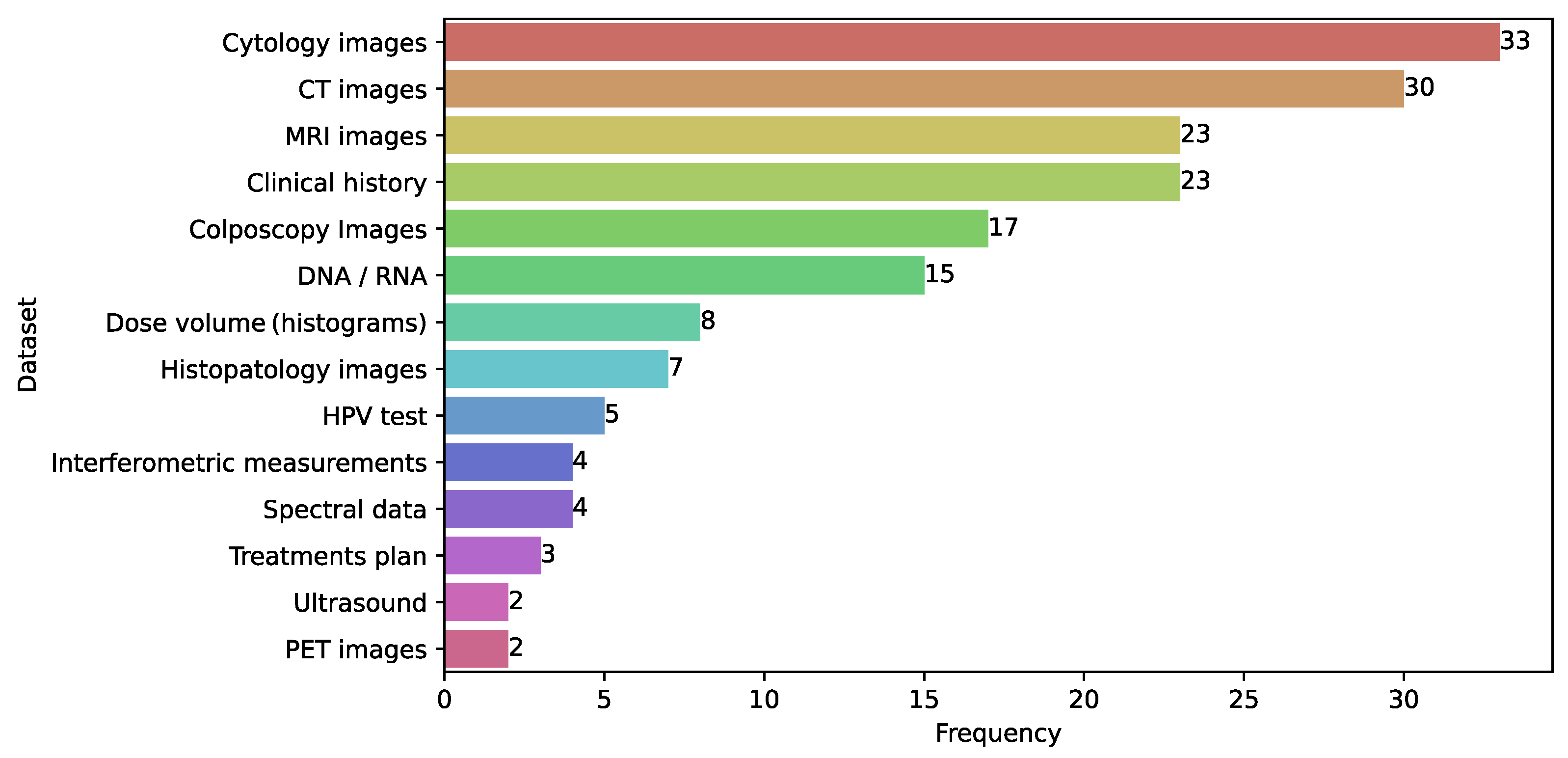
3.12. Databases
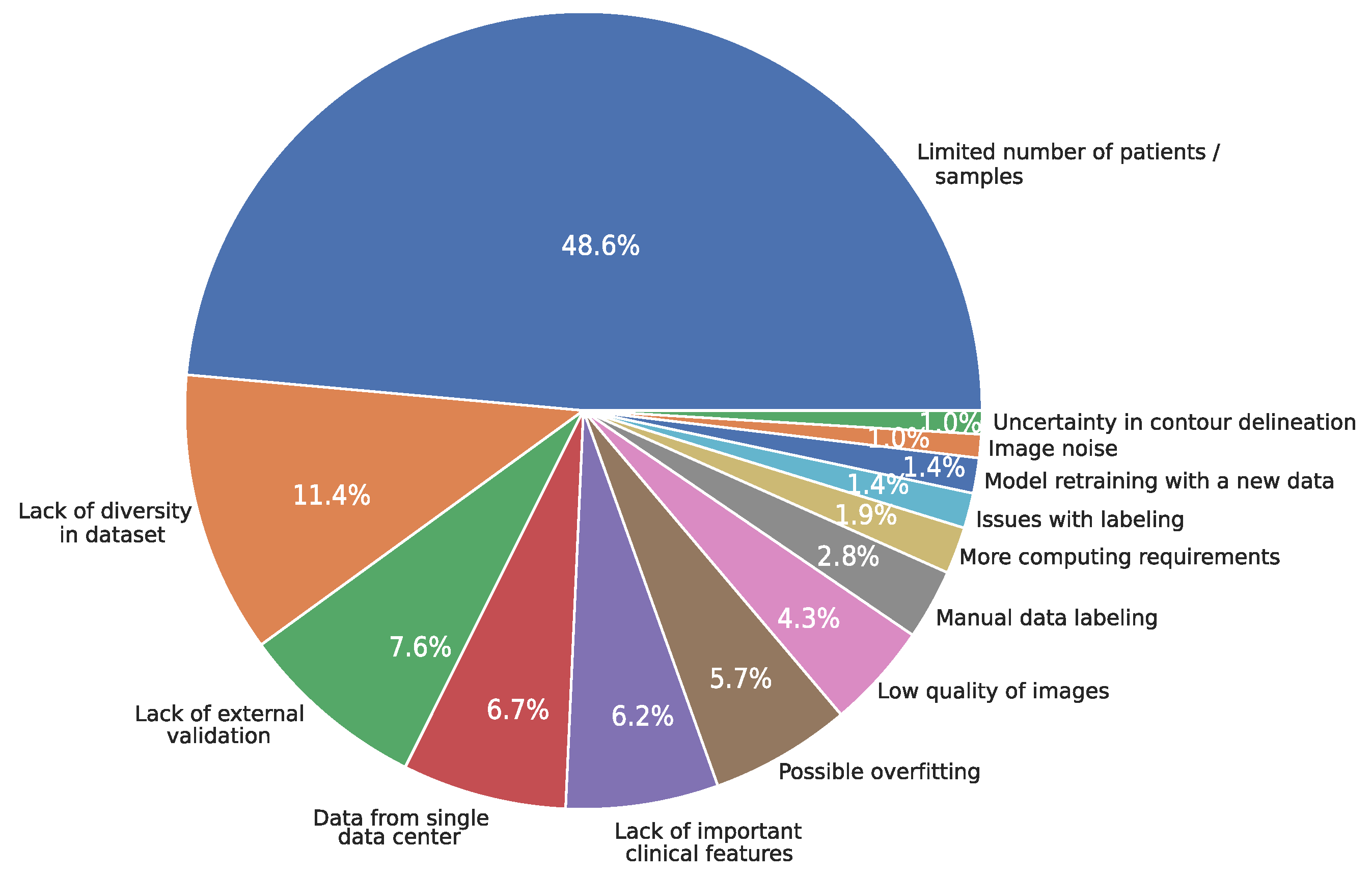
3.13. Limitations
3.14. Reproducibility
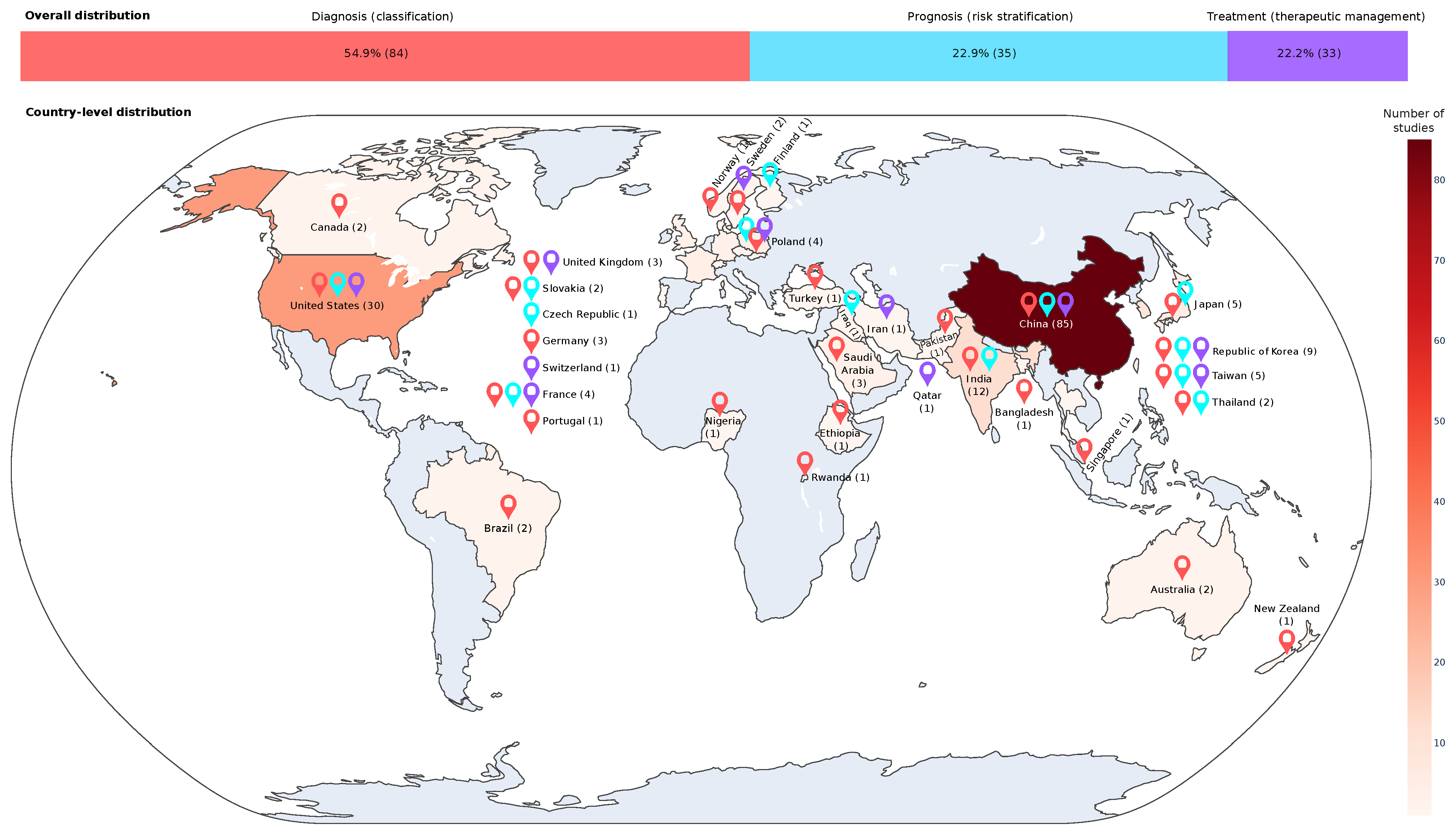
3.15. Distribution of Publications by Country
3.16. Study Design Characteristics
3.17. Study Population
3.18. Medical Specialist Involvement in Predictive Model Development
4. Discussion
4.1. Computational Challenges of Using ML and DL in CC
4.1.1. Data Availability
4.1.2. Data Leakage
4.1.3. Limited External Validation
4.1.4. Limited Evaluation of Model Performance
4.1.5. Complex Data
4.1.6. Privacy Issues
4.1.7. Lack of Explainability
4.1.8. Lack of Reproducibility
4.2. Clinical Implementation Challenges of Using ML and DL in CC
4.2.1. Representativeness of Clinical Stages of CC in the Training of DL-Based Models
4.2.2. Privacy Concerns and Data Security in Health Care
4.2.3. Integration of AI with Clinical Workflows for Real-Time Decision-Making
4.2.4. Ethical and Regulatory Considerations
4.2.5. Public Perspectives on Using AI in Diagnoses Decisions
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | Accuracy |
| AI | Artificial intelligence |
| AUC | Area Under (AUC) the Receiver Operating Characteristic (ROC) curve |
| CC | Cervical cancer |
| CLF | Classifier |
| CNN | Convolutional neural network |
| CT | Computed tomography |
| CTV | Clinical target volume |
| DCNN | Deep convolutional neural networks |
| DL | Deep learning |
| DRS | Diffuse reflectance spectroscopy |
| DT | Decision tree |
| FS | Feature selection |
| GPC | Gaussian process classifier |
| GMM | Gaussian mixture model |
| HPV | Human Papilloma Virus |
| KNN | K-Nearest neighbors |
| LDA | Linear discriminant analysis |
| LR | Logistic regression |
| Mask R-CNN | Mask regional CNN |
| MGI | Magnetic resonance imaging |
| ML | Machine Learning |
| MLP | Multi-layer perceptron |
| NB | Naive Bayes |
| OAR | Organ at risk |
| PCA | Principal component analysis |
| RF | Random forest |
| RNN | Recurrent neural network |
| SVM | Support vector machine |
| ViT | Vision transformer |
References
- International Agency for Research on Cancer. Cancer Today. Available online: https://gco.iarc.fr/today/en (accessed on 28 May 2024).
- Momenimovahed, Z.; Mazidimoradi, A.; Maroofi, P.; Allahqoli, L.; Salehiniya, H.; Alkatout, I. Global, regional and national burden, incidence, and mortality of cervical cancer. Cancer Rep. 2022, 6, e1756. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Global Strategy to Accelerate the Elimination of Cervical Cancer as a Public Health Problem; World Health Organization (WHO): Geneva, Switzerland, 2020; Available online: https://www.who.int/publications-detail-redirect/9789240014107 (accessed on 28 May 2024).
