New Generation of Clinical Epigenetics Analysis and Diagnosis for Precision Medicine
Abstract
:1. Introduction
2. Clinical Sampling for Epigenetics and Epigenomics
- 1.
- Clinical sampling from subjects with common diseases, human behavior changes, and development: As shown in Table 1, tissue sampling is not a priority in conducting experimental studies of epigenetics and epigenomics for subjects with non-tumor diseases, human behavior, or children’s development. For instance, biopsies from the heart, pancreas, and brain tissues are very few because those tissues are difficult to acquire for clinical analysis and diagnosis. In human cardiovascular disease (CVD), diabetes, hereditary diseases, immune diseases, and human behavior change, the most common sampling is harvested from blood, saliva, cheek swabs, and follicles of patients, so we perform careful clinical analysis for the tissue specificity of epigenetic mechanisms related to this disease [21,22]. There are two ways to resolve the disadvantages of addressing epigenetics changes: (1) Physicians can try to achieve specific cells such as biopsy or isolate special cells to address specific epigenetics or epigenomics changes. As Table 1 shows, some sampling performance can increase specific epigenetics information, such as aorta biopsy to study CVD, isolating reticulocytes to study sickle cell disease or thalassemia, isolating lymphocytes or macrophages to study immune diseases, or isolating neutrophil for infectious diseases [10,23]. (2) Epigenetic analysis in silico can uncover some specific epigenetic change for patients. For example, we study epigenetic change related to mental health. In that case, the epigenetics results from blood specimens must co-study the variability in epigenetics from immune cells with their responses. Additionally, suppose we used buccal cells from saliva or cheek swabs for epigenetics study. In that case, these cells derived from the primitive germ layer (the ectoderm) may coincide with the epigenetic change of the brain to study children’s development and behavior [24,25].
