The proposed method for multi-level thresholding segmentation and classification of lung X-ray images was implemented and tested on a Windows 11 system with an Intel Core i5-4710MQ CPU (2.5 GHz, 8 CPUs), 8 GB of RAM, and 1 GB of dedicated GPU memory. Both the segmentation and classification models were developed using MATLAB R2020a and Python3.11. The performance of the model was assessed using several evaluation metrics, including accuracy, precision, recall, F1-score, and the effectiveness of the optimization algorithms used during the segmentation process.
4.2. Segmentation Performance
The multi-level thresholding technique used during the segmentation phase played a key role in enhancing the classification model’s accuracy. Segmentation was carried out using three optimization algorithms: AMO, HSA, and EMO. These algorithms helped determine the optimal thresholds for distinguishing various regions within the lung X-ray images.
Visual inspection of the segmented lung images (
Figure 5) demonstrated the superior performance of the EMO algorithm in accurately delineating lung regions, especially in cases with diffuse COVID-19 lesions. EMO consistently outperformed other algorithms, such as AMO and HSA, by capturing subtle differences in pixel intensity and preserving the structural integrity of the lung regions. This precision is crucial for accurately identifying affected areas, aiding in diagnosis and treatment planning. EMO’s robustness in handling complex lung images makes it highly effective for COVID-19 lesion segmentation.
In the discussion of the results obtained from the COVID-19 lung datasets (
Table 5), the performance of the three optimization algorithms AMO, EMO, and HAS was evaluated based on accuracy and precision.
To ensure a comprehensive assessment of the segmentation performance of AMO, EMO, and HSA, we constructed multiple test subsets from a COVID-19 chest X-ray dataset. These subsets introduced intra-class variation in terms of lesion density, image contrast, and severity level to mimic real-world diagnostic diversity. This strategy allowed us to evaluate not just raw performance, but also algorithmic robustness across distinct yet clinically relevant imaging scenarios. EMO consistently achieved superior results, reinforcing its reliability for COVID-19 lung segmentation across a range of radiographic conditions.
EMO consistently demonstrated superior performance across all evaluations, achieving the highest accuracy on the chest X-ray dataset with 99.1% ± 0.074 and maintaining strong precision, notably reaching 99.8% ± 0.021. These results highlight EMO’s effectiveness in accurately segmenting and classifying COVID-19 lung images with minimal false positives.
HSA exhibited remarkable precision, particularly on two subsets of the chest X-ray dataset, achieving an outstanding precision of 99.9% ± 0.045. This indicates its strength in reducing misclassifications. However, its accuracy showed more variability, dropping to 90.34% in one subset, suggesting that while HSA reliably identifies true positives, it may underperform in overall classification when presented with more complex variations.
AMO, although performing well in terms of precision, achieving 99.1% ± 0.05 in one dataset, showed fluctuations in accuracy. For example, it recorded only 80.39% ± 0.0687 accuracy on a more diverse subset, indicating challenges in generalizing across complex imaging patterns.
Overall, EMO proved to be the most balanced algorithm, delivering both high accuracy and precision, making it particularly well-suited for COVID-19 lung image segmentation. HSA, with its superior precision, may be preferable in applications where minimizing false positives is critical. By contrast, AMO showed potential but may require further tuning to improve its performance on heterogeneous datasets.