- Perkins, R.B.; Wentzensen, N.; Guido, R.S.; Schiffman, M. Cervical Cancer Screening: A Review. JAMA 2023, 330, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Mithun, S.; Sherkhane, U.B.; Jaiswar, V.; Osong, B.; Purandare, N.; Kannan, S.; Prabhash, K.; Gupta, S.; Vanneste, B.; et al. Systematic review and meta-analysis of prediction models used in cervical cancer. Artif. Intell. Med. 2023, 139, 102549. [Google Scholar] [CrossRef]
- Monk, B.J.; Enomoto, T.; Kast, W.M.; McCormack, M.; Tan, D.S.P.; Wu, X.; González-Martín, A. Integration of immunotherapy into treatment of cervical cancer: Recent data and ongoing trials. Cancer Treat. Rev. 2022, 106, 102385. [Google Scholar] [CrossRef]
- Gaur, K.; Jagtap, M.M. Role of Artificial Intelligence and Machine Learning in Prediction, Diagnosis, and Prognosis of Cancer. Cureus 2022, 14, e31008. [Google Scholar] [CrossRef]
- Lam, T.Y.T.; Cheung, M.F.K.; Munro, Y.L.; Lim, K.M.; Shung, D.; Sung, J.J.Y. Randomized Controlled Trials of Artificial Intelligence in Clinical Practice: Systematic Review. J. Med. Internet Res. 2022, 24, e37188. [Google Scholar] [CrossRef]
- Arjmand, B.; Hamidpour, S.K.; Tayanloo-Beik, A.; Goodarzi, P.; Aghayan, H.R.; Adibi, H.; Larijani, B. Machine Learning: A New Prospect in Multi-Omics Data Analysis of Cancer. Front. Genet. 2022, 13, 824451. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.; Nejat, P.; Saha, A.; Campbell, C.J.V.; Norgan, A.P.; Lokker, C. Performance of externally validated machine learning models based on histopathology images for the diagnosis, classification, prognosis, or treatment outcome prediction in female breast cancer: A systematic review. J. Pathol. Inform. 2024, 15, 100348. [Google Scholar] [CrossRef]
- Li, G.X.; Chen, Y.P.; Hu, Y.Y.; Zhao, W.J.; Lu, Y.Y.; Wan, F.J.; Wu, Z.J.; Wang, X.Q.; Yu, Q.Y. Machine learning for identifying tumor stemness genes and developing prognostic model in gastric cancer. Aging 2024, 16, 6455–6477. [Google Scholar] [CrossRef]
- Khalighi, S.; Reddy, K.; Midya, A.; Pandav, K.B.; Madabhushi, A.; Abedalthagafi, M. Artificial intelligence in neuro-oncology: Advances and challenges in brain tumor diagnosis, prognosis, and precision treatment. NPJ Precis. Oncol. 2024, 8, 80. [Google Scholar] [CrossRef]
- Fiste, O.; Gkiozos, I.; Charpidou, A.; Syrigos, N.K. Artificial Intelligence-Based Treatment Decisions: A New Era for NSCLC. Cancers 2024, 16, 831. [Google Scholar] [CrossRef] [PubMed]
- Huan, Q.; Cheng, S.; Ma, H.F.; Zhao, M.; Chen, Y.; Yuan, X. Machine learning-derived identification of prognostic signature for improving prognosis and drug response in patients with ovarian cancer. J. Cell. Mol. Med. 2024, 28, e18021. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.T.; But, B.; Choi, H.C.W.; de Bree, R.; Lee, A.W.M.; Lee, V.H.F.; López, F.; Mäkitie, A.A.; Rodrigo, J.P.; Saba, N.F.; et al. Application of Artificial Intelligence for Nasopharyngeal Carcinoma Management – A Systematic Review. Cancer Manag. Res. 2022, 14, 339–366. [Google Scholar] [CrossRef]
- Mitchell, T. Machine Learning, 2nd ed.; McGraw Hill: Shelter Island, NY, USA, 1997. [Google Scholar]
- Chollet, F. Deep Learning with Python, 2nd ed.; Manning Publications: Shelter Island, NY, USA, 2021. [Google Scholar]
- Aurelien, G. Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems|Guide Books|ACM Digital Library, 2nd ed.; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2019. [Google Scholar]
- Painuli, D.; Mishra, D.; Bhardwaj, S.; Aggarwal, M. Forecast and prediction of COVID-19 using machine learning. In Data Science for COVID-19; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Murphy, K.P. Probabilistic Machine Learning: An Introduction; MIT Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Imagawa, K.; Shiomoto, K. Performance change with the number of training data: A case study on the binary classification of COVID-19 chest X-ray by using convolutional neural networks. Comput. Biol. Med. 2022, 142, 105251. [Google Scholar] [CrossRef]
- Shanthi, P.B.; Faruqi, F.; Hareesha, K.S.; Kudva, R. Deep Convolution Neural Network for Malignancy Detection and Classification in Microscopic Uterine Cervix Cell Images. Asian Pac. J. Cancer Prev. 2019, 20, 3447–3456. [Google Scholar] [CrossRef]
- Reijtenbagh, D.; Godart, J.; de Leeuw, A.; Seppenwoolde, Y.; Jürgenliemk-Schulz, I.; Mens, J.W.; Nout, R.; Hoogeman, M. Multi-center analysis of machine-learning predicted dose parameters in brachytherapy for cervical cancer. Radiother. Oncol. 2022, 170, 169–175. [Google Scholar] [CrossRef]
- Gençtav, A.; Aksoy, S.; Önder, S. Unsupervised segmentation and classification of cervical cell images. Pattern Recognit. 2012, 45, 4151–4168. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, C.; Gao, R.; Yan, Z.; Zhu, Z.; Yang, B.; Chen, C.; Lv, X.; Li, H.; Huang, Z. Rapid identification of cervical adenocarcinoma and cervical squamous cell carcinoma tissue based on Raman spectroscopy combined with multiple machine learning algorithms. Photodiagnosis Photodyn. Ther. 2021, 33, 102104. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.U.; Polosukhin, I. Attention is All you Need. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30, pp. 6000–6010. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Briefings Bioinform. 2017, 19, 1236–1246. [Google Scholar] [CrossRef]
- Adlung, L.; Cohen, Y.; Mor, U.; Elinav, E. Machine learning in clinical decision making. Med 2021, 2, 642–665. [Google Scholar] [CrossRef] [PubMed]
- Sambyal, D.; Sarwar, A. Recent developments in cervical cancer diagnosis using deep learning on whole slide images: An Overview of models, techniques, challenges and future directions. Micron 2023, 173, 103520. [Google Scholar] [CrossRef]
- Yoganathan, S.A.; Paul, S.N.; Paloor, S.; Torfeh, T.; Chandramouli, S.H.; Hammoud, R.; Al-Hammadi, N. Automatic segmentation of magnetic resonance images for high-dose-rate cervical cancer brachytherapy using deep learning. Med. Phys. 2022, 49, 1571–1584. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, J.; Von Elm, E.; Altman, D.; Gøtzsche, P.; Mulrow, C.; Pocock, S.; Poole, C.; Schlesselman, J.; Egger, M. Mejorar la comunicación de estudios observacionales en epidemiología (STROBE): Explicación y elaboración. Gac. Sanit. 2009, 23, 158e1–158e28. [Google Scholar] [CrossRef] [PubMed]
- Dixon-Woods, M.; Agarwal, S.; Jones, D.; Young, B.; Sutton, A. Synthesising qualitative and quantitative evidence: A review of possible methods. J. Health Serv. Res. Policy 2005, 10, 45–53. [Google Scholar] [CrossRef]
- Kim, Y.J.; Ju, W.; Nam, K.H.; Kim, S.N.; Kim, Y.J.; Kim, K.G. RGB Channel Superposition Algorithm with Acetowhite Mask Images in a Cervical Cancer Classification Deep Learning Model. Sensors 2022, 22, 3564. [Google Scholar] [CrossRef]
- Yuan, Z.; Wang, Y.; Hu, P.; Zhang, D.; Yan, B.; Lu, H.M.; Zhang, H.; Yang, Y. Accelerate treatment planning process using deep learning generated fluence maps for cervical cancer radiation therapy. Med. Phys. 2022, 49, 2631–2641. [Google Scholar] [CrossRef]
- Kruczkowski, M.; Drabik-Kruczkowska, A.; Marciniak, A.; Tarczewska, M.; Kosowska, M.; Szczerska, M. Predictions of cervical cancer identification by photonic method combined with machine learning. Sci. Rep. 2022, 12, 3762. [Google Scholar] [CrossRef]
- Fu, L.; Xia, W.; Shi, W.; Cao, G.X.; Ruan, Y.T.; Zhao, X.Y.; Liu, M.; Niu, S.M.; Li, F.; Gao, X. Deep learning based cervical screening by the cross-modal integration of colposcopy, cytology, and HPV test. Int. J. Med. Inform. 2022, 159, 104675. [Google Scholar] [CrossRef]
- Ma, C.Y.; Zhou, J.Y.; Xu, X.T.; Guo, J.; Han, M.F.; Gao, Y.Z.; Du, H.; Stahl, J.N.; Maltz, J.S. Deep learning-based auto-segmentation of clinical target volumes for radiotherapy treatment of cervical cancer. J. Appl. Clin. Med. Phys. 2022, 23, e13470. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, C.; Rahaman, M.M.; Jiang, T.; Sun, H.; Wu, X.; Hu, W.; Chen, H.; Sun, C.; Yao, Y.; et al. Is the aspect ratio of cells important in deep learning? A robust comparison of deep learning methods for multi-scale cytopathology cell image classification: From convolutional neural networks to visual transformers. Comput. Biol. Med. 2022, 141, 105026. [Google Scholar] [CrossRef]
- Nambu, Y.; Mariya, T.; Shinkai, S.; Umemoto, M.; Asanuma, H.; Sato, I.; Hirohashi, Y.; Torigoe, T.; Fujino, Y.; Saito, T. A screening assistance system for cervical cytology of squamous cell atypia based on a two-step combined CNN algorithm with label smoothing. Cancer Med. 2022, 11, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Drokow, E.K.; Baffour, A.A.; Effah, C.Y.; Agboyibor, C.; Akpabla, G.S.; Sun, K. Building a predictive model to assist in the diagnosis of cervical cancer. Future Oncol. 2022, 18, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liang, T.; Peng, Y.; Peng, G.; Sun, L.; Li, L.; Dong, H. Segmentation of acetowhite region in uterine cervical image based on deep learning. Technol. Health Care Off. J. Eur. Soc. Eng. Med. 2022, 30, 469–482. [Google Scholar] [CrossRef]
- Mehmood, M.; Rizwan, M.; Gregus Ml, M.; Abbas, S. Machine Learning Assisted Cervical Cancer Detection. Front. Public Health 2021, 9, 788376. [Google Scholar] [CrossRef]
- Ali, M.M.; Ahmed, K.; Bui, F.M.; Paul, B.K.; Ibrahim, S.M.; Quinn, J.M.W.; Moni, M.A. Machine learning-based statistical analysis for early stage detection of cervical cancer. Comput. Biol. Med. 2021, 139, 104985. [Google Scholar] [CrossRef]
- Chu, R.; Zhang, Y.; Qiao, X.; Xie, L.; Chen, W.; Zhao, Y.; Xu, Y.; Yuan, Z.; Liu, X.; Yin, A.; et al. Risk Stratification of Early-Stage Cervical Cancer with Intermediate-Risk Factors: Model Development and Validation Based on Machine Learning Algorithm. Oncologist 2021, 26, e2217–e2226. [Google Scholar] [CrossRef]
- Dong, Y.; Wan, J.; Wang, X.; Xue, J.H.; Zou, J.; He, H.; Li, P.; Hou, A.; Ma, H. A Polarization-Imaging- Based Machine Learning Framework for Quantitative Pathological Diagnosis of Cervical Precancerous Lesions. IEEE Trans. Med. Imaging 2021, 40, 3728–3738. [Google Scholar] [CrossRef]
- Fick, R.H.J.; Tayart, B.; Bertrand, C.; Lang, S.C.; Rey, T.; Ciompi, F.; Tilmant, C.; Farre, I.; Hadj, S.B. A Partial Label-Based Machine Learning Approach For Cervical Whole-Slide Image Classification: The Winning TissueNet Solution. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Mexico, Mexico, 1–5 November 2021; Volume 2021, pp. 2127–2131. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, B. Segmentation of Overlapping Cervical Cells with Mask Region Convolutional Neural Network. Comput. Math. Methods Med. 2021, 2021, 3890988. [Google Scholar] [CrossRef]
- Cheng, S.; Liu, S.; Yu, J.; Rao, G.; Xiao, Y.; Han, W.; Zhu, W.; Lv, X.; Li, N.; Cai, J.; et al. Robust whole slide image analysis for cervical cancer screening using deep learning. Nat. Commun. 2021, 12, 5639. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, M.M.; Li, C.; Yao, Y.; Kulwa, F.; Wu, X.; Li, X.; Wang, Q. DeepCervix: A deep learning-based framework for the classification of cervical cells using hybrid deep feature fusion techniques. Comput. Biol. Med. 2021, 136, 104649. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.R.; Kim, Y.J.; Ju, W.; Nam, K.; Kim, S.; Kim, K.G. Comparison of machine and deep learning for the classification of cervical cancer based on cervicography images. Sci. Rep. 2021, 11, 16143. [Google Scholar] [CrossRef]
- Tian, R.; Zhou, P.; Li, M.; Tan, J.; Cui, Z.; Xu, W.; Wei, J.; Zhu, J.; Jin, Z.; Cao, C.; et al. DeepHPV: A deep learning model to predict human papillomavirus integration sites. Briefings Bioinform. 2021, 22, bbaa242. [Google Scholar] [CrossRef]
- Da-ano, R.; Lucia, F.; Masson, I.; Abgral, R.; Alfieri, J.; Rousseau, C.; Mervoyer, A.; Reinhold, C.; Pradier, O.; Schick, U.; et al. A transfer learning approach to facilitate ComBat-based harmonization of multicentre radiomic features in new datasets. PLoS ONE 2021, 16, e0253653. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, M.; Chandra Joshi, R.; Kushwah, A.S.; Gupta, M.K.; Banerjee, M.; Burget, R.; Dutta, M.K. Cytokine gene variants and socio-demographic characteristics as predictors of cervical cancer: A machine learning approach. Comput. Biol. Med. 2021, 134, 104559. [Google Scholar] [CrossRef]
- Shi, J.; Ding, X.; Liu, X.; Li, Y.; Liang, W.; Wu, J. Automatic clinical target volume delineation for cervical cancer in CT images using deep learning. Med. Phys. 2021, 48, 3968–3981. [Google Scholar] [CrossRef]
- Ding, D.; Lang, T.; Zou, D.; Tan, J.; Chen, J.; Zhou, L.; Wang, D.; Li, R.; Li, Y.; Liu, J.; et al. Machine learning-based prediction of survival prognosis in cervical cancer. BMC Bioinform. 2021, 22, 331. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; Ong, K.; Zhang, W.; Li, W.; Li, L.; Young, D.; Su, Y.; Shang, B.; Peng, L.; et al. Hybrid AI-assistive diagnostic model permits rapid TBS classification of cervical liquid-based thin-layer cell smears. Nat. Commun. 2021, 12, 3541. [Google Scholar] [CrossRef]
- Chandran, V.; Sumithra, M.G.; Karthick, A.; George, T.; Deivakani, M.; Elakkiya, B.; Subramaniam, U.; Manoharan, S. Diagnosis of Cervical Cancer based on Ensemble Deep Learning Network using Colposcopy Images. BioMed Res. Int. 2021, 2021, 5584004. [Google Scholar] [CrossRef]
- Jiang, X.; Li, J.; Kan, Y.; Yu, T.; Chang, S.; Sha, X.; Zheng, H.; Luo, Y.; Wang, S. MRI Based Radiomics Approach With Deep Learning for Prediction of Vessel Invasion in Early-Stage Cervical Cancer. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Christley, S.; Ostmeyer, J.; Quirk, L.; Zhang, W.; Sirak, B.; Giuliano, A.R.; Zhang, S.; Monson, N.; Tiro, J.; Lucas, E.; et al. T Cell Receptor Repertoires Acquired via Routine Pap Testing May Help Refine Cervical Cancer and Precancer Risk Estimates. Front. Immunol. 2021, 12, 624230. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Shen, Y.; Lu, Y.; Deng, J.; Wright, J.D.; Zhang, Y.; Huang, Q.; Wang, D.; Jing, N.; Liang, X.; et al. Quantitative analysis of abnormalities in gynecologic cytopathology with deep learning. Lab. Investig. J. Tech. Methods Pathol. 2021, 101, 513–524. [Google Scholar] [CrossRef]