- 2.
- Clinical sampling from patients with tumor diseases: As discussed above, tumor tissue sampling for specific epigenetic analysis is more advanced than that from subjects with non-tumor diseases. For example, we can isolate tumor cells from tumor tissues, separate tumor cells from circulating tumor cells (CTC), or culture primary tumor cells from tumor tissues in a clinical laboratory [12,26].
- (1)
- Isolating tumor cells from tumor tissue, CTC, and cultured tumor cells
- (2)
- Liquid tissue
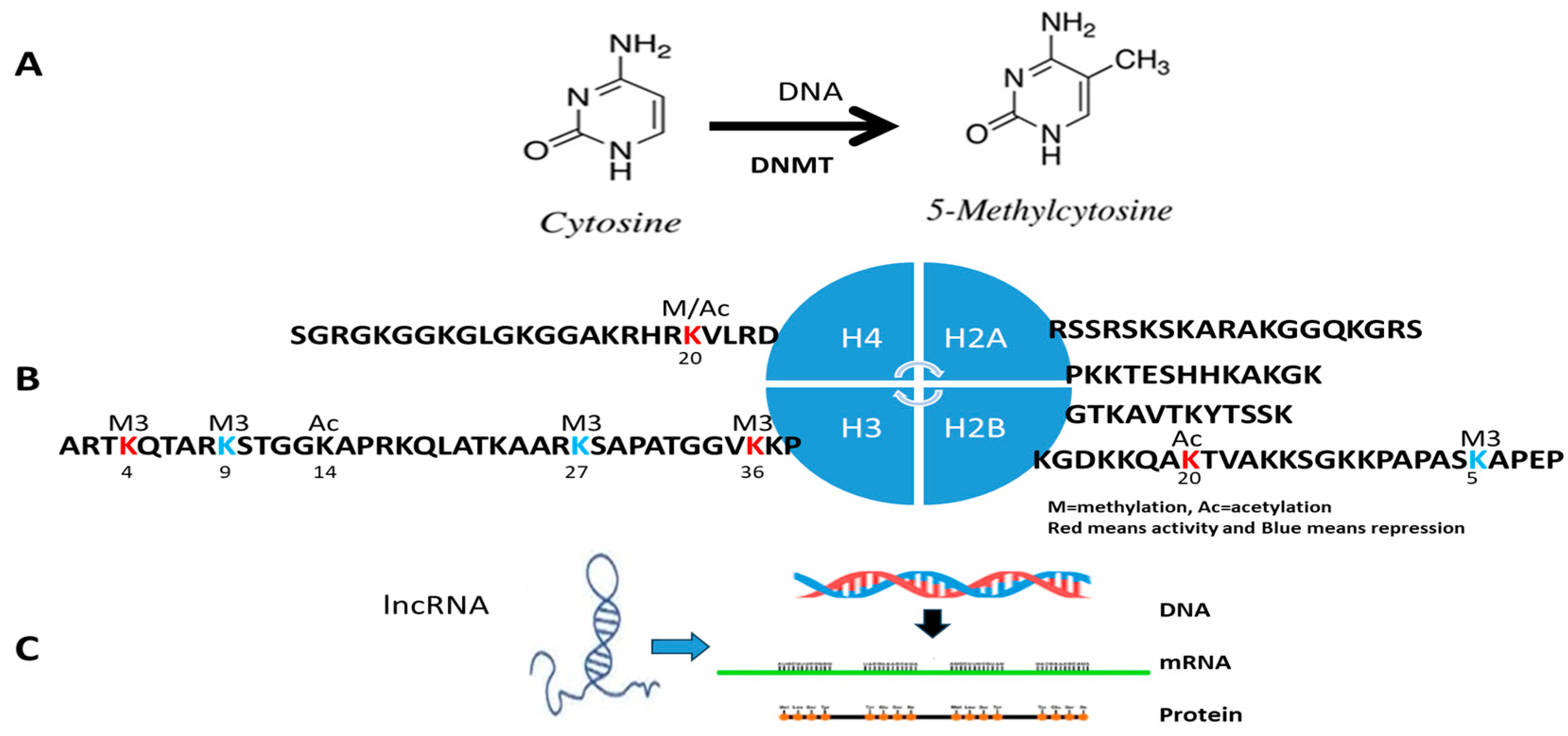
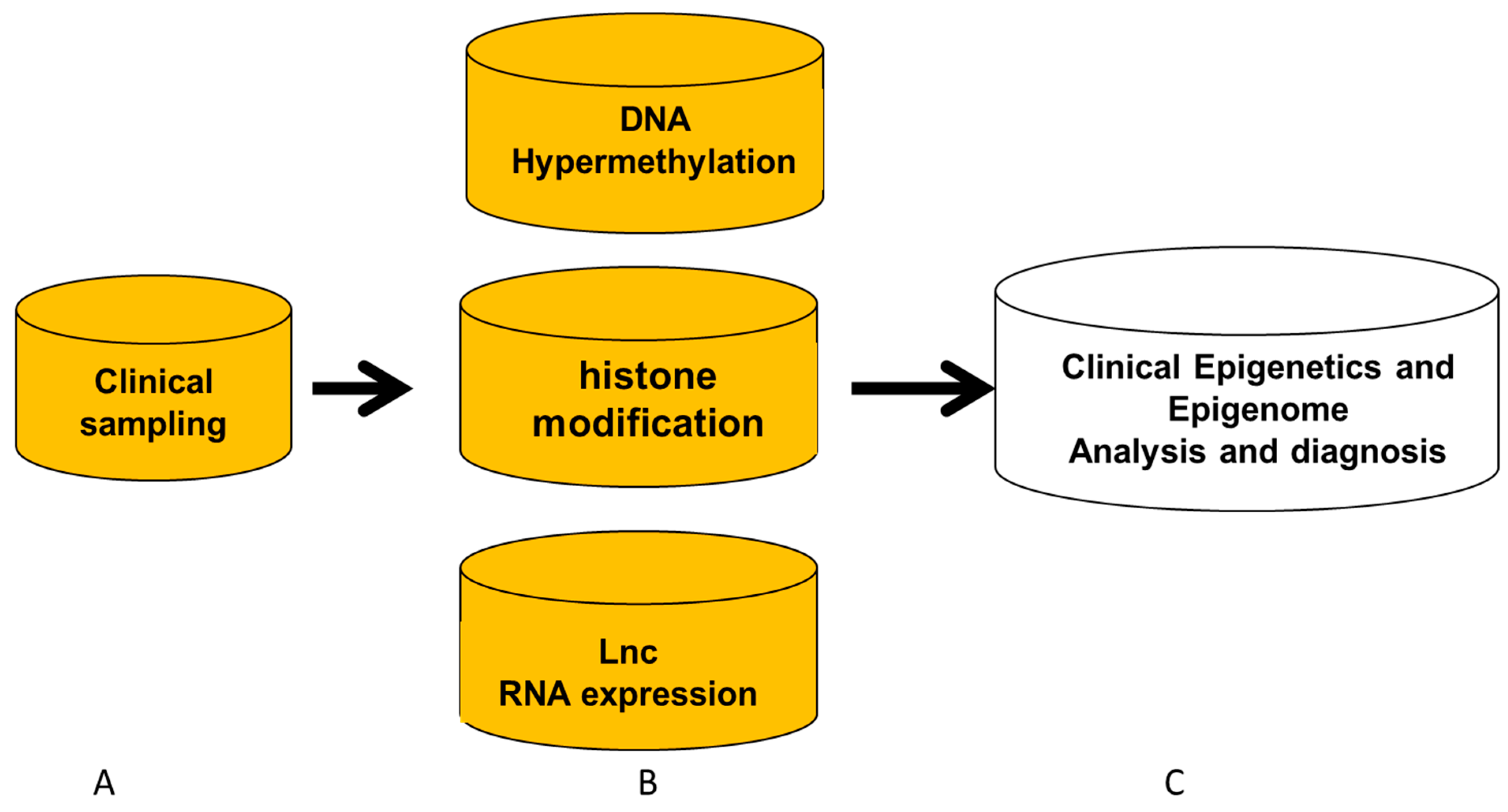
3. Detection Procedure of Clinical Epigenetics and Genomics
- 1.
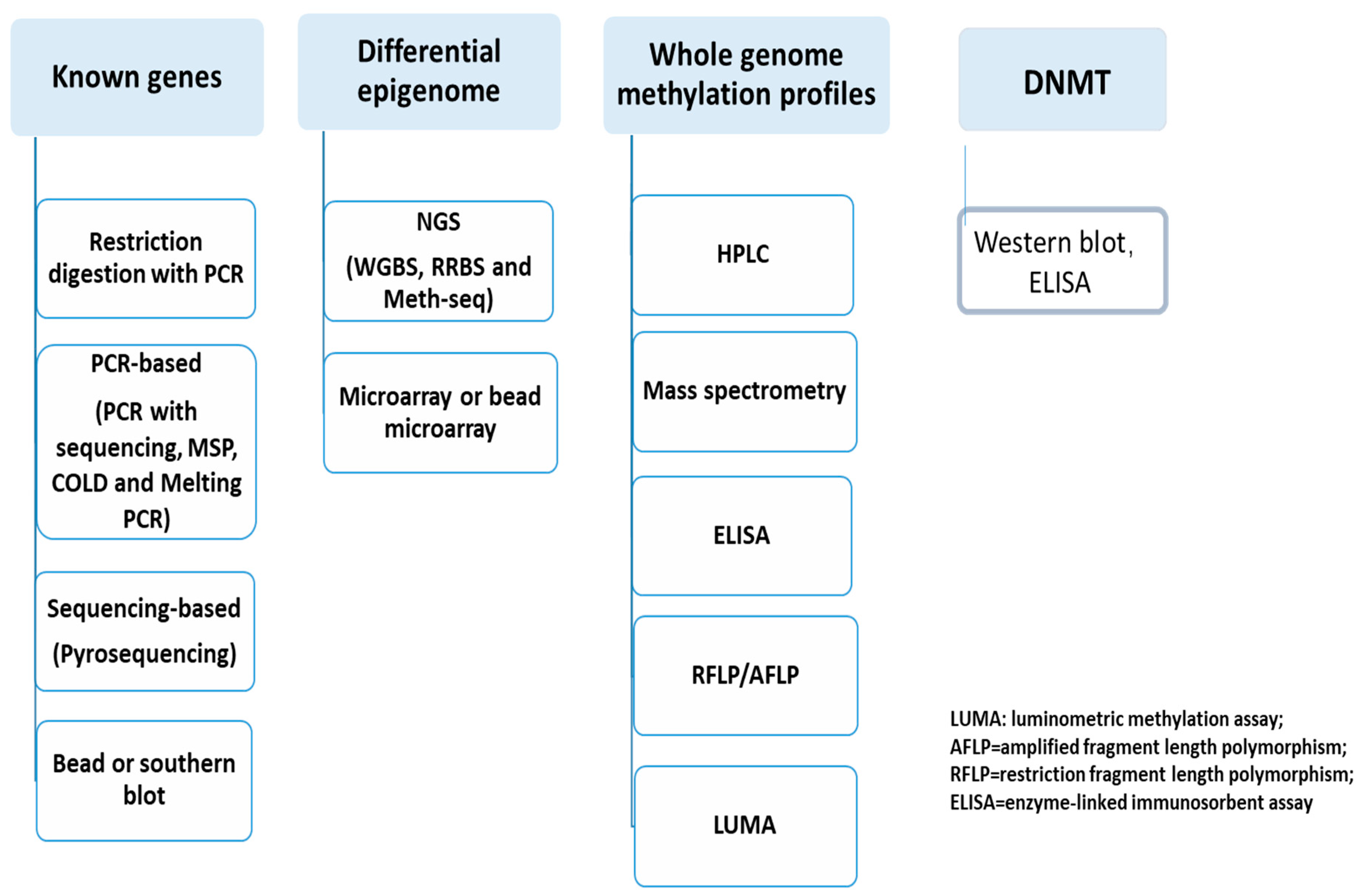
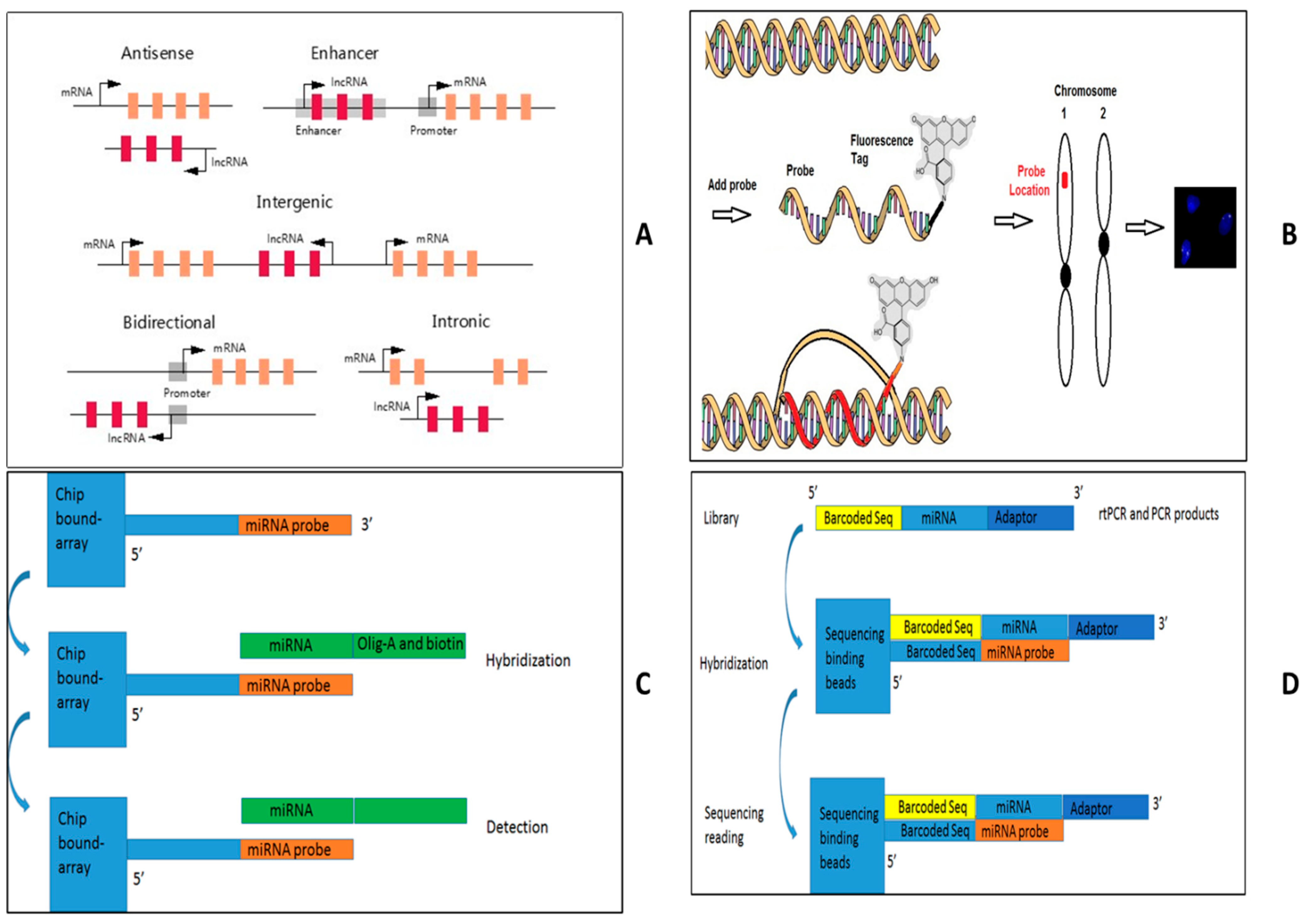
- Clinical IVD for DNA methylation and DNMT
- (1)
- Known-genes epigenetics
- (2)
- Unknown genes or epigenomic aberrance
- (3)
- DNMT measurement
- 2.