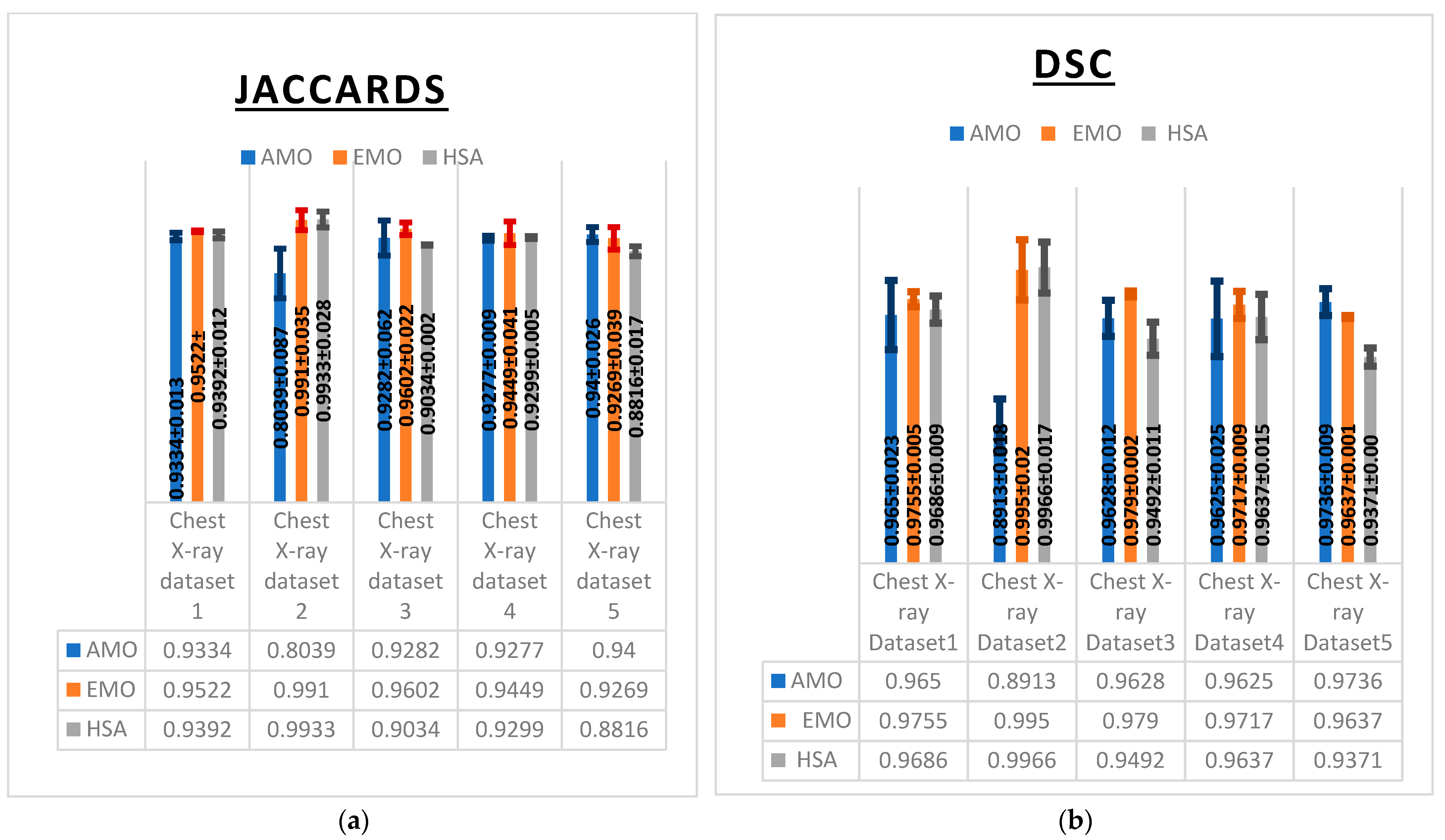
These observations were further supported by multiple evaluation metrics including the Dice similarity coefficient (DSC), Jaccard index (IoU) (
Figure 6), F1-score, specificity, and sensitivity (
Table 6) which collectively reflected consistent performance trends across the algorithms AMO, EMO, and HSA.
EMO exhibited superior performance in segmentation accuracy, as reflected by its consistently high Dice similarity coefficient (DSC) values (
Figure 6a), achieving 0.995 on Chest X-ray Dataset 2 and 0.979 on Chest X-ray Dataset 3. HSA also demonstrated strong performance, slightly outperforming EMO on Chest X-ray Dataset 2 with a DSC of 0.9966. By contrast, AMO yielded lower DSC values, particularly on Chest X-ray Dataset 2, where it achieved only 0.8913, indicating challenges in segmenting certain image types.
The Jaccard index (IoU) results further confirmed the strength of EMO and HSA (
Figure 6b). EMO achieved an impressive 0.9910 on Chest X-ray Dataset 2 and 0.9602 on Chest X-ray Dataset 3, demonstrating its accurate segmentation capabilities. HSA performed comparably, with a value of 0.9933 on Chest X-ray Dataset 2. However, AMO showed a significant drop in IoU on the same dataset, reaching only 0.8039, which reflects its difficulty in accurately identifying lung regions.