- Rigaud, B.; Anderson, B.M.; Yu, Z.H.; Gobeli, M.; Cazoulat, G.; Söderberg, J.; Samuelsson, E.; Lidberg, D.; Ward, C.; Taku, N.; et al. Automatic Segmentation Using Deep Learning to Enable Online Dose Optimization During Adaptive Radiation Therapy of Cervical Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2021, 109, 1096–1110. [Google Scholar] [CrossRef]
- Urushibara, A.; Saida, T.; Mori, K.; Ishiguro, T.; Sakai, M.; Masuoka, S.; Satoh, T.; Masumoto, T. Diagnosing uterine cervical cancer on a single T2-weighted image: Comparison between deep learning versus radiologists. Eur. J. Radiol. 2021, 135, 109471. [Google Scholar] [CrossRef]
- Wentzensen, N.; Lahrmann, B.; Clarke, M.A.; Kinney, W.; Tokugawa, D.; Poitras, N.; Locke, A.; Bartels, L.; Krauthoff, A.; Walker, J.; et al. Accuracy and Efficiency of Deep-Learning-Based Automation of Dual Stain Cytology in Cervical Cancer Screening. J. Natl. Cancer Inst. 2021, 113, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chang, Y.; Peng, Z.; Lv, Y.; Shi, W.; Wang, F.; Pei, X.; Xu, X.G. Evaluation of deep learning-based auto-segmentation algorithms for delineating clinical target volume and organs at risk involving data for 125 cervical cancer patients. J. Appl. Clin. Med. Phys. 2020, 21, 272–279. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Guan, H.; Zhen, H.; Sun, Y.; Chen, Q.; Chen, Y.; Wang, S.; Qiu, J. Development and validation of a deep learning algorithm for auto-delineation of clinical target volume and organs at risk in cervical cancer radiotherapy. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2020, 153, 172–179. [Google Scholar] [CrossRef]
- Mao, X.; Pineau, J.; Keyes, R.; Enger, S.A. RapidBrachyDL: Rapid Radiation Dose Calculations in Brachytherapy Via Deep Learning. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, 802–812. [Google Scholar] [CrossRef]
- Xue, Z.; Novetsky, A.P.; Einstein, M.H.; Marcus, J.Z.; Befano, B.; Guo, P.; Demarco, M.; Wentzensen, N.; Long, L.R.; Schiffman, M.; et al. A demonstration of automated visual evaluation of cervical images taken with a smartphone camera. Int. J. Cancer 2020, 147, 2416–2423. [Google Scholar] [CrossRef]
- Bao, H.; Bi, H.; Zhang, X.; Zhao, Y.; Dong, Y.; Luo, X.; Zhou, D.; You, Z.; Wu, Y.; Liu, Z.; et al. Artificial intelligence-assisted cytology for detection of cervical intraepithelial neoplasia or invasive cancer: A multicenter, clinical-based, observational study. Gynecol. Oncol. 2020, 159, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.J.; Choi, Y.J.; Lee, M.J.; Kim, J.H.; Son, G.H.; Park, S.H.; Kim, H.B.; Joo, Y.J.; Cho, H.Y.; Kyung, M.S.; et al. Classification of cervical neoplasms on colposcopic photography using deep learning. Sci. Rep. 2020, 10, 13652. [Google Scholar] [CrossRef]
- Ju, Z.; Wu, Q.; Yang, W.; Gu, S.; Guo, W.; Wang, J.; Ge, R.; Quan, H.; Liu, J.; Qu, B. Automatic segmentation of pelvic organs-at-risk using a fusion network model based on limited training samples. Acta Oncol. 2020, 59, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Kanai, R.; Ohshima, K.; Ishii, K.; Sonohara, M.; Ishikawa, M.; Yamaguchi, M.; Ohtani, Y.; Kobayashi, Y.; Ota, H.; Kimura, F. Discriminant analysis and interpretation of nuclear chromatin distribution and coarseness using gray-level co-occurrence matrix features for lobular endocervical glandular hyperplasia. Diagn. Cytopathol. 2020, 48, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Yao, Y.; Cheng, B.; Cheng, Y.; Li, Y.; Li, Y.; Liu, X.; Cheng, X.; Xie, X.; Wu, J.; et al. The application of deep learning based diagnostic system to cervical squamous intraepithelial lesions recognition in colposcopy images. Sci. Rep. 2020, 10, 11639. [Google Scholar] [CrossRef]
- Hu, L.; Horning, M.P.; Banik, D.; Ajenifuja, O.K.; Adepiti, C.A.; Yeates, K.; Mtema, Z.; Wilson, B.; Mehanian, C. Deep learning-based image evaluation for cervical precancer screening with a smartphone targeting low resource settings—Engineering approach. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; Volume 2020, pp. 1944–1949. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, S.; Zhang, S.; Wang, M.; Ding, Y.; Fang, J.; Wu, Q.; Qian, W.; Liu, Z.; Sun, K.; et al. Development of a Deep Learning Model to Identify Lymph Node Metastasis on Magnetic Resonance Imaging in Patients With Cervical Cancer. JAMA Netw. Open 2020, 3, e2011625. [Google Scholar] [CrossRef]
- Wang, T.; Gao, T.; Guo, H.; Wang, Y.; Zhou, X.; Tian, J.; Huang, L.; Zhang, M. Preoperative prediction of parametrial invasion in early-stage cervical cancer with MRI-based radiomics nomogram. Eur. Radiol. 2020, 30, 3585–3593. [Google Scholar] [CrossRef]
- Kudva, V.; Prasad, K.; Guruvare, S. Hybrid Transfer Learning for Classification of Uterine Cervix Images for Cervical Cancer Screening. J. Digit. Imaging 2020, 33, 619–631. [Google Scholar] [CrossRef]
- Ijaz, M.F.; Attique, M.; Son, Y. Data-Driven Cervical Cancer Prediction Model with Outlier Detection and Over-Sampling Methods. Sensors 2020, 20, 2809. [Google Scholar] [CrossRef]
- Bae, J.K.; Roh, H.J.; You, J.S.; Kim, K.; Ahn, Y.; Askaruly, S.; Park, K.; Yang, H.; Jang, G.J.; Moon, K.H.; et al. Quantitative Screening of Cervical Cancers for Low-Resource Settings: Pilot Study of Smartphone-Based Endoscopic Visual Inspection After Acetic Acid Using Machine Learning Techniques. JMIR mHealth uHealth 2020, 8, e16467. [Google Scholar] [CrossRef]
- Sornapudi, S.; Brown, G.T.; Xue, Z.; Long, R.; Allen, L.; Antani, S. Comparing Deep Learning Models for Multi-cell Classification in Liquid- based Cervical Cytology Image. AMIA Annu. Symp. Proc. 2020, 2019, 820–827. [Google Scholar] [PubMed]
- Shao, J.; Zhang, Z.; Liu, H.; Song, Y.; Yan, Z.; Wang, X.; Hou, Z. DCE-MRI pharmacokinetic parameter maps for cervical carcinoma prediction. Comput. Biol. Med. 2020, 118, 103634. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Yokota, H.; Watanabe Nemoto, M.; Horikoshi, T.; Matsushima, J.; Uno, T. A multi-scanner study of MRI radiomics in uterine cervical cancer: Prediction of in-field tumor control after definitive radiotherapy based on a machine learning method including peritumoral regions. Jpn. J. Radiol. 2020, 38, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Lin, C.H.; Lu, H.Y.; Chiang, H.J.; Wang, H.K.; Huang, Y.T.; Ng, S.H.; Hong, J.H.; Yen, T.C.; Lai, C.H.; et al. Deep learning for fully automated tumor segmentation and extraction of magnetic resonance radiomics features in cervical cancer. Eur. Radiol. 2020, 30, 1297–1305. [Google Scholar] [CrossRef]
- Jihong, C.; Penggang, B.; Xiuchun, Z.; Kaiqiang, C.; Wenjuan, C.; Yitao, D.; Jiewei, Q.; Kerun, Q.; Jing, Z.; Tianming, W. Automated Intensity Modulated Radiation Therapy Treatment Planning for Cervical Cancer Based on Convolution Neural Network. Technol. Cancer Res. Treat. 2020, 19, 1533033820957002. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Xiao, B.; Wang, S.; Miao, Z.; Sun, Y.; Zhang, F. Segmentation of organs-at-risk in cervical cancer CT images with a convolutional neural network. Phys. Medica 2020, 69, 184–191. [Google Scholar] [CrossRef]
- Shen, W.C.; Chen, S.W.; Wu, K.C.; Hsieh, T.C.; Liang, J.A.; Hung, Y.C.; Yeh, L.S.; Chang, W.C.; Lin, W.C.; Yen, K.Y.; et al. Prediction of local relapse and distant metastasis in patients with definitive chemoradiotherapy-treated cervical cancer by deep learning from [18F]-fluorodeoxyglucose positron emission tomography/computed tomography. Eur. Radiol. 2019, 29, 6741–6749. [Google Scholar] [CrossRef]
- Pathania, D.; Landeros, C.; Rohrer, L.; D’Agostino, V.; Hong, S.; Degani, I.; Avila-Wallace, M.; Pivovarov, M.; Randall, T.; Weissleder, R.; et al. Point-of-care cervical cancer screening using deep learning-based microholography. Theranostics 2019, 9, 8438–8447. [Google Scholar] [CrossRef]
- Aljakouch, K.; Hilal, Z.; Daho, I.; Schuler, M.; Krauß, S.D.; Yosef, H.K.; Dierks, J.; Mosig, A.; Gerwert, K.; El-Mashtoly, S.F. Fast and Noninvasive Diagnosis of Cervical Cancer by Coherent Anti-Stokes Raman Scattering. Anal. Chem. 2019, 91, 13900–13906. [Google Scholar] [CrossRef]
- Dong, N.; Zhao, L.; Wu, A. Cervical cell recognition based on AGVF-Snake algorithm. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 2031–2041. [Google Scholar] [CrossRef]
- Hu, L.; Bell, D.; Antani, S.; Xue, Z.; Yu, K.; Horning, M.P.; Gachuhi, N.; Wilson, B.; Jaiswal, M.S.; Befano, B.; et al. An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening. J. Natl. Cancer Inst. 2019, 111, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Du, H.; Guan, H.; Zhang, X. Multiscale Time-Sharing Elastography Algorithms and Transfer Learning of Clinicopathological Features of Uterine Cervical Cancer for Medical Intelligent Computing System. J. Med. Syst. 2019, 43, 310. [Google Scholar] [CrossRef] [PubMed]
- Geetha, R.; Sivasubramanian, S.; Kaliappan, M.; Vimal, S.; Annamalai, S. Cervical Cancer Identification with Synthetic Minority Oversampling Technique and PCA Analysis using Random Forest Classifier. J. Med. Syst. 2019, 43, 286. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Gonzalez, Y.; Klages, P.; Qin, N.; Jung, H.; Chen, L.; Nguyen, D.; Jiang, S.B.; Jia, X. Intelligent inverse treatment planning via deep reinforcement learning, a proof-of-principle study in high dose-rate brachytherapy for cervical cancer. Phys. Med. Biol. 2019, 64, 115013. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Z.; Du, B.; He, J.; Li, G.; Chen, D. Binary tree-like network with two-path Fusion Attention Feature for cervical cell nucleus segmentation. Comput. Biol. Med. 2019, 108, 223–233. [Google Scholar] [CrossRef]
- Chen, L.; Shen, C.; Zhou, Z.; Maquilan, G.; Albuquerque, K.; Folkert, M.R.; Wang, J. Automatic PET Cervical Tumor Segmentation by Combining Deep Learning and Anatomic Prior. Phys. Med. Biol. 2019, 64, 085019. [Google Scholar] [CrossRef]
- Matsuo, K.; Purushotham, S.; Jiang, B.; Mandelbaum, R.S.; Takiuchi, T.; Liu, Y.; Roman, L.D. Survival outcome prediction in cervical cancer: Cox models vs deep-learning model. Am. J. Obstet. Gynecol. 2019, 220, 381.e1–381.e14. [Google Scholar] [CrossRef]
- Araújo, F.H.D.; Silva, R.R.V.; Ushizima, D.M.; Rezende, M.T.; Carneiro, C.M.; Campos Bianchi, A.G.; Medeiros, F.N.S. Deep learning for cell image segmentation and ranking. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc. 2019, 72, 13–21. [Google Scholar] [CrossRef]
- Kan, Y.; Dong, D.; Zhang, Y.; Jiang, W.; Zhao, N.; Han, L.; Fang, M.; Zang, Y.; Hu, C.; Tian, J.; et al. Radiomic signature as a predictive factor for lymph node metastasis in early-stage cervical cancer. J. Magn. Reson. Imaging JMRI 2019, 49, 304–310. [Google Scholar] [CrossRef]
- Zhen, X.; Chen, J.; Zhong, Z.; Hrycushko, B.; Zhou, L.; Jiang, S.; Albuquerque, K.; Gu, X. Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: A feasibility study. Phys. Med. Biol. 2017, 62, 8246–8263. [Google Scholar] [CrossRef]
- Xie, Y.; Xing, F.; Shi, X.; Kong, X.; Su, H.; Yang, L. Efficient and robust cell detection: A structured regression approach. Med. Image Anal. 2018, 44, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Kudva, V.; Prasad, K.; Guruvare, S. Automation of Detection of Cervical Cancer Using Convolutional Neural Networks. Crit. Rev. Biomed. Eng. 2018, 46, 135–145. [Google Scholar] [CrossRef]
- Wei, L.; Gan, Q.; Ji, T. Cervical cancer histology image identification method based on texture and lesion area features. Comput. Assist. Surg. 2017, 22, 186–199. [Google Scholar] [CrossRef]
- Guo, P.; Banerjee, K.; Joe Stanley, R.; Long, R.; Antani, S.; Thoma, G.; Zuna, R.; Frazier, S.R.; Moss, R.H.; Stoecker, W.V. Nuclei-Based Features for Uterine Cervical Cancer Histology Image Analysis With Fusion-Based Classification. IEEE J. Biomed. Health Inform. 2016, 20, 1595–1607. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wu, A.; Song, J.; Sun, X.; Dong, N. Automatic screening of cervical cells using block image processing. BioMed. Eng. Online 2016, 15, 14. [Google Scholar] [CrossRef]
- Weegar, R.; Kvist, M.; Sundström, K.; Brunak, S.; Dalianis, H. Finding Cervical Cancer Symptoms in Swedish Clinical Text using a Machine Learning Approach and NegEx. AMIA Annu. Symp. Proc. 2015, 2015, 1296–1305. [Google Scholar] [PubMed]
- Kahng, J.; Kim, E.H.; Kim, H.G.; Lee, W. Development of a cervical cancer progress prediction tool for human papillomavirus-positive Koreans: A support vector machine-based approach. J. Int. Med. Res. 2015, 43, 518–525. [Google Scholar] [CrossRef]
- Mu, W.; Chen, Z.; Liang, Y.; Shen, W.; Yang, F.; Dai, R.; Wu, N.; Tian, J. Staging of cervical cancer based on tumor heterogeneity characterized by texture features on 18F-FDG PET images. Phys. Med. Biol. 2015, 60, 5123. [Google Scholar] [CrossRef]
- Zhi, L.; Carneiro, G.; Bradley, A.P. An improved joint optimization of multiple level set functions for the segmentation of overlapping cervical cells. IEEE Trans. Image Process. 2015, 24, 1261–1272. [Google Scholar] [CrossRef]
- Dong, Y.; Kuang, Q.; Dai, X.; Li, R.; Wu, Y.; Leng, W.; Li, Y.; Li, M. Improving the Understanding of Pathogenesis of Human Papillomavirus 16 via Mapping Protein-Protein Interaction Network. BioMed Res. Int. 2015, 2015, 890381. [Google Scholar] [CrossRef]
- Mariarputham, E.J.; Stephen, A. Nominated Texture Based Cervical Cancer Classification. Comput. Math. Methods Med. 2015, 2015, 586928. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Yan, W.; Feng, Y.; Yunzhi, L.; Zhang, Y.; Wang, D.; Chen, W.; Peng, J.; Guo, C.; Chen, Z.; et al. An MRI-based machine learning radiomics can predict short-term response to neoadjuvant chemotherapy in patients with cervical squamous cell carcinoma: A multicenter study. Cancer Med. 2023, 12, 19383–19393. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.; Hoong, K.; Kleijn, W.B.; Doronin, A.; Rehbinder, J.; Vizet, J.; Pierangelo, A.; Novikova, T. Polarimetric imaging for cervical pre-cancer screening aided by machine learning: Ex vivo studies. J. Biomed. Opt. 2023, 28, 102904. [Google Scholar] [CrossRef]