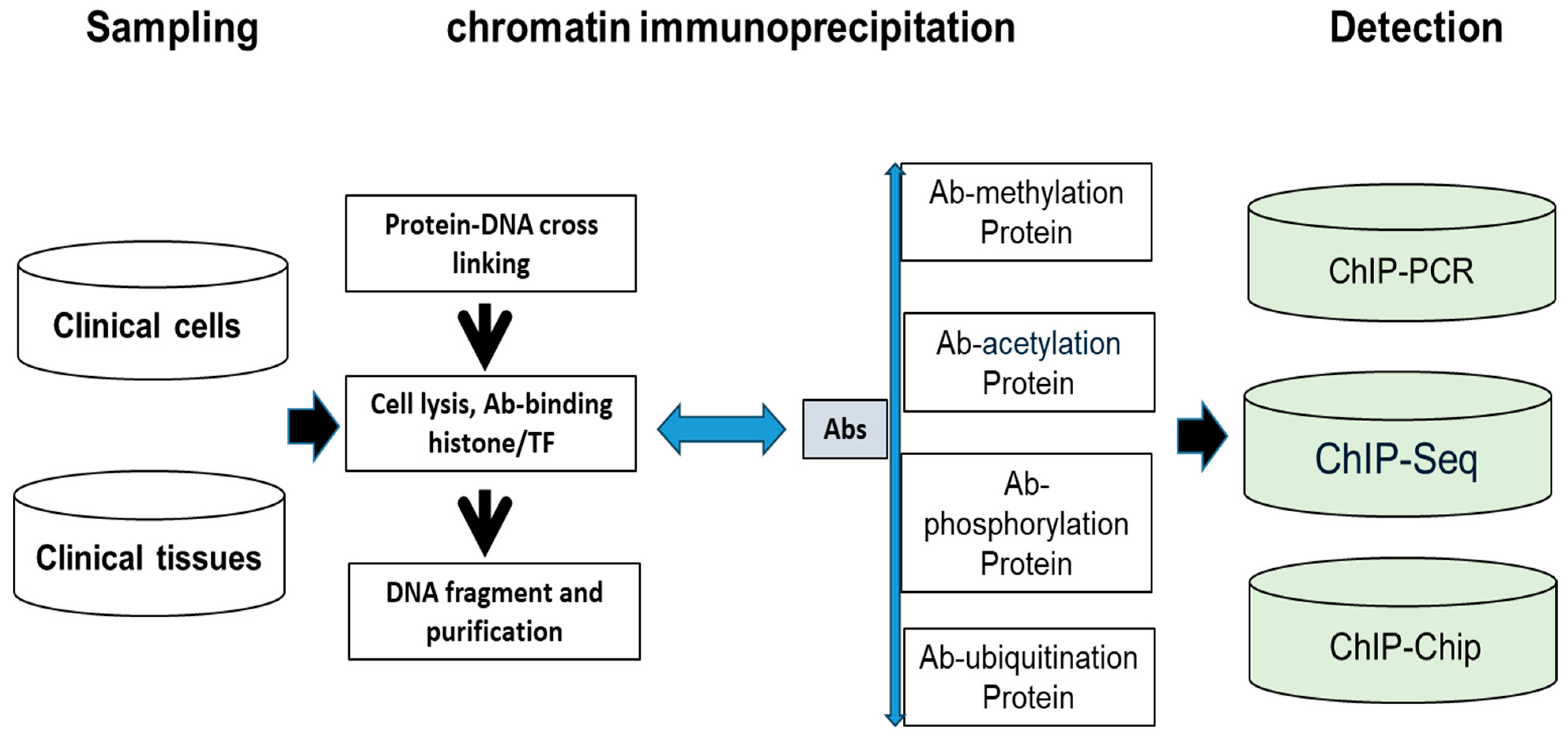
- Clinical IVD procedure for histone modification
- 3.
- Clinical IVD procedures for lncRNA
4. R&D of Clinical Epigenetics Diagnosis
- 1.
- Technology development
- (1)
- Single-cell technologies enable profiling DNA methylation (DNAm) at cytosines in individual cells with a bioinformatic program, a transformer-based deep learning model for inputting DNAm states at each CpG site for single-cell analysis. Moreover, single-cell lncRNA technologies have been successfully used in animal models, so the techniques will be developed to detect lncRNA for clinical specimens from patients [65,66].
- (2)
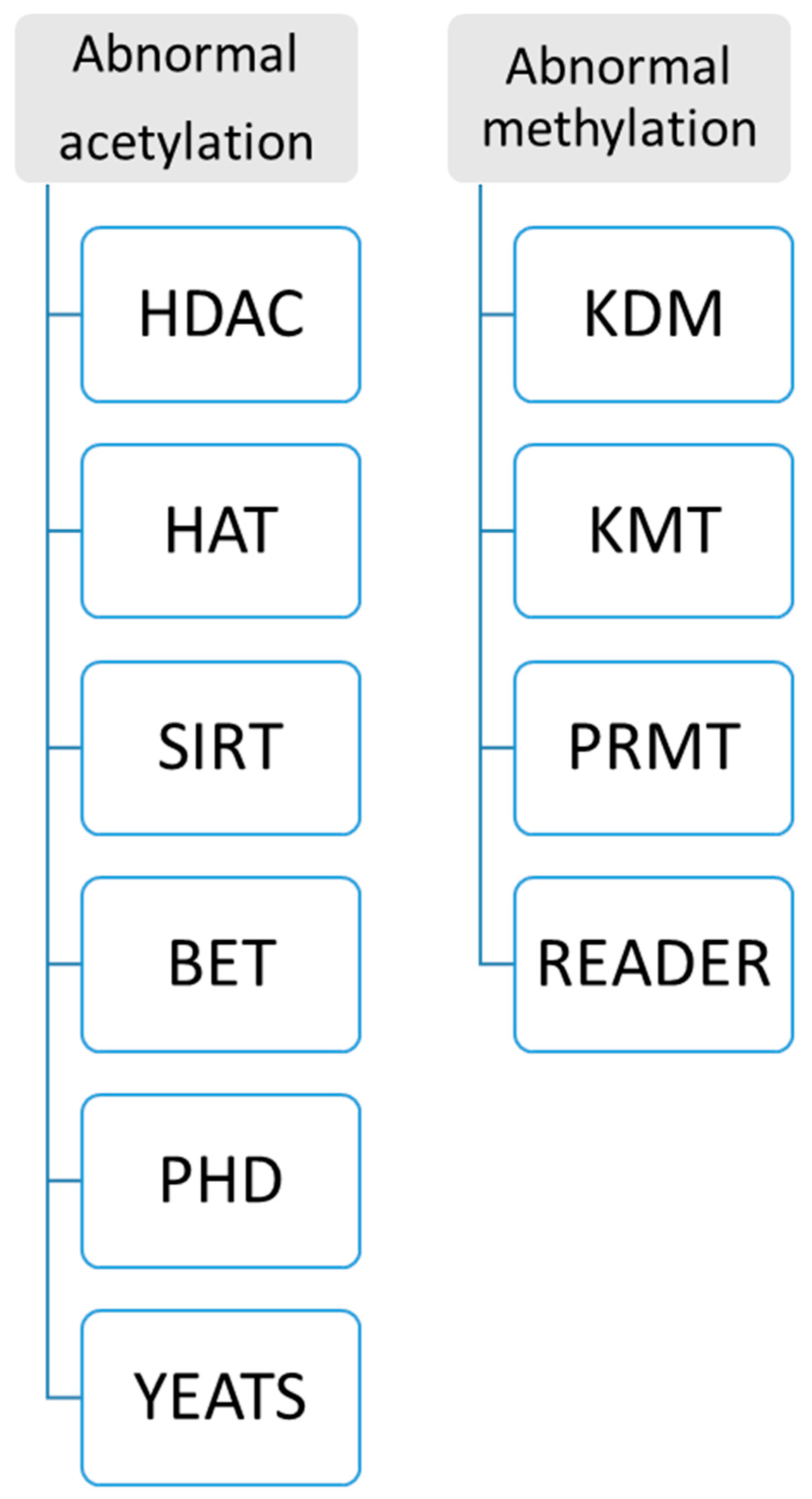
- New enzyme and protein domains discovered in epigenetics change can support precision medicine. Because abnormal histone modification is a very complicated mechanism, a novel R&D technique should be developed by different enzymes and targeting domains. Currently, two kinds of assays for abnormal histone modification have been used in clinical patients as Figure 6: (A) abnormal acetylation histone modification and (B) abnormal histone methylation modification. For example, abnormal histone acetylation with the enzyme or domain has been discovered to develop detection of HDAC (histone deacetylase), SIRT (class III histone deacetylases) activity, KAT activity (lysine acetyltransferase enzymes), BET (bromodomain and extra-terminal involved in acetylated histones) assay, plant homeodomain (PHD) finger detection, YEATS (domain acylated histones). Abnormal methylation, as shown in Figure 6, includes lysine methyltransferases (KMTs) and demethylases (KDMs), and Protein Arginine Methyltransferase (PRMT). Now, more and more enzymes and methods such as acetyltransferase 1 (HAT1), General Control Non-Repressible 5 (GNC5), CREB-binding protein (CBP), P300/CBP-binding protein (PCAF), MYST family, P300, TAFII250, and Rtt109 are increasingly reported in histone modification, which can increase clinical epigenetics analysis and diagnosis to increase epigenetics therapeutic potential [67,68,69,70,71,72,73,74].