The F1-score results also highlighted EMO’s balanced performance, particularly on Chest X-ray Dataset 2, where it achieves 99.55%. HSA followed closely, with strong scores on Chest X-ray Datasets 2 and 3. However, its performance declined slightly on Chest X-ray Dataset 5, where it achieved 93.71%, indicating a reduced ability to handle more complex cases. While AMO achieved a solid F1-score of 96.56% on Chest X-ray Dataset 1, it fell behind the other algorithms on most datasets, particularly on Chest X-ray Dataset 2, where it reached only 90% (
Table 6).
In terms of specificity, HSA excelled, achieving 99% on Chest X-ray Datasets 1 and 4, highlighting its effectiveness in minimizing false positives. EMO also performed well in this metric, reaching 99.07% on Chest X-ray Dataset 3. Although AMO maintained acceptable specificity across most datasets, it showed a slight drop on Chest X-ray Dataset 2 with 97%, indicating a relatively higher rate of false positives.
The recall (Sensitivity) results again placed EMO in the lead, achieving 99.1% on Chest X-ray Dataset 2 and 98.35% on Chest X-ray Dataset 1, confirming its high sensitivity in detecting lung abnormalities. HSA performed similarly, achieving 99.3% on Chest X-ray Dataset 2. However, AMO showed a notable decrease in recall on Chest X-ray Dataset 2, with only 80.39%, revealing its limitation in correctly identifying true positive cases in this dataset.
Overall, EMO proved to be the most balanced algorithm, consistently achieving high performance across all metrics and datasets. HSA also demonstrated strong performance, particularly in precision and specificity, but showed slight variability in handling complex cases. AMO, while effective on some datasets, exhibited noticeable performance drops, particularly on Dataset 2, highlighting the need for further optimization to improve its consistency in segmentation and classification tasks.