- Tian, M.; Wang, H.; Liu, X.; Ye, Y.; Ouyang, G.; Shen, Y.; Li, Z.; Wang, X.; Wu, S. Delineation of clinical target volume and organs at risk in cervical cancer radiotherapy by deep learning networks. Med. Phys. 2023, 50, 6354–6365. [Google Scholar] [CrossRef]
- Kaur, M.; Singh, D.; Kumar, V.; Lee, H.N. MLNet: Metaheuristics-Based Lightweight Deep Learning Network for Cervical Cancer Diagnosis. IEEE J. Biomed. Health Inform. 2023, 27, 5004–5014. [Google Scholar] [CrossRef]
- Ji, J.; Zhang, W.; Dong, Y.; Lin, R.; Geng, Y.; Hong, L. Automated cervical cell segmentation using deep ensemble learning. BMC Med. Imaging 2023, 23, 137. [Google Scholar] [CrossRef]
- Liu, Q.; Jiang, N.; Hao, Y.; Hao, C.; Wang, W.; Bian, T.; Wang, X.; Li, H.; Zhang, Y.; Kang, Y.; et al. Identification of lymph node metastasis in pre-operation cervical cancer patients by weakly supervised deep learning from histopathological whole-slide biopsy images. Cancer Med. 2023, 12, 17952–17966. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Liu, J.; Ma, C.; Chen, C.; Lv, X.; Chen, C. Early screening of cervical cancer based on tissue Raman spectroscopy combined with deep learning algorithms. Photodiagnosis Photodyn. Ther. 2023, 42, 103557. [Google Scholar] [CrossRef]
- Wu, G.; Li, C.; Yin, L.; Wang, J.; Zheng, X. Compared between support vector machine (SVM) and deep belief network (DBN) for multi-classification of Raman spectroscopy for cervical diseases. Photodiagnosis Photodyn. Ther. 2023, 42, 103340. [Google Scholar] [CrossRef]
- Ince, O.; Uysal, E.; Durak, G.; Onol, S.; Donmez Yilmaz, B.; Erturk, S.M.; Onder, H. Prediction of carcinogenic human papillomavirus types in cervical cancer from multiparametric magnetic resonance images with machine learning-based radiomics models. Diagn. Interv. Radiol. 2023, 29, 460–468. [Google Scholar] [CrossRef]
- Kurita, Y.; Meguro, S.; Tsuyama, N.; Kosugi, I.; Enomoto, Y.; Kawasaki, H.; Uemura, T.; Kimura, M.; Iwashita, T. Accurate deep learning model using semi-supervised learning and Noisy Student for cervical cancer screening in low magnification images. PLoS ONE 2023, 18, e0285996. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yang, C.; Song, S.; Wang, R.; Gu, L.; Chen, Z. Classification of multiple cancer types by combination of plasma-based near-infrared spectroscopy analysis and machine learning modeling. Anal. Biochem. 2023, 669, 115120. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, Y.; Chen, Z.; Wang, Z.; Zhou, Q.; Qiu, J. Tissues margin-based analytical anisotropic algorithm boosting method via deep learning attention mechanism with cervical cancer. Int. J. Comput. Assist. Radiol. Surg. 2023, 18, 953–959. [Google Scholar] [CrossRef]
- Devi, S.; Gaikwad, S.R.; R, H. Prediction and Detection of Cervical Malignancy Using Machine Learning Models. Asian Pac. J. Cancer Prev. APJCP 2023, 24, 1419–1433. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Pu, X.; Chen, Z.; Li, L.; Zhao, K.N.; Liu, H.; Zhu, H. Application of EfficientNet-B0 and GRU-based deep learning on classifying the colposcopy diagnosis of precancerous cervical lesions. Cancer Med. 2023, 12, 8690–8699. [Google Scholar] [CrossRef]
- Salehi, M.; Vafaei Sadr, A.; Mahdavi, S.R.; Arabi, H.; Shiri, I.; Reiazi, R. Deep Learning-based Non-rigid Image Registration for High-dose Rate Brachytherapy in Inter-fraction Cervical Cancer. J. Digit. Imaging 2023, 36, 574–587. [Google Scholar] [CrossRef]
- Yu, W.; Lu, Y.; Shou, H.; Xu, H.; Shi, L.; Geng, X.; Song, T. A 5-year survival status prognosis of nonmetastatic cervical cancer patients through machine learning algorithms. Cancer Med. 2023, 12, 6867–6876. [Google Scholar] [CrossRef]
- Liu, S.; Chu, R.; Xie, J.; Song, K.; Su, X. Differentiating single cervical cells by mitochondrial fluorescence imaging and deep learning-based label-free light scattering with multi-modal static cytometry. Cytom. Part A J. Int. Soc. Anal. Cytol. 2023, 103, 240–250. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Y.; Tu, Y.; Xie, H.; Chen, Y.; Luo, L.; Zhou, P.; Tang, Q. Evaluation of auto-segmentation for brachytherapy of postoperative cervical cancer using deep learning-based workflow. Phys. Med. Biol. 2023, 68, 055012. [Google Scholar] [CrossRef]
- Yu, W.; Xiao, C.; Xu, J.; Jin, J.; Jin, X.; Shen, L. Direct Dose Prediction With Deep Learning for Postoperative Cervical Cancer Underwent Volumetric Modulated Arc Therapy. Technol. Cancer Res. Treat. 2023, 22, 15330338231167039. [Google Scholar] [CrossRef]
- Huang, M.; Feng, C.; Sun, D.; Cui, M.; Zhao, D. Segmentation of Clinical Target Volume From CT Images for Cervical Cancer Using Deep Learning. Technol. Cancer Res. Treat. 2023, 22, 15330338221139164. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Ma, H.; Fan, Y.; Liu, Y.; Zhang, H.; Cao, C.; Yu, H. A deep learning-based method for cervical transformation zone classification in colposcopy images. Technol. Health Care Off. J. Eur. Soc. Eng. Med. 2023, 31, 527–538. [Google Scholar] [CrossRef]
- Chen, C.; Cao, Y.; Li, W.; Liu, Z.; Liu, P.; Tian, X.; Sun, C.; Wang, W.; Gao, H.; Kang, S.; et al. The pathological risk score: A new deep learning-based signature for predicting survival in cervical cancer. Cancer Med. 2022, 12, 1051–1063. [Google Scholar] [CrossRef]
- He, S.; Xiao, B.; Wei, H.; Huang, S.; Chen, T. SVM classifier of cervical histopathology images based on texture and morphological features. Technol. Health Care Off. J. Eur. Soc. Eng. Med. 2023, 31, 69–80. [Google Scholar] [CrossRef]
- Qian, W.; Li, Z.; Chen, W.; Yin, H.; Zhang, J.; Xu, J.; Hu, C. RESOLVE-DWI-based deep learning nomogram for prediction of normal-sized lymph node metastasis in cervical cancer: A preliminary study. BMC Med. Imaging 2022, 22, 221. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Wang, L.; Li, R.; Liu, Q.; Xu, Y.; Wei, Y.; Jiao, X.; Li, X.; Zhang, Y.; Zhang, Y.; et al. Development of a Novel Deep Learning-Based Prediction Model for the Prognosis of Operable Cervical Cancer. Comput. Math. Methods Med. 2022, 2022, 4364663. [Google Scholar] [CrossRef]
- Ji, M.; Zhong, J.; Xue, R.; Su, W.; Kong, Y.; Fei, Y.; Ma, J.; Wang, Y.; Mi, L. Early Detection of Cervical Cancer by Fluorescence Lifetime Imaging Microscopy Combined with Unsupervised Machine Learning. Int. J. Mol. Sci. 2022, 23, 11476. [Google Scholar] [CrossRef]
- Ming, Y.; Dong, X.; Zhao, J.; Chen, Z.; Wang, H.; Wu, N. Deep learning-based multimodal image analysis for cervical cancer detection. Methods 2022, 205, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, Y.; Xie, H.; Luo, L.; Tang, Q. Evaluation of auto-segmentation for EBRT planning structures using deep learning-based workflow on cervical cancer. Sci. Rep. 2022, 12, 13650. [Google Scholar] [CrossRef]
- Ma, C.Y.; Zhou, J.Y.; Xu, X.T.; Qin, S.B.; Han, M.F.; Cao, X.H.; Gao, Y.Z.; Xu, L.; Zhou, J.J.; Zhang, W.; et al. Clinical evaluation of deep learning-based clinical target volume three-channel auto-segmentation algorithm for adaptive radiotherapy in cervical cancer. BMC Med. Imaging 2022, 22, 123. [Google Scholar] [CrossRef]
- Chang, X.; Cai, X.; Dan, Y.; Song, Y.; Lu, Q.; Yang, G.; Nie, S. Self-supervised learning for multi-center magnetic resonance imaging harmonization without traveling phantoms. Phys. Med. Biol. 2022, 67, 145004. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Chu, X.; Guo, B.; Pan, Q.; Ji, S.; Lou, W.; Lv, C.; Xie, G.; Hua, K. Scrutinizing high-risk patients from ASC-US cytology via a deep learning model. Cancer Cytopathol. 2022, 130, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Qilin, Z.; Peng, B.; Ang, Q.; Weijuan, J.; Ping, J.; Hongqing, Z.; Bin, D.; Ruijie, Y. The feasibility study on the generalization of deep learning dose prediction model for volumetric modulated arc therapy of cervical cancer. J. Appl. Clin. Med. Phys. 2022, 23, e13583. [Google Scholar] [CrossRef]
- Xue, Y.; Zheng, X.; Wu, G.; Wang, J. Rapid diagnosis of cervical cancer based on serum FTIR spectroscopy and support vector machines. Lasers Med. Sci. 2023, 38, 276. [Google Scholar] [CrossRef]
- Gay, S.S.; Kisling, K.D.; Anderson, B.M.; Zhang, L.; Rhee, D.J.; Nguyen, C.; Netherton, T.; Yang, J.; Brock, K.; Jhingran, A.; et al. Identifying the optimal deep learning architecture and parameters for automatic beam aperture definition in 3D radiotherapy. J. Appl. Clin. Med. Phys. 2023, 24, e14131. [Google Scholar] [CrossRef]
- Shen, Y.; Tang, X.; Lin, S.; Jin, X.; Ding, J.; Shao, M. Automatic dose prediction using deep learning and plan optimization with finite-element control for intensity modulated radiation therapy. Med. Phys. 2024, 51, 545–555. [Google Scholar] [CrossRef]
- Aljrees, T. Improving prediction of cervical cancer using KNN imputer and multi-model ensemble learning. PLoS ONE 2024, 19, e0295632. [Google Scholar] [CrossRef] [PubMed]
- Munshi, R.M. Novel ensemble learning approach with SVM-imputed ADASYN features for enhanced cervical cancer prediction. PLoS ONE 2024, 19, e0296107. [Google Scholar] [CrossRef]
- Jeong, S.; Yu, H.; Park, S.H.; Woo, D.; Lee, S.J.; Chong, G.O.; Han, H.S.; Kim, J.C. Comparing deep learning and handcrafted radiomics to predict chemoradiotherapy response for locally advanced cervical cancer using pretreatment MRI. Sci. Rep. 2024, 14, 1180. [Google Scholar] [CrossRef]
- Stegmüller, T.; Abbet, C.; Bozorgtabar, B.; Clarke, H.; Petignat, P.; Vassilakos, P.; Thiran, J.P. Self-supervised learning-based cervical cytology for the triage of HPV-positive women in resource-limited settings and low-data regime. arXiv 2023, arXiv:2302.05195. [Google Scholar] [CrossRef]
- Wu, K.C.; Chen, S.W.; Hsieh, T.C.; Yen, K.Y.; Chang, C.J.; Kuo, Y.C.; Chang, R.F.; Chia-Hung, K. Early prediction of distant metastasis in patients with uterine cervical cancer treated with definitive chemoradiotherapy by deep learning using pretreatment [18 F]fluorodeoxyglucose positron emission tomography/computed tomography. Nucl. Med. Commun. 2024, 45, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, D.; Xu, C.; Peng, S.; Deng, L.; Liu, M.; Wu, Y. Clinical target volume (CTV) automatic delineation using deep learning network for cervical cancer radiotherapy: A study with external validation. J. Appl. Clin. Med. Phys. 2025, 26, e14553. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Rixin, S.; Linrui, L.; Zhihui, Q.; Long, L.; Yu, Z. Machine learning-based radiomics for predicting outcomes in cervical cancer patients undergoing concurrent chemoradiotherapy. Comput. Biol. Med. 2024, 177, 108593. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shen, C.; Zhou, Z.; Maquilan, G.; Thomas, K.; Folkert, M.R.; Albuquerque, K.; Wang, J. Accurate segmenting of cervical tumors in PET imaging based on similarity between adjacent slices. Comput. Biol. Med. 2018, 97, 30–36. [Google Scholar] [CrossRef]
- Chen, X.; Liu, W.; Thai, T.C.; Castellano, T.; Gunderson, C.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Developing a new radiomics-based CT image marker to detect lymph node metastasis among cervical cancer patients. Comput. Methods Programs Biomed. 2020, 197, 105759. [Google Scholar] [CrossRef]
- Zheng, C.; Shen, Q.; Zhao, L.; Wang, Y. Utilising deep learning networks to classify ZEB2 expression images in cervical cancer. Br. J. Hosp. Med. 2024, 85, 1–13. [Google Scholar] [CrossRef]
- Brenes, D.; Salcedo, M.P.; Coole, J.B.; Maker, Y.; Kortum, A.; Schwarz, R.A.; Carns, J.; Vohra, I.S.; Possati-Resende, J.C.; Antoniazzi, M.; et al. Multiscale Optical Imaging Fusion for Cervical Precancer Diagnosis: Integrating Widefield Colposcopy and High-Resolution Endomicroscopy. IEEE Trans. Bio-Med. Eng. 2024, 71, 2547–2556. [Google Scholar] [CrossRef]
- Shandilya, G.; Gupta, S.; Almogren, A.; Bharany, S.; Altameem, A.; Rehman, A.U.; Hussen, S. Enhancing advanced cervical cell categorization with cluster-based intelligent systems by a novel integrated CNN approach with skip mechanisms and GAN-based augmentation. Sci. Rep. 2024, 14, 29040. [Google Scholar] [CrossRef]
- Mathivanan, S.K.; Francis, D.; Srinivasan, S.; Khatavkar, V.; P, K.; Shah, M.A. Enhancing cervical cancer detection and robust classification through a fusion of deep learning models. Sci. Rep. 2024, 14, 10812. [Google Scholar] [CrossRef]
- Wang, C.W.; Liou, Y.A.; Lin, Y.J.; Chang, C.C.; Chu, P.H.; Lee, Y.C.; Wang, C.H.; Chao, T.K. Artificial intelligence-assisted fast screening cervical high grade squamous intraepithelial lesion and squamous cell carcinoma diagnosis and treatment planning. Sci. Rep. 2021, 11, 16244. [Google Scholar] [CrossRef]
- Dong, B.; Xue, H.; Li, Y.; Li, P.; Chen, J.; Zhang, T.; Chen, L.; Pan, D.; Liu, P.; Sun, P. Classification and diagnosis of cervical lesions based on colposcopy images using deep fully convolutional networks: A man-machine comparison cohort study. Fundam. Res. 2025, 5, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Wang, C.; Yang, M.; Yang, J.; Xu, X.; Shen, L.; Yang, Z.; Xing, H.; Ou, C.Q. Use of Virus Genotypes in Machine Learning Diagnostic Prediction Models for Cervical Cancer in Women With High-Risk Human Papillomavirus Infection. JAMA Netw. Open 2023, 6, e2326890. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhu, G.; Zhou, Y.; Yang, B.; Wang, J.; Wang, Z.; Wang, T. Predictive models for personalized precision medical intervention in spontaneous regression stages of cervical precancerous lesions. J. Transl. Med. 2024, 22, 686. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, T.; Yan, D.; Zhao, H.; Wang, M.; Liu, T.; Fan, X.; Xu, X. Vaginal microbial profile of cervical cancer patients receiving chemoradiotherapy: The potential involvement of Lactobacillus iners in recurrence. J. Transl. Med. 2024, 22, 575. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Han, Y.; Jiang, M.; Yu, Y.; Xu, H.; Yu, L.; Cui, J.; Liu, B.; Chen, F.; et al. Development of models for cervical cancer screening: Construction in a cross-sectional population and validation in two screening cohorts in China. BMC Med. 2021, 19, 197. [Google Scholar] [CrossRef]