- 2.
- Product development
- 3.
- Clinical development
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, X.; Xu, Z.; Li, B. Editorial: Epigenetics in cancer: Mechanisms and drug development-volume II. Front. Genet. 2023, 14, 1242960. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Liu, H.; Xu, B.; Qi, Y.; Xu, K.; Wu, X.; He, X.; Qin, Y.; Chen, Z.J. Epidemiology, genetic etiology and intervention of premature ovarian insufficiency. Endocr. Rev. 2025, bnaf011. [Google Scholar] [CrossRef]
- Cintron, M.A.; Baumer, Y.; Pang, A.P.; Peterson, E.M.A.; Ortiz-Whittingham, L.R.; Jacobs, J.A.; Sharda, S.; Potharaju, K.A.; Baez, A.S.; Gutierrez-Huerta, C.A.; et al. Associations between the neural-hematopoietic-inflammatory axis and DNA methylation of stress-related genes in human leukocytes: Data from the Washington, D.C. cardiovascular health and needs assessment. Brain Behav. Immun. Health 2025, 45, 100976. [Google Scholar] [CrossRef]
- Nagao, Y.; Fujimoto, M.; Tian, Y.; Kameyama, S.; Hattori, K.; Hidese, S.; Kunugi, H.; Kanai, Y.; Arai, E. Genome-wide DNA methylation profiling of blood samples from patients with major depressive disorder: Correlation with symptom heterogeneity. J. Psychiatry Neurosci. 2025, 50, E112–E124. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Luo, X.; Liang, Y.; Jiang, X.; Yang, W.; Yan, J.; Qi, K.; Li, P. Epigenetic modifications of nuclear and mitochondrial DNA are associated with the disturbance of serum iron biomarkers among the metabolically unhealthy obesity school-age children. Nutr. J. 2025, 24, 51. [Google Scholar] [CrossRef]
- Dorscheid, D.; Gauvreau, G.M.; Georas, S.N.; Hiemstra, P.S.; Varricchi, G.; Lambrecht, B.N.; Marone, G. Airway epithelial cells as drivers of severe asthma pathogenesis. Mucosal Immunol. 2025, 18, 524–536. [Google Scholar] [CrossRef]
- Brunty, S.; Wagner, K.; Fleshman, T.; Ruley, M.; Mitchell, B.; Santanam, N. Dual targeting of CXCR4 and EZH2 in endometriosis. iScience 2025, 28, 112143. [Google Scholar] [CrossRef] [PubMed]
- Arruda, B.P.; Martins, P.P.; Kihara, A.H.; Takada, S.H. Perinatal asphyxia and Alzheimer’s disease: Is there a correlation? Front. Pediatr. 2025, 13, 1567719. [Google Scholar] [CrossRef]
- Janik, K.; Jin, L.Q.; Kyzy, K.Z.; Kaminski, R.; Smith, G.M.; Krynska, B. Neural tube defects induce abnormal astrocyte development by activation and epigenetic permissiveness of STAT3. Exp. Neurol. 2025, 389, 115231. [Google Scholar] [CrossRef]
- Jaiswal, A.; Halasz, L.; Williams, D.L.; Osborne, T. Setdb2 Regulates Inflammatory Trigger-Induced Trained Immunity of Macrophages Through Two Different Epigenetic Mechanisms. bioRxiv 2025, 19, 644013. [Google Scholar] [CrossRef]
- Yuan, H.; Huang, Y.; Tao, S.; Li, B.; Xu, Z.; Qi, Y.; Wu, B.; Luo, H.; Zhu, X. Epigenetics in Cancer: Mechanisms and Drug Development. Front. Genet. 2022, 13, 831704. [Google Scholar] [CrossRef] [PubMed]
- Li, B. Therapeutic Targeting of Tumorigenesis and Tumor Disease—For Clinical Analysis of Epigenetics and Epigenome. Int. J. Sci. Res. Sci. Technol. 2017, 1, 1–12. [Google Scholar] [CrossRef]
- Banimohammad, M.; Khalafi, P.; Gholamin, D.; Bangaleh, Z.; Akhtar, N.; Solomon, A.D.; Prabhakar, P.K.; Sanami, S.; Prakash, A.; Pazoki-Toroudi, H. Exploring recent advances in signaling pathways and hallmarks of uveal melanoma: A comprehensive review. Explor. Target. Anti-Tumor Ther. 2025, 6, 1002306. [Google Scholar] [CrossRef]
- Wang, F.; Wen, J.; Liu, J.; Xin, L.; Fang, Y.; Sun, Y.; He, M. Demethylase FTO mediates m6A modification of ENST00000619282 to promote apoptosis escape in rheumatoid arthritis and the intervention effect of Xinfeng Capsule. Front. Immunol. 2025, 16, 1556764. [Google Scholar] [CrossRef]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumor immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Karnik, R.; Gu, H.; Ziller, M.J.; Clement, K.; Tsankov, A.M.; Akopian, V.; Gifford, C.A.; Donaghey, J.; Galonska, C.; et al. Targeted disruption of DNMT1, DNMT3A, and DNMT3B in human embryonic stem cells. Nat. Genet. 2015, 47, 469–478. [Google Scholar] [CrossRef]
- Varela, M.A.; Roberts, T.C.; Wood, M.J. Epigenetics and ncRNAs in brain function and disease: Mechanisms and prospects for therapy. Neurother. J. Am. Soc. Exper. Neurother. 2013, 10, 621–631. [Google Scholar] [CrossRef]
- Pal, S.; Tyler, J.K. Epigenetics and aging. Sci. Adv. 2016, 2, e1600584. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Raffington, L.; Schwaba, T.; Aikins, M.; Richter, D.; Wagner, G.G.; Harden, K.P.; Belsky, D.W.; Tucker-Drob, E.M. Associations of socioeconomic disparities with buccal DNA-methylation measures of biological aging. Clin. Epigenetics 2023, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Herzog, C.M.S.; Theeuwes, B.; Jones, A.; Evans, I.; Bjørge, L.; Zikan, M.; Cibula, D.; Harbeck, N.; Colombo, N.; Pashayan, N.; et al. Systems epigenetic approach towards non-invasive breast cancer detection. Nat. Commun. 2025, 16, 3082. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.; Pennell, D. The role of endomyocardial biopsy in the management of cardiovascular disease: A scientific statement from the American Heart Association, the American College of Cardiology, and the European Society of Cardiology. Eur. Heart J. 2008, 29, 1696. [Google Scholar] [CrossRef]
- Liu, Y.-D.; Teng, X.-M.; Bai, D.-D.; Xing, F.-Y.; Chang, Q.-R.; Li, J.-L.; Gao, S.-R.; Liu, W.-Q.; Guo, Y. Inhibition of aging-induced DNA hypermethylation by si-Dnmt3a/3b in pre-implantation embryos improves aberrant social behavior in offspring. Int. J. Biol. Macromol. 2025, 307, 142130. [Google Scholar] [CrossRef]
- Lardenoije, R.; Smulders, M.N.C.A.; Morin, E.L.; Howell, B.R.; Guzman, D.; Meyer, J.S.; Ressler, K.J.; Sánchez, M.; Klengel, T. A cross-generational methylomic signature of infant maltreatment in newborn rhesus macaques. Biol. Psychiatry, 2025; in press. [Google Scholar] [CrossRef]
- Ying, X.; Li, B. Machine-learning Modeling for Personalized Immunotherapy- An Evaluation Module. Biomed. J. Sci. Tech. Res. 2022, 47, 38211–38216. [Google Scholar]
- Ying, X.N.; Luo, H.; Zhang, Y.F.; Lu, J.; Li, W.Q.; Li, B. Pathway-Based Approaches for Analysis of RNA-seq with SNPs-A Case Report Without Discovering Targeting Drugs for Metastatic Lung Cancer. Sch. J. Appl. Med. Sci. 2019, 447–450. [Google Scholar]
- Huang, X.H.; Lu, J.; Zhang, Y.F.; Qiu, J.; Yao, G.R.; Song, P.T.; Li, B. Prediction, Prevention, Prognostic and Personalized Therapy of Ovarian Cancer- Biomarkers and Precision Medicine. Biomed. J. Sci. Tech. Res. 2024, 8, 223–231. [Google Scholar] [CrossRef]
- Liatsou, E.; Kollias, I.; Trapali, M.; Tsilimigras, D.I.; Gavriatopoulou, M.; Ntanasis-Stathopoulos, I. Liquid Biopsies in the Early Diagnosis, Prognosis, and Tailored Treatment of Colorectal Cancer. Cancers 2025, 17, 927. [Google Scholar] [CrossRef]
- Domaszewska-Szostek, A.; Krzyżanowska, M.; Polak, A.; Puzianowska-Kuźnicka, M. Effectiveness of Extracellular Vesicle Application in Skin. Aging Treatment and Regeneration: Do We Have Enough Evidence from Clinical Trials? Int. J. Mol. Sci. 2025, 26, 2354. [Google Scholar] [CrossRef]
- Avram, G.-E.; Marcu, A.; Moatar, A.; Samoila, C.; Podariu, A.; Seclaman, E.; Marian, C. Changes in global DNA methylation and hydroxymethylation in oral mucosa according to tobacco smoke exposure. J. Int. Med. Res. 2020, 48, 0300060520954677. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.; Willemsen, G.; de Geus, E.J.; Boomsma, D.I.; Neale, M.C.; Consortium, B. Effects of smoking on genome-wide DNA methylation profiles: A study of discordant and concordant monozygotic twin pairs. eLife 2023, 12, e83286. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Greene, R.M.; Pisano, M.M. Cigarette smoke induces proteasomal-mediated degradation of DNA methyltransferases and methyl CpG-/CpG domain-binding proteins in embryonic orofacial cells. Reprod. Toxicol. 2015, 58, 140–148. [Google Scholar] [CrossRef]
- Liu, F.; Killian, J.K.; Yang, M.; Walker, R.L.; Hong, J.A.; Zhang, M.; Davis, S.; Zhang, Y.; Hussain, M.; Xi, S.; et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 2010, 29, 3650–3664. [Google Scholar] [CrossRef]
- Ko, H.; Ahn, H.J.; Kim, Y.I. Methylation and mutation of the inhibin-α gene in human melanoma cells and regulation of PTEN expression and AKT/PI3K signaling by a demethylating agent. Oncol. Rep. 2022, 47, 37. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Yang, L.; Li, N.; Hu, Y.; Hu, Q.; Zhou, Z.; Cong, X. The effect of 1,25(OH)2D3 on Dickkopf-1 methylation in colorectal cancer. Clin. Epigenetics 2025, 17, 52. [Google Scholar] [CrossRef]
- Handoko; Adham, M.; Rachmadi, L.; Tobing, D.L.; Asmarinah; Fadilah; Dai, W.; Lee, A.W.M.; Gondhowiardjo, S. First Indonesian Nasopharyngeal Cancer Whole Epigenome Sequencing Identify Tumour Suppressor CpG Methylation. Biologics 2025, 19, 1–13. [Google Scholar] [CrossRef]
- Van Campen, H.; Bishop, J.V.; Brink, Z.; Engle, T.E.; Gonzalez-Berrios, C.L.; Georges, H.M.; Kincade, J.N.; Murtazina, D.A.; Hansen, T.R. Epigenetic Modifications of White Blood Cell DNA Caused by Transient Fetal Infection with Bovine Viral Diarrhea Virus. Viruses 2024, 16, 721. [Google Scholar] [CrossRef] [PubMed]
- Aleman, A.; Adrien, L.; Lopez-Serra, L.; Cordon-Cardo, C.; Esteller, M.; Belbin, T.J.; Sanchez-Carbayo, M. Editorial Expression of Concern: Identification of DNA hypermethylation of SOX9 in association with bladder cancer progression using CpG microarrays. Br. J. Cancer 2024, 131, 1946. [Google Scholar] [CrossRef]
- Brueckner, B.; Boy, R.G.; Siedlecki, P.; Musch, T.; Kliem, H.C.; Zielenkiewicz, P.; Suhai, S.; Wiessler, M.; Lyko, F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005, 65, 6305–6311. [Google Scholar] [CrossRef]