- Namalinzi, F.; Galadima, K.R.; Nalwanga, R.; Sekitoleko, I.; Uwimbabazi, L.F.R. Prediction of precancerous cervical cancer lesions among women living with HIV on antiretroviral therapy in Uganda: A comparison of supervised machine learning algorithms. BMC Women’s Health 2024, 24, 393. [Google Scholar] [CrossRef]
- Liu, S.; Zhou, Y.; Wang, C.; Shen, J.; Zheng, Y. Prediction of lymph node status in patients with early-stage cervical cancer based on radiomic features of magnetic resonance imaging (MRI) images. BMC Med. Imaging 2023, 23, 101. [Google Scholar] [CrossRef]
- Senthilkumar, G.; Ramakrishnan, J.; Frnda, J.; Ramachandran, M.; Gupta, D.; Tiwari, P.; Shorfuzzaman, M.; Mohammed, M.A. Incorporating Artificial Fish Swarm in Ensemble Classification Framework for Recurrence Prediction of Cervical Cancer. IEEE Access 2021, 9, 83876–83886. [Google Scholar] [CrossRef]
- Suvanasuthi, R.; Therasakvichya, S.; Kanchanapiboon, P.; Promptmas, C.; Chimnaronk, S. Analysis of precancerous lesion-related microRNAs for early diagnosis of cervical cancer in the Thai population. Sci. Rep. 2025, 15, 142. [Google Scholar] [CrossRef]
- Du, S.; Zhao, Y.; Lv, C.; Wei, M.; Gao, Z.; Meng, X. Applying Serum Proteins and MicroRNA as Novel Biomarkers for Early-Stage Cervical Cancer Detection. Sci. Rep. 2020, 10, 9033. [Google Scholar] [CrossRef]
- Liu, Y.; Dong, T.f.; Li, P.j.; Chen, L.b.; Song, T. MRI-based radiomics features for the non-invasive prediction of FIGO stage in cervical carcinoma: A multi-center study. Magn. Reson. Imaging 2024, 110, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Lei, X.; Zhang, L.; Zheng, Q.; Jin, J.; Xie, C.; Jin, X.; Ai, Y. The Influence of Different Ultrasonic Machines on Radiomics Models in Prediction Lymph Node Metastasis for Patients with Cervical Cancer. Technol. Cancer Res. Treat. 2022, 21, 15330338221118412. [Google Scholar] [CrossRef]
- Ye, Y.; Li, M.; Pan, Q.; Fang, X.; Yang, H.; Dong, B.; Yang, J.; Zheng, Y.; Zhang, R.; Liao, Z. Machine learning-based classification of deubiquitinase USP26 and its cell proliferation inhibition through stabilizing KLF6 in cervical cancer. Comput. Biol. Med. 2024, 168, 107745. [Google Scholar] [CrossRef]
- Guo, L.; Wang, W.; Xie, X.; Wang, S.; Zhang, Y. Machine learning-based models for genomic predicting neoadjuvant chemotherapeutic sensitivity in cervical cancer. Biomed. Pharmacother. 2023, 159, 114256. [Google Scholar] [CrossRef]
- Ma, J.H.; Huang, Y.; Liu, L.Y.; Feng, Z. An 8-gene DNA methylation signature predicts the recurrence risk of cervical cancer. J. Int. Med. Res. 2021, 49, 03000605211018443. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, K.; Agarwal, P.; Ahuja, V.; Mir, B.A.; Yuriy, S.; Altuwairiqi, M.; Nuagah, S.J. Biomedical Application of Identified Biomarkers Gene Expression Based Early Diagnosis and Detection in Cervical Cancer with Modified Probabilistic Neural Network. Contrast Media Mol. Imaging 2022, 2022, 4946154. [Google Scholar] [CrossRef] [PubMed]
- Monthatip, K.; Boonnag, C.; Muangmool, T.; Charoenkwan, K. A machine learning-based prediction model of pelvic lymph node metastasis in women with early-stage cervical cancer. J. Gynecol. Oncol. 2023, 35, e17. [Google Scholar] [CrossRef]
- Félix, M.M.; Tavares, M.V.; Santos, I.P.; Batista de Carvalho, A.L.M.; Batista de Carvalho, L.A.E.; Marques, M.P.M. Cervical Squamous Cell Carcinoma Diagnosis by FTIR Microspectroscopy. Molecules 2024, 29, 922. [Google Scholar] [CrossRef]
- Kawahara, D.; Nishibuchi, I.; Kawamura, M.; Yoshida, T.; Koh, I.; Tomono, K.; Sekine, M.; Takahashi, H.; Kikuchi, Y.; Kudo, Y.; et al. Radiomic Analysis for Pretreatment Prediction of Recurrence Post-Radiotherapy in Cervical Squamous Cell Carcinoma Cancer. Diagnostics 2022, 12, 2346. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, W.; Jin, H.; Yu, B.; Zhang, H.; Ding, B.; Chen, X.; Zhang, Y.; Xia, Q.; Meng, D.; et al. Transcriptional and post-transcriptional regulation of CARMN and its anti-tumor function in cervical cancer through autophagic flux blockade and MAPK cascade inhibition. J. Exp. Clin. Cancer Res. 2024, 43, 305. [Google Scholar] [CrossRef]
- Park, S.H.; Hahm, M.H.; Bae, B.K.; Chong, G.O.; Jeong, S.Y.; Na, S.; Jeong, S.; Kim, J.C. Magnetic resonance imaging features of tumor and lymph node to predict clinical outcome in node-positive cervical cancer: A retrospective analysis. Radiat. Oncol. 2020, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Wu, H.y.; Liang, X.W.; Chen, J.L.; Li, J.; Zhang, S.; Liu, Z. Deep-learning-based radiomics of intratumoral and peritumoral MRI images to predict the pathological features of adjuvant radiotherapy in early-stage cervical squamous cell carcinoma. BMC Women’s Health 2024, 24, 182. [Google Scholar] [CrossRef]
- Cai, Z.; Li, S.; Xiong, Z.; Lin, J.; Sun, Y. Multimodal MRI-based deep-radiomics model predicts response in cervical cancer treated with neoadjuvant chemoradiotherapy. Sci. Rep. 2024, 14, 19090. [Google Scholar] [CrossRef]
- William, W.; Ware, A.; Basaza-Ejiri, A.H.; Obungoloch, J. A review of image analysis and machine learning techniques for automated cervical cancer screening from pap-smear images. Comput. Methods Programs Biomed. 2018, 164, 15–22. [Google Scholar] [CrossRef]
- Al Mudawi, N.; Alazeb, A. A Model for Predicting Cervical Cancer Using Machine Learning Algorithms. Sensors 2022, 22, 4132. [Google Scholar] [CrossRef]
- Rhee, D.J.; Jhingran, A.; Rigaud, B.; Netherton, T.; Cardenas, C.E.; Zhang, L.; Vedam, S.; Kry, S.; Brock, K.K.; Shaw, W.; et al. Automatic contouring system for cervical cancer using convolutional neural networks. Med. Phys. 2020, 47, 5648–5658. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Qin, L.H.; Xie, Y.E.; Liao, J.Y. Deep learning in CT image segmentation of cervical cancer: A systematic review and meta-analysis. Radiat. Oncol. 2022, 17, 175. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, M.; Akbari, A.; Asadi, F.; Emami, H. Cervical cancer survival prediction by machine learning algorithms: A systematic review. BMC Cancer 2023, 23, 341. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Pal, S.; Lee, S.S. From machine learning to deep learning: Advances of the recent data-driven paradigm shift in medicine and healthcare. Curr. Res. Biotechnol. 2024, 7, 100164. [Google Scholar] [CrossRef]
- Rahman, A.; Debnath, T.; Kundu, D.; Khan, M.S.I.; Aishi, A.A.; Sazzad, S.; Sayduzzaman, M.; Band, S.S.; Rahman, A.; Debnath, T.; et al. Machine learning and deep learning-based approach in smart healthcare: Recent advances, applications, challenges and opportunities. AIMS Public Health 2024, 11, 58–109. [Google Scholar] [CrossRef]
- Pandey, M.; Fernandez, M.; Gentile, F.; Isayev, O.; Tropsha, A.; Stern, A.C.; Cherkasov, A. The transformational role of GPU computing and deep learning in drug discovery. Nat. Mach. Intell. 2022, 4, 211–221. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, K.; Li, J.; Zhu, Y.; Zhang, Y. Various Frameworks and Libraries of Machine Learning and Deep Learning: A Survey. Arch. Comput. Methods Eng. 2024, 31, 1–24. [Google Scholar] [CrossRef]
- Mienye, I.D.; Swart, T.G. A Comprehensive Review of Deep Learning: Architectures, Recent Advances, and Applications. Information 2024, 15, 755. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Bai, J.; Al-Sabaawi, A.; Santamaría, J.; Albahri, A.S.; Al-dabbagh, B.S.N.; Fadhel, M.A.; Manoufali, M.; Zhang, J.; Al-Timemy, A.H.; et al. A survey on deep learning tools dealing with data scarcity: Definitions, challenges, solutions, tips, and applications. J. Big Data 2023, 10, 46. [Google Scholar] [CrossRef]
- Hakami, A. Strategies for overcoming data scarcity, imbalance, and feature selection challenges in machine learning models for predictive maintenance. Sci. Rep. 2024, 14, 9645. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, B.; Hevia-Montiel, N.; Perez-Gonzalez, J.; Haro, P. Weighted–VAE: A deep learning approach for multimodal data generation applied to experimental T. cruzi infection. PLoS ONE 2025, 20, e0315843. [Google Scholar] [CrossRef] [PubMed]
- Apicella, A.; Isgrò, F.; Prevete, R. Don’t Push the Button! Exploring Data Leakage Risks in Machine Learning and Transfer Learning. arXiv 2024, arXiv:2401.13796. [Google Scholar] [CrossRef]
- Yoshizawa, R.; Yamamoto, K.; Ohtsuki, T. Investigation of Data Leakage in Deep-Learning-Based Blood Pressure Estimation Using Photoplethysmogram/Electrocardiogram. IEEE Sens. J. 2023, 23, 13311–13318. [Google Scholar] [CrossRef]
- Tampu, I.E.; Eklund, A.; Haj-Hosseini, N. Inflation of test accuracy due to data leakage in deep learning-based classification of OCT images. Sci. Data 2022, 9, 580. [Google Scholar] [CrossRef]
- Kapoor, S.; Narayanan, A. Leakage and the reproducibility crisis in machine-learning-based science. Patterns 2023, 4, 100804. [Google Scholar] [CrossRef]
- Xu, Y.; Goodacre, R. On Splitting Training and Validation Set: A Comparative Study of Cross-Validation, Bootstrap and Systematic Sampling for Estimating the Generalization Performance of Supervised Learning. J. Anal. Test. 2018, 2, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Oeding, J.F.; Krych, A.J.; Pearle, A.D.; Kelly, B.T.; Kunze, K.N. Medical Imaging Applications Developed Using Artificial Intelligence Demonstrate High Internal Validity Yet Are Limited in Scope and Lack External Validation. Arthrosc. J. Arthrosc. Relat. Surg. 2025, 41, 455–472. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.S.; Amorim-Lopes, M. Externally validated and clinically useful machine learning algorithms to support patient-related decision-making in oncology: A scoping review. BMC Med. Res. Methodol. 2025, 25, 45. [Google Scholar] [CrossRef] [PubMed]
- Shaharuddin, S.; Abdul Maulud, K.N.; Syed Abdul Rahman, S.A.F.; Che Ani, A.I.; Pradhan, B. The role of IoT sensor in smart building context for indoor fire hazard scenario: A systematic review of interdisciplinary articles. Internet Things 2023, 22, 100803. [Google Scholar] [CrossRef]
- Woldaregay, A.Z.; Årsand, E.; Walderhaug, S.; Albers, D.; Mamykina, L.; Botsis, T.; Hartvigsen, G. Data-driven modeling and prediction of blood glucose dynamics: Machine learning applications in type 1 diabetes. Artif. Intell. Med. 2019, 98, 109–134. [Google Scholar] [CrossRef]
- Lawan, A.A.; Cavus, N.; Yunusa, R.; Abdulrazak, U.I.; Tahir, S. Chapter 12—Fundamentals of machine-learning modeling for behavioral screening and diagnosis of autism spectrum disorder. In Neural Engineering Techniques for Autism Spectrum Disorder; El-Baz, A.S., Suri, J.S., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 253–268. [Google Scholar] [CrossRef]
- Viering, T.; Loog, M. The Shape of Learning Curves: A Review. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 7799–7819. [Google Scholar] [CrossRef]
- Taylor, M.J. Legal Bases For Disclosing Confidential Patient Information for Public Health: Distinguishing Between Health Protection And Health Improvement. Med. Law Rev. 2015, 23, 348–374. [Google Scholar] [CrossRef] [PubMed]
- Sharif, M.I.; Mehmood, M.; Uddin, M.P.; Siddique, K.; Akhtar, Z.; Waheed, S. Federated Learning for Analysis of Medical Images: A Survey. J. Comput. Sci. 2024, 20, 1610–1621. [Google Scholar] [CrossRef]
- Liu, X.; Xie, L.; Wang, Y.; Zou, J.; Xiong, J.; Ying, Z.; Vasilakos, A.V. Privacy and Security Issues in Deep Learning: A Survey. IEEE Access 2021, 9, 4566–4593. [Google Scholar] [CrossRef]
- Golla, G. Security and Privacy Challenges in Deep Learning Models. arXiv 2023, arXiv:2311.13744. [Google Scholar] [CrossRef]
- Meng, H.; Wagner, C.; Triguero, I. Explaining time series classifiers through meaningful perturbation and optimisation. Inf. Sci. 2023, 645, 119334. [Google Scholar] [CrossRef]
- Teng, Q.; Liu, Z.; Song, Y.; Han, K.; Lu, Y. A survey on the interpretability of deep learning in medical diagnosis. Multimed. Syst. 2022, 28, 2335–2355. [Google Scholar] [CrossRef] [PubMed]
- Miller, T. Explanation in artificial intelligence: Insights from the social sciences. Artif. Intell. 2019, 267, 1–38. [Google Scholar] [CrossRef]
- Beam, A.L.; Manrai, A.K.; Ghassemi, M. Challenges to the Reproducibility of Machine Learning Models in Health Care. JAMA 2020, 323, 305–306. [Google Scholar] [CrossRef]
- Mislan, K.A.S.; Heer, J.M.; White, E.P. Elevating The Status of Code in Ecology. Trends Ecol. Evol. 2016, 31, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Torres-Poveda, K.; Piña-Sánchez, P.; Vallejo-Ruiz, V.; Lizano, M.; Cruz-Valdez, A.; Juárez-Sánchez, P.; Garza-Salazar, J.d.l.; Manzo-Merino, J. Molecular Markers for the Diagnosis of High-Risk Human Papillomavirus Infection and Triage of Human Papillomavirus-Positive Women. Rev. Investig. Clín. 2020, 72, 198–212. [Google Scholar] [CrossRef]
- Goodman, A. Deep-Learning-Based Evaluation of Dual Stain Cytology for Cervical Cancer Screening: A New Paradigm. J. Natl. Cancer Inst. 2021, 113, 1451–1452. [Google Scholar] [CrossRef]
- Desai, K.T.; Befano, B.; Xue, Z.; Kelly, H.; Campos, N.G.; Egemen, D.; Gage, J.C.; Rodriguez, A.C.; Sahasrabuddhe, V.; Levitz, D.; et al. The development of “automated visual evaluation” for cervical cancer screening: The promise and challenges in adapting deep-learning for clinical testing: Interdisciplinary principles of automated visual evaluation in cervical screening. Int. J. Cancer 2022, 150, 741–752. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Considerations for the Use of Artificial Intelligence to Support Regulatory Decision-Making for Drug and Biological Products. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/considerations-use-artificial-intelligence-support-regulatory-decision-making-drug-and-biological (accessed on 28 May 2025).
- World Health Organization (WHO). Atlas of Visual Inspection of the Cervix with Acetic Acid for Screening, Triage, and Assessment for Treatment. Available online: https://screening.iarc.fr/atlasvia.php (accessed on 28 May 2025).
- Li, J.; Hu, P.; Gao, H.; Shen, N.; Hua, K. Classification of cervical lesions based on multimodal features fusion. Comput. Biol. Med. 2024, 177, 108589. [Google Scholar] [CrossRef]
- Mennella, C.; Maniscalco, U.; De Pietro, G.; Esposito, M. Ethical and regulatory challenges of AI technologies in healthcare: A narrative review. Heliyon 2024, 10, e26297. [Google Scholar] [CrossRef]
- Wu, T.; Lucas, E.; Zhao, F.; Basu, P.; Qiao, Y. Artificial intelligence strengthens cervical cancer screening - present and future. Cancer Biol. Med. 2024, 21, 864–879. [Google Scholar] [CrossRef] [PubMed]

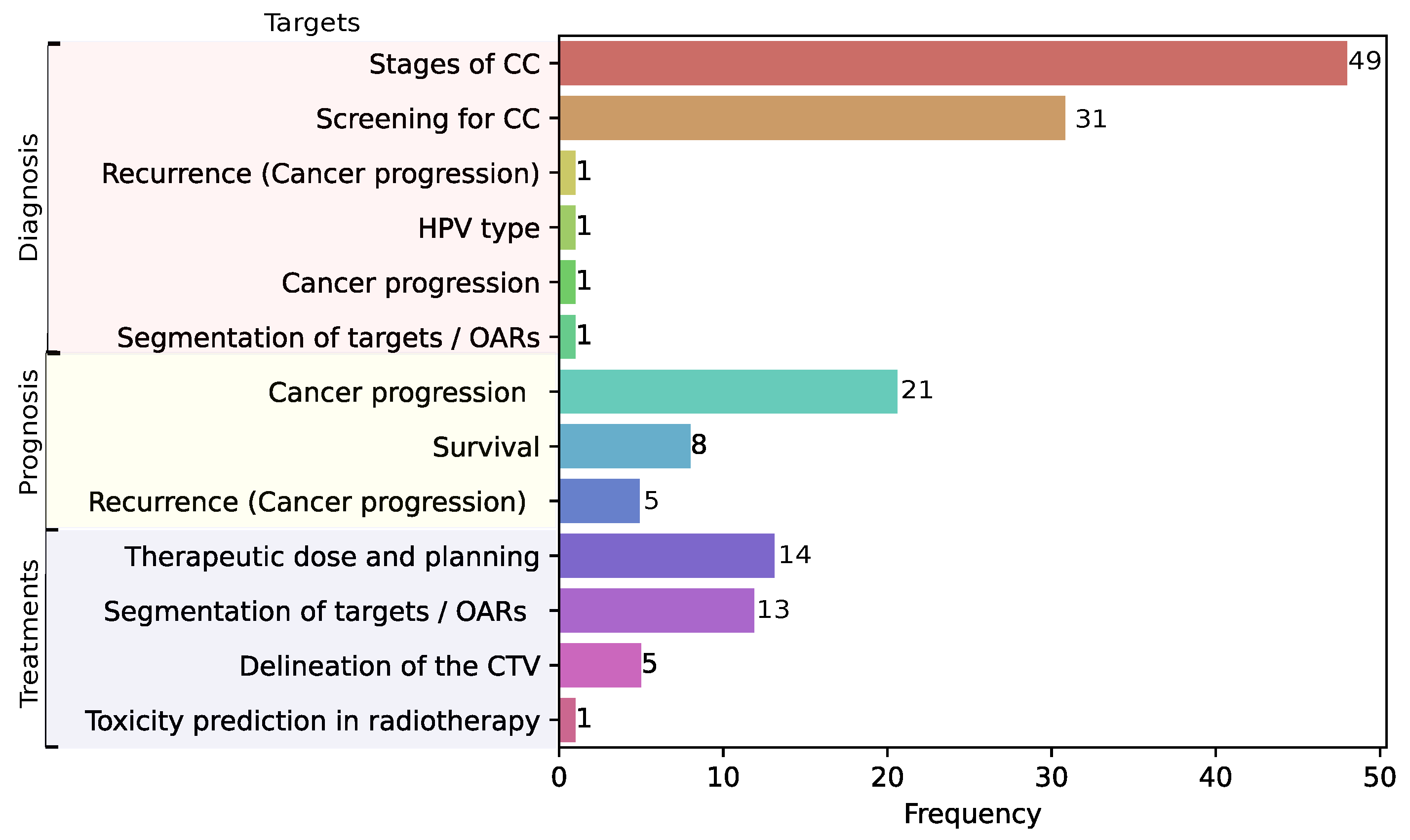
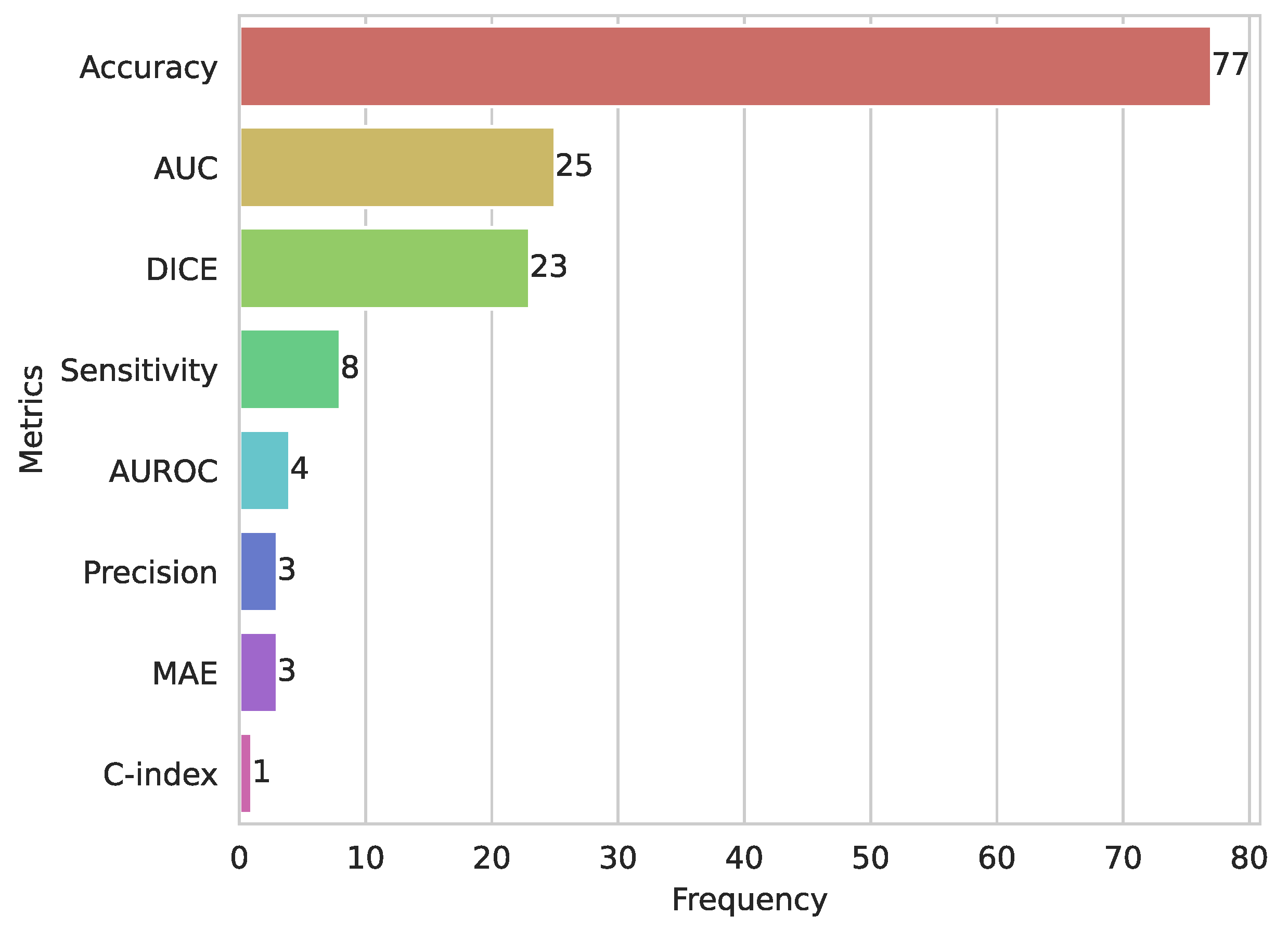


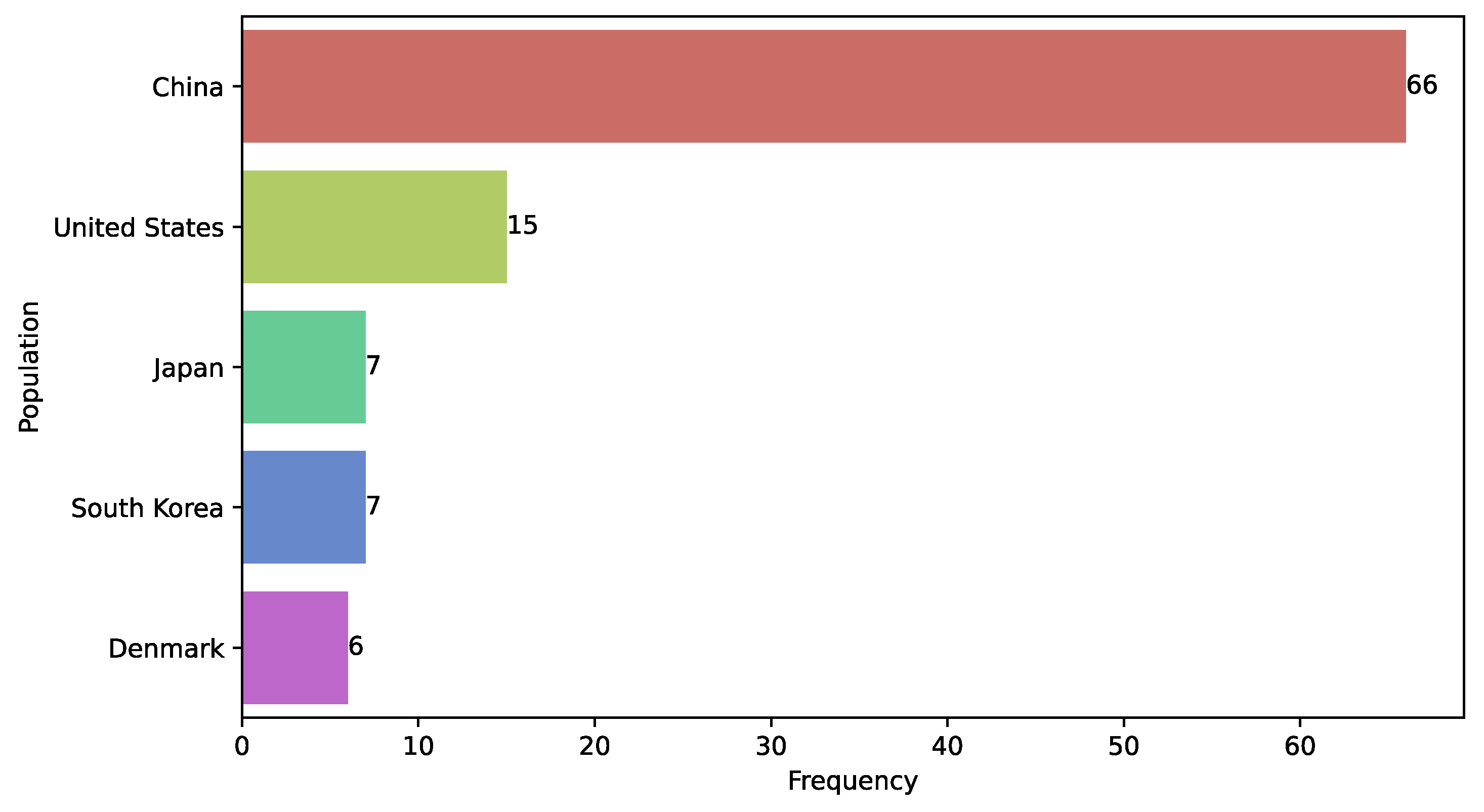


| Section | Question |
|---|---|
| Introduction | Q1
Is the scientific background adequately described? Q2 Are the goals clearly outlined? |
| Methods | Q3 What is the study design? (1 cross-sectional; 2 case–control; 3 cohort study; 4 clinical trial) Q4 Are the inclusion criteria and participant selection clearly outlined? Q5 Sample size (0 if <20, 1 if between 20 and 100, 2 if >100) Q6 Is the method (validity) explained? Q7 Are the statistical analyses suitable? |
| Results | Q8 Are subjects’ characteristics provided? Q9 Are the results understandable? |
| Discussion | Q10 Are the study results compared and discussed in relation to other studies published in the literature? Q11 Are study limitations discussed? |
| Ref. | Q1 | Q2 | Q3 | Q4 | Q5 | Q6 | Q7 | Q8 | Q9 | Q10 | Q11 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kim et al. 2022 [36] | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 17 |
| Yuan et al. 2022 [37] | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 10 |
| Kruczkowski et al. 2022 [38] | 2 | 2 | 1 | 2 | 2 | 1 | 0 | 1 | 1 | 1 | 0 | 13 |
| Yoganathan et al [32] | 1 | 2 | 1 | 1 | 1 | 2 | 0 | 2 | 2 | 2 | 0 | 14 |
| Fu et al. 2022 [39] | 1 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 1 | 0 | 16 |
| Ma et al. 2022 [40] | 2 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 20 |
| Liu et al. 2022 [41] | 1 | 2 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 11 |
| Nambu et al. 2022 [42] | 2 | 2 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 18 |
| Drokow et al. 2022 [43] | 1 | 2 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 11 |
| Liu et al. 2022 [44] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 2 | 17 |
| Mehmood et al. 2021 [45] | 2 | 2 | 1 | 0 | 2 | 1 | 1 | 0 | 2 | 1 | 0 | 12 |
| Ali et al. 2021 [46] | 1 | 2 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 11 |
| Chu et al. 2021 [47] | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 23 |
| Dong et al. 2021 [48] | 1 | 2 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 11 |
| Fick et al. 2021 [49] | 1 | 2 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 11 |
| Chen et al. 2021 [50] | 2 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 2 | 0 | 13 |
| Cheng et al. 2021 [51] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 0 | 15 |
| Rahaman et al. 2021 [52] | 2 | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 14 |
| Park et al. 2021 [53] | 2 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 19 |
| Tian et al. 2021 [54] | 2 | 2 | 1 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | 0 | 10 |
| Da-ano et al. 2021 [55] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 18 |
| Kaushik et al. 2021 [56] | 2 | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 12 |
| Shi et al. 2021 [57] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Ding et al. 2021 [58] | 2 | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 12 |
| Zhu et al. 2021 [59] | 2 | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 12 |
| Chandran et al. 2021 [60] | 2 | 2 | 1 | 0 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 15 |
| Jian et al. 2021 [61] | 2 | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Christley et al. 2021 [62] | 2 | 2 | 3 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 17 |
| Ke et al. 2021 [63] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Rigaud et al. 2021 [64] | 2 | 2 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 16 |
| Urushibara et al. 2021 [65] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Wentzensen et al. 2021 [66] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Wang et al. 2020 [67] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Liu et al. 2020 [68] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Mao et al. 2020 [69] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Xue et al. 2020 [70] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Bao et al. 2020 [71] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Cho et al. 2020 [72] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Ju et al. 2020 [73] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Kanai et al. 2020 [74] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Yuan et al. 2020 [75] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Hu et al. 2020 [76] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Wu et al. 2020 [77] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Wang et al. 2020 [78] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 16 |
| Kudva et al. 2020 [79] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Ijaz et al. 2020 [80] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Bae et al. 2020 [81] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Sornapudi et al. 2020 [82] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Shao et al. 2020 [83] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Takada et al. 2020 [84] | 2 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 16 |
| Lin et al. 2020 [85] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Jihong et al- 2020 [86] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Liu et al. 2020 [87] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Shen et al. 2019 [88] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Pathania et al. 2019 [89] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Aljakouch et al. 2019 [90] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Shanthi et al. 2019 [22] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Dong et al. 2019 [91] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Hu et al. 2019 [92] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Dong et al. 2019 [93] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Geetha et al. 2019 [94] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Shen et al. 2019 [95] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Zhang et al. 2019 [96] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Chen et al. 2019 [97] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Matsuo et al. 2019 [98] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 16 |
| Araujo et al. 2019 [99] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Kan et al. 2019 [100] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 16 |
| Zhen et al. 2017 [101] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 16 |
| Xie et al. 2018[102] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Kudva et al. 2018 [103] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Wei et al. 2017 [104] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Guo et al. 2016 [105] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Zhao et al. 2016 [106] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Weegar et al. 2015 [107] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 16 |
| Kahng et al. 2015 [108] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 16 |
| Mu et al. 2015 [109] | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 12 |
| Zhi et al. 2015 [110] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Dong et al. 2015 [111] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Mariarputham et al. 2015 [112] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Xin et al. 2023 [113] | 2 | 2 | 4 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 17 |
| Robinson et al. 2023 [114] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Tian et al. 2023 [115] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Kaur et al. 2023 [116] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Ji et al. 2023 [117] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Liu et al. 2023 [118] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Kang et al. 2023 [119] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Wu et al. 2023 [120] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Ince et al. 2023 [121] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Kurita et al. 2023 [122] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Zhu et al. 2023 [123] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Yang et al. 2023 [124] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Devi et al. 2023 [125] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Chen et al. 2023 [126] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Salehi et al. 2023 [127] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Yu et al. 2023 [128] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 16 |
| Liu et al. 2023 [129] | 2 | 2 | 4 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 17 |
| Wang et al. 2023 [130] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Yu et al. 2023 [131] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Huang et al. 2023 [132] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Cao et al. 2023 [133] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Chen et al. 2022 [134] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| He et al. 2023 [135] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Qian et al. 2022 [136] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Dong et al. 2022 [137] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Ji et al. 2022 [138] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Ming et al. 2022 [139] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Wang et al. 2022 [140] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Ma et al. 2022 [141] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Chang et al. 2022 [142] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Tao et al. 2022 [143] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Qilin et al. 2022 [144] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Xue et al. 2023 [145] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Gay et al. 2023 [146] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Shen et al. 2024 [147] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
| Aljrees et al. 2024 [148] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Munshi et al. 2024 [149] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Jeong et al. 2024 [150] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Stegmuller et al. 2023 [151] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 13 |
| Wu et al. 2024 [152] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Wu et al. 2025 [153] | 2 | 2 | 4 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 18 |