- Stenzig, J.; Hirt, M.N.; Löser, A.; Bartholdt, L.M.; Hensel, J.-T.; Werner, T.R.; Riemenschneider, M.; Indenbirken, D.; Guenther, T.; Müller, C.; et al. DNA methylation in an engineered heart tissue model of cardiac hypertrophy: Common signatures and effects of DNA methylation inhibitors. Basic Res. Cardiol. 2016, 111, 9. [Google Scholar] [CrossRef] [PubMed]
- Assis, R.I.F.; Wiench, M.; Silvério, K.G.; da Silva, R.A.; Feltran, G.d.S.; Sallum, E.A.; Casati, M.Z.; Nociti, F.H.; Andia, D.C.; Branco, M. RG108 increases NANOG and OCT4 in bone marrow-derived mesenchymal cells through global changes in DNA modifications and epigenetic activation. PLoS ONE 2018, 13, e0207873. [Google Scholar] [CrossRef]
- Motzer, R.J.; Penkov, K.; Haanen, J.; Rini, B.; Albiges, L.; Campbell, M.T.; Venugopal, B.; Kollmannsberger, C.; Negrier, S.; Uemura, M.; et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how, and why? Mol. Cell 2016, 49, 825–837. [Google Scholar] [CrossRef]
- Russ, B.E.; Olshansky, M.; Li, J.; Nguyen, M.L.; Gearing, L.J.; Nguyen, T.H.; Olson, M.R.; McQuilton, H.A.; Nüssing, S.; Khoury, G.; et al. Regulation of H3K4me3 at transcriptional enhancers characterizes the acquisition of virus-specific CD8+ T cell-lineage-specific function. Cell Rep. 2017, 21, 3624–3636. [Google Scholar] [CrossRef]
- Zhang, J.A.; Mortazavi, A.; Williams, B.A.; Wold, B.J.; Rothenberg, E.V. Dynamic Transformations of Genome-wide Epigenetic Marking and Transcriptional Control Establish T Cell Identity. Cell 2012, 149, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, S.; Meng, N.; He, Y.; Lu, R.; Yan, G.-R. ncRNA-encoded peptides or proteins and cancer. Mol. Ther. 2019, 27, 1718–1725. [Google Scholar] [CrossRef]
- Vos, P.D.; Leedman, P.J.; Filipovska, A.; Rackham, O. Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer. Cell Mol. Life Sci. 2019, 76, 3745–3752. [Google Scholar] [CrossRef]
- Wang, N.; Yu, Y.; Xu, B.; Zhang, M.; Li, Q.; Miao, L. Pivotal prognostic and diagnostic role of the long non-coding RNA colon cancer-associated transcript 1 expression in human cancer (Review). Mol. Med. Rep. 2019, 19, 771–782. [Google Scholar] [CrossRef]
- Zhao, W.; An, Y.; Liang, Y.; Xie, X.W. Role of HOTAIR long non-coding RNA in metastatic progression of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1930–1936. [Google Scholar]
- Dragomir, M.P.; Kopetz, S.; A Ajani, J.; Calin, G.A. Non-coding RNAs in GI cancers: From cancer hallmarks to clinical utility. Gut 2020, 69, 748–763. [Google Scholar] [CrossRef] [PubMed]
- El Fatimy, R.; Subramanian, S.; Uhlmann, E.J.; Krichevsky, A.M. Genome editing reveals glioblastoma addiction to microRNA-10b. Mol. Ther. 2017, 25, 368–378. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by non-coding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Balas, M.M.; Johnson, A.M. Exploring the mechanisms behind long non-coding RNAs and cancer. Non-Coding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, K.-Y.; Lin, Y.-C.; Gupta, S.K.; Chang, N.; Yen, L.; Sun, H.S.; Tsai, S.-J. Non-coding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017, 77, 2339–2350. [Google Scholar] [CrossRef]
- Huang, H.; Jin, H.; Lei, R.; He, Z.; He, S.; Chen, J.; Saw, P.E.; Qiu, Z.; Ren, G.; Nie, Y. lncRNA-WAL Promotes Triple-Negative Breast Cancer Aggression by Inducing β-Catenin Nuclear Translocation. Mol. Cancer Res. 2024, 22, 1036–1050. [Google Scholar] [CrossRef] [PubMed]
- De Felice, B.; De Luca, P.; Montanino, C.; Mallardo, M.; Babino, G.; Mattera, E.; Sorbo, R.; Ragozzino, G.; Argenziano, G.; Daniele, A.; et al. LncRNA microarray profiling identifies novel circulating lncRNAs in hidradenitis suppurativa. Mol. Med. Rep. 2024, 30, 112. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Lin, B.; Liu, Z.; Xu, F.; Song, D.; Che, K.; Chen, D.; Su, Y.; Li, W.; et al. A maternal low-protein diet impaired glucose metabolism and altered the lncRNA profiles of islets in adult offspring. J. Nutr. Biochem. 2024, 128, 109618. [Google Scholar] [CrossRef]
- Yang, R.; Liu, N.; Li, T.; Liu, F.; Zhang, J.; Zhao, H.; Zou, L.; He, X. LncRNA AC142119.1 facilitates the progression of neuroblastoma by epigenetically initiating the transcription of MYCN. J. Transl. Med. 2023, 21, 659. [Google Scholar] [CrossRef]
- Bahia, R.K.; Lopez, C.; Nardocci, G.; Davie, J.R. Differential H3K4me3 Domains in Normal and Colorectal Cancer Cells Reveal Novel Epigenetic Targets. Int. J. Mol. Sci. 2025, 26, 2546. [Google Scholar] [CrossRef]
- Fedyuk, V.; Erez, N.; Furth, N.; Beresh, O.; Andreishcheva, E.; Shinde, A.; Jones, D.; Zakai, B.B.; Mavor, Y.; Peretz, T.; et al. Multiplexed, single-molecule, epigenetic analysis of plasma-isolated nucleosomes for cancer diagnostics. Nat. Biotechnol. 2022, 41, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Sadeh, R.; Sharkia, I.; Fialkoff, G.; Rahat, A.; Gutin, J.; Chappleboim, A.; Nitzan, M.; Fox-Fisher, I.; Neiman, D.; Meler, G.; et al. ChIP-seq of plasma cell-free nucleosomes identifies gene expression programs of the cells of origin. Nat. Biotechnol. 2021, 39, 586–598. [Google Scholar] [CrossRef]