| Xin et al. 2024 [154] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Chen et al. 2018 [155] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 14 |
| Chen et al. 2020 [156] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Matsuo et al. 2019 [98] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Zheng et al. 2024 [157] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Brenes et al. 2024 [158] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Shandilya et al. 2024 [159] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Mathivanan et al. 2024 [160] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Wang et al. 2021 [161] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Dong et al. 2025 [162] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Xiao et al. 2023 [163] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| He et al. 2024 [164] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Wang et al. 2024 [165] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Wu et al. 2021 [166] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Namalinzi et al. 2024 [167] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Liu et al. 2023 [168] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Senthilkumar et al. 2021 [169] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Suvanasuthi et al. 2025 [170] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 14 |
| Du et al. 2020 [171] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Liu et al. 2024 [172] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Yi et al. 2022 [173] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Ye et al. 2024 [174] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Guo et al. 2023 [175] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Hang et al. 2021 [176] | 2 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 16 |
| Ramesh et al. 2022 [177] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Monthatip et al. 2023 [178] | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 15 |
| Felix et al. 2024 [179] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 14 |
| Kawahara et al. 2022 [180] | 2 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 16 |
| Zhang et al. 2024 [181] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 14 |
| Park et al. 2020 [182] | 2 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 16 |
| Kruczkowski et al. 2022 [38] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 14 |
| Zhang et al. 2024 [183] | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 14 |
| Cai et al. 2024 [184] | 2 | 2 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 17 |
| Ref. | Year | Clinical Application | Prediction Task | Target | Datasets | No. Folds for CV | Best Performance on Test Set | Extval | |
|---|---|---|---|---|---|---|---|---|---|
| Model | Metric | ||||||||
| [170] | 2025 | Diagnosis | Classification | Screening for CC | DNA | 10 | RF | ACC = 90.9% | - |
| [148] | 2024 | Diagnosis | Classification | Screening for CC | Clinical history | 5 | KNN | ACC = 99% | - |
| [149] | 2024 | Diagnosis | Classification | Screening for CC | Clinical history | 5 | SVM | ACC = 99% | - |
| [154] | 2024 | Prognosis | Classification | Survival | MRI images | - | RF | ACC = 86% | - |
| [164] | 2024 | Prognosis | Classification | Cancer progression | Clinical history | 10 | RF | ACC = 86% | - |
| [165] | 2024 | Treatment | Classification | Recurrence (Cancer progression) | DNA | 10 | RF | ACC = 84% | - |
| [167] | 2024 | Diagnosis | Classification | Screening for CC | Clinical history | - | RF | ACC = 90% | - |
| [172] | 2024 | Prognosis | Classification | Screening for CC | MRI images | 10 | SVM | AUC = 76% | Yes |
| [174] | 2024 | Treatment | Classification | Therapeutic dose and planning | DNA | 10 | RF | ACC = 96% | - |
| [179] | 2024 | Diagnosis | Classification | Stages of CC | Dose volume | 5 | SVM | ACC = 96% | - |
| [181] | 2024 | Prognosis | Classification | Stages of CC | DNA | - | Lightgbm | AUC = 98.7% | - |
| [113] | 2023 | Prognosis | Classification | Cancer progression | MRI images | 5 | SVM | ACC = 90% | Yes |
| [121] | 2023 | Diagnosis | Classification | Stages of CC | MRI images | 5 | SVM | ACC > 83% | - |
| [123] | 2023 | Diagnosis | Classification | Stages of CC | Spectral data | 10 | SVM | ACC = 95% | - |
| [124] | 2023 | Treatment | Classification | Therapeutic dose and planning | CT images | - | AAA | ACC = 73% | - |
| [125] | 2023 | Diagnosis | Classification | Stages of CC | Clinical history | 5 | LR DT | ACC > 88% | - |
| [128] | 2023 | Prognosis | Regression | Survival | Histopatology images Clinical history | 5 | XGBoost | AUC = 83% | - |
| [135] | 2023 | Diagnosis | Classification | Stages of CC | Histopathology images | - | SVM | ACC > 87% | - |
| [168] | 2023 | Prognosis | Classification | Cancer progression | MRI images Clinical history | 5 | MNB | ACC = 77% | - |
| [175] | 2023 | Treatment | Classification | Cancer progression | DNA | - | RF | - | - |
| [178] | 2023 | Prognosis | Classification | Cancer progression | CT images Clinical history | 10 | SVM | ACC = 90.1% | - |
| [38] | 2022 | Diagnosis | Classification | Stages of CC | Interferometry | 3 | NB | ACC = 92% | - |
| [39] | 2022 | Diagnosis | Classification | Screening for CC | Colposcopy images Cytology images HPV test | - | MLR | AUC = 92.1% | - |
| [138] | 2022 | Diagnosis | Classification | Stages of CC | Fluorescence images | - | KNN | SEN = 90% | - |
| [173] | 2022 | Prognosis | Classification | Cancer progression | Ultrasound | - | SVM | ACC = 85% | - |
| [177] | 2022 | Diagnosis | Classification | Stages of CC | DNA | - | SVM | ACC = 91.5% | - |
| [180] | 2022 | Prognosis | Classification | Recurrence (Cancer progression) | MRI images | 5 | LASSO | ACC = 93.1% | - |
| [38] | 2022 | Prognosis | Classification | Stages of CC | Dose volume | 3 | NB | ACC = 92% | - |
| [46] | 2021 | Diagnosis | Classification | Screening for CC | Biopsy results Cytology images | - | RT RF KNN | ACC = 95.5% | - |
| [47] | 2021 | Prognosis | Classification | Cancer progression | Histopathology images Clinical history | 5 | LR SVM | AUC = 88% | - |
| [55] | 2021 | Prognosis | Classification | Surv forest | MRI images CT images Clinical history | 10 | RF | AUC > 84% | - |
| [56] | 2021 | Prognosis | Classification | Cancer progression | DNA | 5 | Ridge | ACC = 84.7% | - |
| [58] | 2021 | Prognosis | Classification | Survival | DNA | 10 | SVM | AUC > 91% | - |
| [62] | 2021 | Diagnosis | Classification | Cancer progression | DNA | LOOCV | LR | ACC = 95% | - |
| [166] | 2021 | Diagnosis | Classification | Screening for CC | Ultrasound | - | LR | AUC = 91% | Yes |
| [176] | 2021 | Prognosis | Classification | Recurrence (Cancer progression) | DNA | - | SVM | ACC = 83% | Yes |
| [74] | 2020 | Diagnosis | Classification | Stages of CC | Cytology images | 10 | LSVM | ACC = 84% | - |
| [78] | 2020 | Prognosis | Classification | Cancer progression | MRI images | 10 | SVM | C-index 0.96 | - |
| [80] | 2020 | Diagnosis | Classification | Screening for CC | Clinical history | 10 | RF | ACC > 95% | - |
| [81] | 2020 | Diagnosis | Classification | Stages of CC | Colposcopy images | 10 | KNN | ACC = 80% | - |
| [156] | 2020 | Prognosis | Classification | Cancer progression | CT iimages | - | SVM | ACC = 76% | - |
| [171] | 2020 | Diagnosis | Classification | Screening for CC | DNA | 10 | DT | SEN = 88.6% | Yes |
| [182] | 2020 | Prognosis | Classification | Survival | MRI images | - | SF | AUC = 79.6% | - |
| [94] | 2019 | Diagnosis | Classification | Screening for CC | Clinical history | 10 | RF | ACC > 94% | - |
| [100] | 2019 | Prognosis | Classification | Cancer progression | MRI images Clinical history | - | SVM | AUROC = 75% | - |
| [104] | 2017 | Diagnosis | Classification | Stages of CC | Histopathology images | - | SVM | ACC > 90% | - |
| [105] | 2016 | Diagnosis | Classification | Stages of CC | Histopathology images | 10 | SVM | ACC = 88.5% | - |
| [106] | 2016 | Diagnosis | Classification | Screening for CC | Cytology images | 10 | SVM | ACC = 98% | - |
| [108] | 2015 | Prognosis | Classification | Cancer progression | Clinical history | 10 | SVM | ACC = 74% | - |
| [109] | 2015 | Treatment | Image segmentation | Segmentation of targets/OARs | CT images | LOOCV | SVM | DSC = 91.78 | - |
| [111] | 2015 | Prognosis | Classification | Cancer progression | DNA | 5 | SVM | ACC = 80% | - |
| [112] | 2015 | Diagnosis | Classification | Stages of CC | Cytology images | 10 | SVM | Precision >87% | - |
| Ref. | Year | Clinical Application | Prediction Task | Target | Datasets | No. Folds For CV | Best Performance on Test Set | Externalval | |
|---|---|---|---|---|---|---|---|---|---|
| Model | Metric | ||||||||
| [147] | 2024 | Treatment | Classification | Therapeutic dose and planning | CT images Treatment plan Dose volume | - | U-Net | AUC = 94% | - |
| [150] | 2024 | Prognosis | Classification | Cancer progression | MRI images | 5 | CNN | ACC = 78% | - |
| [152] | 2024 | Prognosis | Regression | Cancer progression | CT images | 5 | DNN | ACC = 75% | - |
| [153] | 2024 | Treatment | Image Segmentation | Delineation of the CTV | CT images | - | ResCANet | DSC = 74.8 | Yes |
| [157] | 2024 | Diagnosis | Classification | Stages of CC | DNA | - | EfficientNet DenseNet InceptionNet | ACC = 94.4% | - |
| [158] | 2024 | Diagnosis | Image Segmentation | Screening for CC | Colposcopy images | - | Efficient U-Net LSTM-Attention | ACC = 87% | - |
| [159] | 2024 | Diagnosis | Classification | Stages of CC | Cytology images | - | CNN | ACC = 99.11% | - |
| [183] | 2024 | Treatment | Classification | Therapeutic dose and planning | MRI images | 10 | ResNet101 MLP | AUC = 87% | - |
| [114] | 2023 | Diagnosis | Image Segmentation | Stages of CC | Interferometric Measurements | LOOCV | CNN | ACC = 81% | - |
| [115] | 2023 | Treatment | Image Segmentation | Delineation of the CTV | CT images | 5 | AFN | DSC > 88 | - |
| [116] | 2023 | Diagnosis | Classification | Stages of CC | Cytology images | - | MLNet | ACC > 99% | Yes |
| [117] | 2023 | Diagnosis | Image Segmentation | Screening for CC | Cytology images | - | CNN | DSC = 0.94 | - |
| [118] | 2023 | Treatment | Classification | Therapeutic dose and planning | Histopathology images | - | ViT RNN | ACC = 90% | Yes |
| [119] | 2023 | Diagnosis | Classification | Stages of CC | Spectral data | 5 | CNN | ACC = 94% | - |
| [120] | 2023 | Diagnosis | Classification | Stages of CC | Spectral data | LOOCV | DBN | ACC = 93% | - |
| [122] | 2023 | Diagnosis | Classification | Stages of CC | Cytology images | - | CNN | ACC = 87% | - |
| [126] | 2023 | Diagnosis | Classification | Stages of CC | Colposcopy images | - | Efficient Net GRU | ACC = 91% | - |
| [127] | 2023 | Treatment | Image Segmentation | Therapeutic dose and planning | CT images | - | CNN | Jaccard > 0.86 | - |
| [129] | 2023 | Diagnosis | Classification | Screening for CC | Histopatology images | 5 | CNN | ACC = 76% | - |
| [130] | 2023 | Treatment | Image Segmentation | Segmentation of targets/OARs | CT images | - | CNN | DSC = 0.77 | Yes |
| [131] | 2023 | Treatment | Regression | Therapeutic dose and planning | CT images | - | 3DResUnet | Dose = 5% | - |
| [132] | 2023 | Diagnosis | Image Segmentation | Segmentation of targets/OARs | CT images | 4 | CNN | DSC = 0.80 | - |
| [133] | 2023 | Diagnosis | Classification | Stages of CC | Colposcopy images | - | CNN | ACC > 88% | - |