- Khan, D.H.; Healy, S.; He, S.; Lichtensztejn, D.; Klewes, L.; Sharma, K.L.; Lau, V.; Mai, S.; Delcuve, G.P.; Davie, J.R. Mitogen-induced distinct epialleles are phosphorylated at either H3S10 or H3S28, depending on H3K27 acetylation. Mol. Biol. Cell 2017, 28, 817–824. [Google Scholar] [CrossRef]
- Khan, D.H.; Gonzalez, C.; Cooper, C.; Sun, J.-M.; Chen, H.Y.; Healy, S.; Xu, W.; Smith, K.T.; Workman, J.L.; Leygue, E.; et al. RNA-dependent dynamic histone acetylation regulates MCL1 alternative splicing. Nucleic Acids Res. 2013, 42, 1656–1670. [Google Scholar] [CrossRef]
- Moeng, S.; Chamorro-Parejo, A.D.; Jeon, M.S.; Cai, J.J.; Ramos, K.S. Single-Cell RNA Sequencing Reveals Extensive Heterogeneity and Unique Gene Trajectories in Non-Transformed and Transformed Human Lung Epithelial Cells: Insights into the Role of LncRNAs in Tumor Heterogeneity. Int. J. Mol. Sci. 2025, 26, 1690. [Google Scholar] [CrossRef]
- Qin, H.; Qi, T.; Xu, J.; Wang, T.; Zeng, H.; Yang, J.; Yu, F. Integration of ubiquitination-related genes in predictive signatures for prognosis and immunotherapy response in sarcoma. Front. Oncol. 2024, 14, 1446522. [Google Scholar] [CrossRef] [PubMed]
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.-F.; Yao, T.-P. HDAC6 is a microtubule-associated deacetylase. Nature 2002, 417, 455–458. [Google Scholar] [CrossRef]
- Wang, N.; Wu, R.; Tang, D.; Kang, R. The BET family in immunity and disease. Signal Transduct. Target. Ther. 2021, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, metabolism, and cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef]
- Dong, Y.; Tu, R.; Liu, H.; Qing, G. Regulation of cancer cell metabolism: Oncogenic MYC in the driver’s seat. Signal Transduct. Target. Ther. 2020, 5, 124. [Google Scholar] [CrossRef]
- Shu, S.; Lin, C.Y.; He, H.H.; Witwicki, R.M.; Tabassum, D.P.; Roberts, J.M.; Janiszewska, M.; Huh, S.J.; Liang, Y.; Ryan, J.; et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016, 529, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Asangani, I.A.; Dommeti, V.L.; Wang, X.; Malik, R.; Cieslik, M.; Yang, R.; Escara-Wilke, J.; Wilder-Romans, K.; Dhanireddy, S.; Engelke, C.; et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014, 510, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Hah, N.; Yu, R.T.; Sherman, M.H.; Benner, C.; Leblanc, M.; He, M.; Liddle, C.; Downes, M.; Evans, R.M. BRD4 is a novel therapeutic target for liver fibrosis. Proc. Natl. Acad. Sci. USA 2015, 112, 15713–15718. [Google Scholar] [CrossRef] [PubMed]
- Alberto-Aguilar, D.R.; Hernández-Ramírez, V.I.; Osorio-Trujillo, J.C.; Gallardo-Rincón, D.; Toledo-Leyva, A.; Talamás-Rohana, P. PHD finger protein 20-like protein 1 (PHF20L1) in ovarian cancer: From its overexpression in tissue to its upregulation by the ascites microenvironment. Cancer Cell Int. 2022, 22, 6. [Google Scholar] [CrossRef]
- Hebert, K.; Bruno, A.; Matta, R.; Horns, J.; Paudel, N.; Das, R.; Hotaling, J.; McCormick, B.; Myers, J.B. Impact of Prostate Cancer-related Genitourinary Radiation Injury on Mental Health Diagnosis and Treatment: Assessment of 55,425 Men. Urology 2024, 183, 228–235. [Google Scholar] [CrossRef]
- Sirajee, R.; El Khatib, S.; Dieleman, L.A.; Salla, M.; Baksh, S. ImmunoMet Oncogenesis: A New Concept to Understand the Molecular Drivers of Cancer. J. Clin. Med. 2025, 14, 1620. [Google Scholar] [CrossRef]
| Diseases | Common Sampling | Specific Sampling |
| CVD | blood (such as WBC), and saliva, urine, follicles | Tissues affected areas (such as biopsy of the aorta) [23] |
| Diabetes | blood (such as WBC), and saliva, urine, follicles | Tissue biopsy for affected areas |
| Immune diseases | blood (such as WBC), and saliva, urine, follicles | Tissue biopsy for affected areas |
| Infection disease | blood (such as WBC), and saliva, urine, follicles | Tissue collection |
| Hereditary diseases | blood (such as WBC), and saliva, urine, follicles | Pathological tissues such as SCD reticulocyte |
| Children behavior | blood (such as WBC), and saliva, urine, follicles | N/A |
| Children development | blood (such as WBC), and saliva, urine, follicles | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, P.; Li, B. New Generation of Clinical Epigenetics Analysis and Diagnosis for Precision Medicine. Diagnostics 2025, 15, 1539. https://doi.org/10.3390/diagnostics15121539
Song P, Li B. New Generation of Clinical Epigenetics Analysis and Diagnosis for Precision Medicine. Diagnostics. 2025; 15(12):1539. https://doi.org/10.3390/diagnostics15121539
Chicago/Turabian StyleSong, Pengtao, and Biaoru Li. 2025. "New Generation of Clinical Epigenetics Analysis and Diagnosis for Precision Medicine" Diagnostics 15, no. 12: 1539. https://doi.org/10.3390/diagnostics15121539
APA StyleSong, P., & Li, B. (2025). New Generation of Clinical Epigenetics Analysis and Diagnosis for Precision Medicine. Diagnostics, 15(12), 1539. https://doi.org/10.3390/diagnostics15121539