| [145] | 2023 | Diagnosis | Classification | Stages of CC | Spectral data | LOOCV | DeepLabv3+ D-LinkNet | DSC = 0.95 | - |
| [146] | 2023 | Treatment | Regression | Therapeutic dose and planning | CT images | - | U-Net | Score > 1% | - |
| [163] | 2023 | Diagnosis | Classification | Stages of CC | Clinical history | - | Stacking models | AUROC = 87% | - |
| [36] | 2022 | Diagnosis | Classification | Stages of CC | Colposcopy images | 5 | ResNet50 | ACC = 81.3% | - |
| [37] | 2022 | Treatment | Regression | Therapeutic dose and planning | CT images | - | ResNet | DSC > 0.94 | - |
| [32] | 2022 | Treatment | Image Segmentation | Segmentation of targets/OARs | MRI images | - | Inception ResNetv2 | DSC = 0.72 | - |
| [40] | 2022 | Treatment | Image Segmentation | Segmentation of targets/OARs | CT images | - | CNN | DSC > 0.70 | - |
| [41] | 2022 | Diagnosis | Classification | Screening for CC | Cytology images | 5 | CNN | ACC = 95.4% | - |
| [42] | 2022 | Diagnosis | Classification | Screening for CC | Cytology images | - | YOLO ResNet | ACC = 90.5% | - |
| [43] | 2022 | Diagnosis | Classification | Screening for CC | Clinical history | 5 | Voting | ACC = 96.6% | - |
| [44] | 2022 | Diagnosis | Image Segmentation | Screening for CC | Colposcopy images | - | DeepLab V3+ | ACC = 91.2% | - |
| [134] | 2022 | Prognosis | Regression | Survival | Histopathology images | - | CNN | AUC = 80% | - |
| [136] | 2022 | Prognosis | Regression | Cancer progression | MRI images PET images | 5 | CNN | AUC = 84% | - |
| [137] | 2022 | Prognosis | Classification | Cancer progression | Clinical history | - | MLP | AUC = 82% | - |
| [139] | 2022 | Diagnosis | Classification | Stages of CC | CT images | 5 | CNN | ACC > 60% | - |
| [140] | 2022 | Treatment | Image Segmentation | Segmentation of targets/OARs | CT images | - | CNN | DSC = 0.88 | - |
| [141] | 2022 | Treatment | Image Segmentation | Delineation of the CTV | CT images | - | VN-Net | DSC = 0.81 | - |
| [142] | 2022 | Diagnosis | Classification | Stages of CC | MRI images | 5 | CycleGAN | AUC = 89% | - |
| [143] | 2022 | Diagnosis | Classification | Stages of CC | Cytology images | - | CNN | SEN = 89% | - |
| [144] | 2022 | Treatment | Regression | Therapeutic dose and planning | Dose volume | - | U-Net | MAE = 2.4 | Yes |
| [161] | 2022 | Treatment | Image Segmentation | Segmentation of targets/OARs | MRI images | - | CNN | Precision = 93% | - |
| [162] | 2022 | Diagnosis | Classification | Stages of CC | Colposcopy images | - | Dense-U-Net | ACC = 89% | Yes |
| [50] | 2021 | Diagnosis | Image Segmentation | Screening for CC | Cytology images | - | CNN | DSC = 0.92 | - |
| [51] | 2021 | Diagnosis | Classification | Stages of CC | Cytology images | - | ResNet RNN CNN | SEN = 95.1% | Yes |
| [52] | 2021 | Diagnosis | Classification | Stages of CC | Cytology images | - | ResNet | ACC > 90% | Yes |
| [53] | 2021 | Diagnosis | Classification | Stages of CC | Colposcopy images | 5 | ResNet | AUC = 97% | - |
| [54] | 2021 | Diagnosis | Classification | HPV type | DNA | - | CNN | AUROC = 85% | Yes |
| [57] | 2021 | Treatment | Image Segmentation | Delineation of the CTV | CT images | 3 | U-Net CNN | DSC = 0.734 | Yes |
| [59] | 2021 | Diagnosis | Image Segmentation | Screening for CC | Cytology images | 10 | YOLO v3 | SEN = 92% | Yes |
| [60] | 2021 | Diagnosis | Classification | Screening for CC | Colposcopy images | - | CNN | ACC = 92% | - |
| [61] | 2021 | Prognosis | Classification | Cancer progression | MRI images | 10 | CNN | AUC = 91% | - |
| [63] | 2021 | Diagnosis | Classification | Screening for CC | Cytology images | 5 | CNN | ACC = 94% | - |
| [64] | 2021 | Treatment | Image Segmentation | Segmentation of targets/OARs | CT images | - | CNN | DSC = 0.85 | Yes |
| [65] | 2021 | Diagnosis | Classification | Stages of CC | MRI images | - | XceptionNet | AUC = 93% | - |
| [66] | 2021 | Diagnosis | Classification | Screening for CC | Cytology images | 5 | CNN | AUC = 77% | - |
| [67] | 2020 | Treatment | Image Segmentation | Delineation of the CTV | CT images | - | CNN | DSC > 0.81 | - |
| [68] | 2020 | Treatment | Image Segmentation | Therapeutic dose and planning | CT images | 5 | CNN | DSC > 0.82 | - |
| [69] | 2020 | Treatment | Image Segmentation | Therapeutic dose and planning | CT images Dose volume | - | CNN | DVH = 0.73 | - |
| [70] | 2020 | Diagnosis | Classification | Screening for CC | Colposcopy images | - | Faster R-CNN | AUC > 90% | - |
| [71] | 2020 | Diagnosis | Classification | Stages of CC | Cytology images | - | CNN | SEN = 100% | - |
| [72] | 2020 | Diagnosis | Classification | Stages of CC | Colposcopy images | 10 | Resnet | AUC = 78% | - |
| [73] | 2020 | Treatment | Image Segmentation | Segmentation of targets/OARs | CT images | - | CNN | DSC > 0.87 | - |
| [75] | 2020 | Diagnosis | Image Segmentation | Stages of CC | Colposcopy images | - | CNN | ACC = 84% | - |
| [76] | 2020 | Diagnosis | Classification | Screening for CC | Colposcopy images | - | RetinaNet | AUC = 95% | - |
| [77] | 2020 | Prognosis | Image Segmentation | Cancer progression | MRI images | - | CNN | AUC = 93% | - |
| [79] | 2020 | Diagnosis | Classification | Screening for CC | Colposcopy images | - | CNN | ACC = 91% | - |
| [82] | 2020 | Diagnosis | Classification | Stages of CC | Cytology images | - | VGG-19 | ACC = 95% | - |
| [83] | 2020 | Diagnosis | Classification | Stages of CC | MRI images Treatment plan | LOOCV | CNN | ACC = 94.3% | - |
| [85] | 2020 | Treatment | Image Segmentation | Therapeutic dose and planning | MRI images | 5 | UNet | SEN = 89% | - |
| [86] | 2020 | Treatment | Classification | Therapeutic dose and planning | Dose volume | - | CNN | p < 0.017 | - |
| [87] | 2020 | Treatment | Image Segmentation | Segmentation of targets/OARs | CT images | - | UNet | DSC > 0.791 | Yes |
| [88] | 2019 | Prognosis | Classification | Cancer progression | CT images | 7 | CNN | ACC = 89% | - |
| [89] | 2019 | Diagnosis | Classification | Stages of CC | Interferometric Measurements | - | CNN | SEN = 100% | - |
| [90] | 2019 | Diagnosis | Classification | Stages of CC | Spectral data | LOOCV | CNN | ACC = 100% | - |
| [22] | 2019 | Diagnosis | Classification | Stages of CC | Cytology images | - | CNN | ACC > 94% | - |
| [91] | 2019 | Diagnosis | Classification | Stages of CC | Cytology images | 10 | AGVFSM | ACC = 99% | - |
| [92] | 2019 | Diagnosis | Classification | Screening for CC | Colposcopy images | - | CNN | AUC = 91% | - |
| [93] | 2019 | Diagnosis | Classification | Stages of CC | CT images | - | CNN | ACC > 90% | - |
| [95] | 2019 | Treatment | Regression | Therapeutic dose and planning | Dose volume Histograms | - | Reinforcement learning | Score > 8.5% | Yes |
| [96] | 2019 | Diagnosis | Image Segmentation | Screening for CC | Cytology images | 5 | CNN | ACC = 91% | - |
| [97] | 2019 | Treatment | Image Segmentation | Segmentation of targets/OARs | CT images | 5 | CNN | DSC = 0.84 | - |
| [98] | 2019 | Prognosis | Regression | Survival | Clinical history | 5 | FFNN | MAE = 29.3 | - |
| [99] | 2019 | Diagnosis | Image Segmentation | Screening for CC | Cytology images | - | CNN | MAP = 0.936 | - |
| [98] | 2019 | Prognosis | Regression | Survival | Clinical history | - | FFNN | MAE = 29.3 | - |
| [102] | 2018 | Diagnosis | Classification | Stages of CC | Cytology images | - | CNN | Precision > 89% | Yes |
| [103] | 2018 | Diagnosis | Classification | Screening for CC | Colposcopy images | - | CNN | ACC = 100% | - |
| [155] | 2018 | Treatment | Image Segmentation | Segmentation of targets/OARs | CT images PET images | - | Graph cut | DSC = 0.83 | - |
| [101] | 2017 | Treatment | Classification | Toxicity prediction in radiotherapy | CT images Treatment plan | 10 | VGG16 | AUC = 89% | - |
| [107] | 2015 | Diagnosis | Classification | Screening for CC | Clinical history | - | NER | F1 = 67% | - |
| [110] | 2015 | Diagnosis | Image Segmentation | Stages of CC | Cytology images | - | GMM | DSC = 0.92 | - |
| Ref. | Year | Clinical Application | Prediction Task | Target | Datasets | No. Folds For CV | Best Performance on Test Set | Externalval | |
|---|---|---|---|---|---|---|---|---|---|
| Model | Metric | ||||||||
| [160] | 2024 | Diagnosis | Classification | Stages of CC | Cytology images | - | ResNet152 LR | ACC = 98% | - |
| [184] | 2024 | Treatment | Image segmentation | Segmentation of targets/OARs | MRI images | - | SVM - RF ResNet50 | ACC = 75% | - |
| [151] | 2023 | Diagnosis | Classification | Stages of CC | Cytology images | 4 | KNN ResNet50 ViT-S/16 | ACC > 83% | - |
| [45] | 2021 | Diagnosis | Classification | Screening for CC | Clinical history | - | RF SNN | ACC = 93.6% | - |
| [48] | 2021 | Diagnosis | Image segmentation | Stages of CC | Histopathology images | 5 | U-Net SVM | ACC = 94.4% | - |
| [49] | 2021 | Diagnosis | Classification | Stages of CC | Histopatology images | 10 | CNN SVM | ACC = 97.4% | - |
| [169] | 2021 | Prognosis | Classification | Recurrence (Cancer progression) | DNA | 10 | SVM RNN | ACC = 92% | - |
| 1. Data availability |
| 2. Data leakage |
| 3. Limited external validation |
| 4. Limited evaluation of model performance |
| 5. Complex data |
| 6. Privacy issues |
| 7. Lack of explainability |
| 8. Lack of reproducibility |
| 1. Representativeness of clinical stages of CC in the training of deep learning-based models. |
| 2. Privacy concerns and data security in health care. |
| 3. Integration of AI with clinical workflows for real-time decision-making. |
| 4. Ethical and regulatory considerations. |
| 5. Public perspectives on using AI in their diagnoses and treatment decisions. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vazquez, B.; Rojas-García, M.; Rodríguez-Esquivel, J.I.; Marquez-Acosta, J.; Aranda-Flores, C.E.; Cetina-Pérez, L.d.C.; Soto-López, S.; Estévez-García, J.A.; Bahena-Román, M.; Madrid-Marina, V.; et al. Machine and Deep Learning for the Diagnosis, Prognosis, and Treatment of Cervical Cancer: A Scoping Review. Diagnostics 2025, 15, 1543. https://doi.org/10.3390/diagnostics15121543
Vazquez B, Rojas-García M, Rodríguez-Esquivel JI, Marquez-Acosta J, Aranda-Flores CE, Cetina-Pérez LdC, Soto-López S, Estévez-García JA, Bahena-Román M, Madrid-Marina V, et al. Machine and Deep Learning for the Diagnosis, Prognosis, and Treatment of Cervical Cancer: A Scoping Review. Diagnostics. 2025; 15(12):1543. https://doi.org/10.3390/diagnostics15121543
Chicago/Turabian StyleVazquez, Blanca, Mariano Rojas-García, Jocelyn Isabel Rodríguez-Esquivel, Janeth Marquez-Acosta, Carlos E. Aranda-Flores, Lucely del Carmen Cetina-Pérez, Susana Soto-López, Jesús A. Estévez-García, Margarita Bahena-Román, Vicente Madrid-Marina, and et al. 2025. "Machine and Deep Learning for the Diagnosis, Prognosis, and Treatment of Cervical Cancer: A Scoping Review" Diagnostics 15, no. 12: 1543. https://doi.org/10.3390/diagnostics15121543
APA StyleVazquez, B., Rojas-García, M., Rodríguez-Esquivel, J. I., Marquez-Acosta, J., Aranda-Flores, C. E., Cetina-Pérez, L. d. C., Soto-López, S., Estévez-García, J. A., Bahena-Román, M., Madrid-Marina, V., & Torres-Poveda, K. (2025). Machine and Deep Learning for the Diagnosis, Prognosis, and Treatment of Cervical Cancer: A Scoping Review. Diagnostics, 15(12), 1543. https://doi.org/10.3390/diagnostics15121543