4.4. Classification Performance
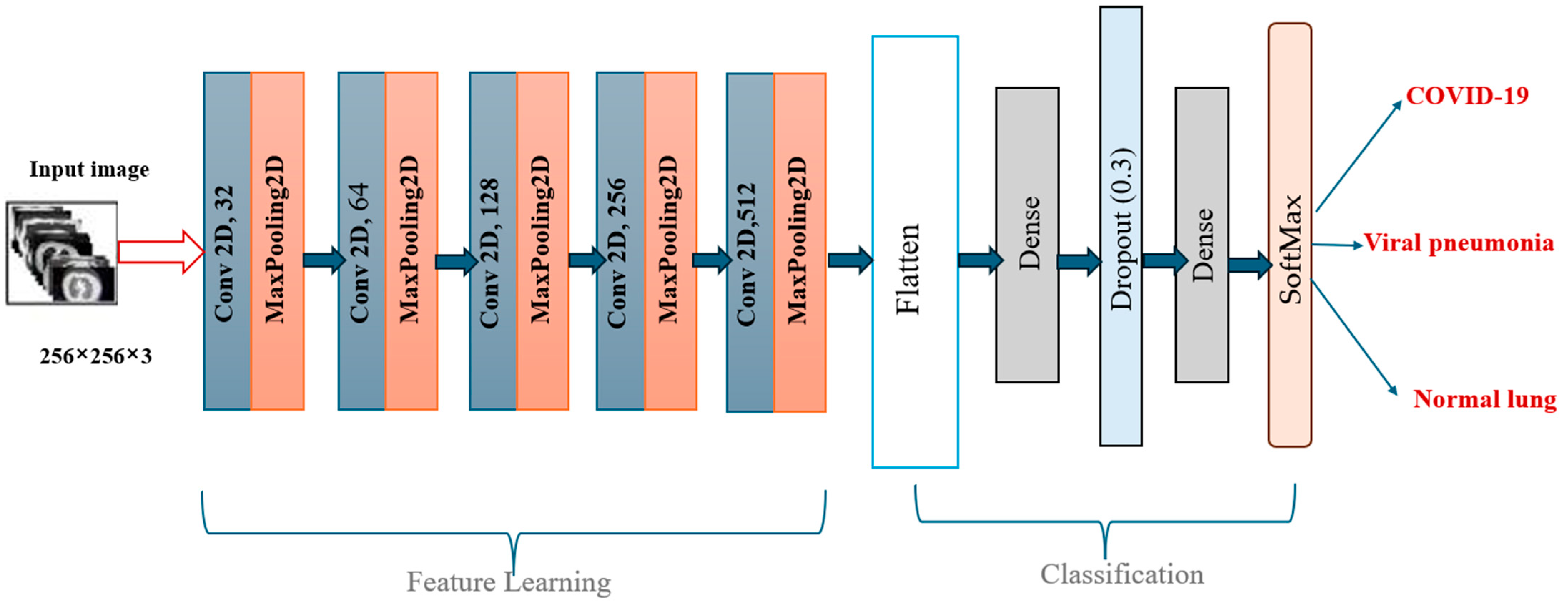
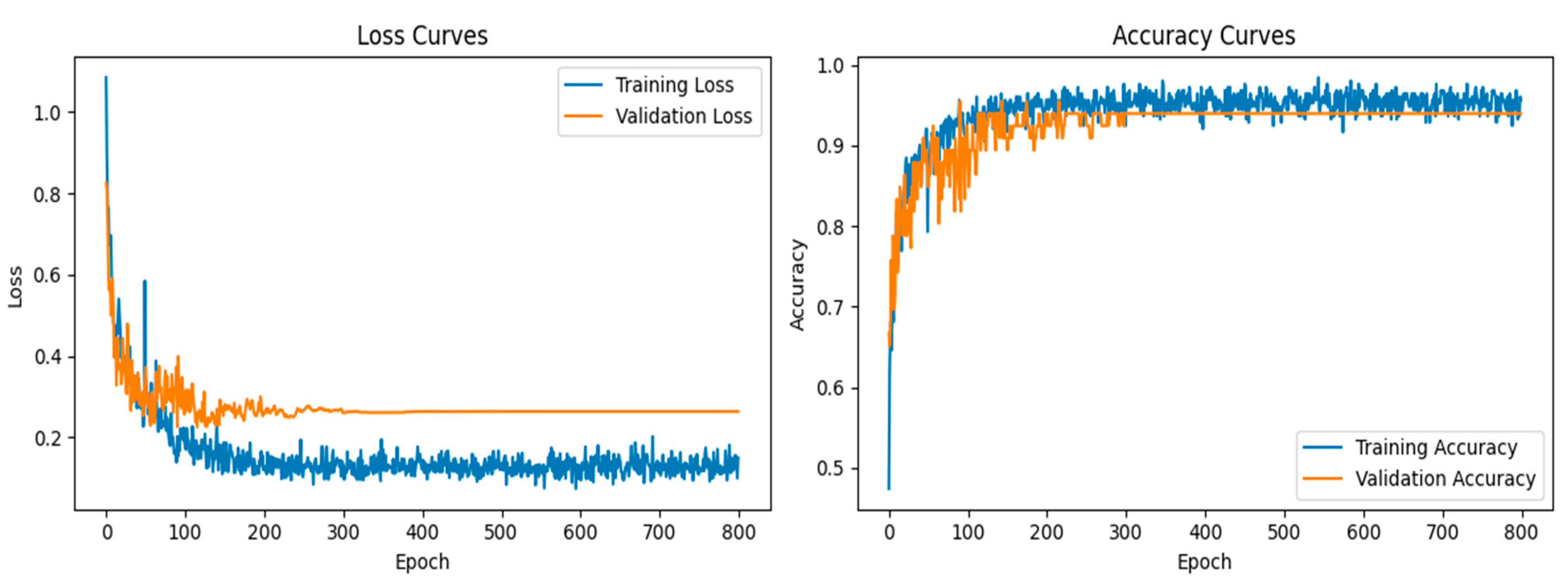
In the classification phase, the proposed CNN model exhibited exceptional performance, as demonstrated by the accuracy and loss curves. The accuracy curves showed rapid improvement during the early epochs, with both the training and validation accuracy curves converging toward 95% after approximately 300 epochs (
Figure 7). This indicated that the model was able to learn the distinguishing features of the lung X-ray images effectively while maintaining a balance between training and validation performance. The loss curves for both training and validation exhibited steady declines, with minimal overfitting observed, as the gap between the training and validation losses remained narrow throughout the training process.
Although the proposed methodology achieved high performance on a dataset of 306 X-ray images, we acknowledge that this represents a relatively small-scale dataset compared to publicly available repositories like Chest X-ray of COVID-19 lung. The segmentation and classification performance could vary when applied to larger and more heterogeneous datasets. However, the modular structure of our pipeline, including image preprocessing, multi-threshold segmentation, and CNN classification, makes it adaptable for generalization. Future work will involve validating the method on more extensive and diverse datasets to further assess its robustness and scalability in real-world diagnostic settings.
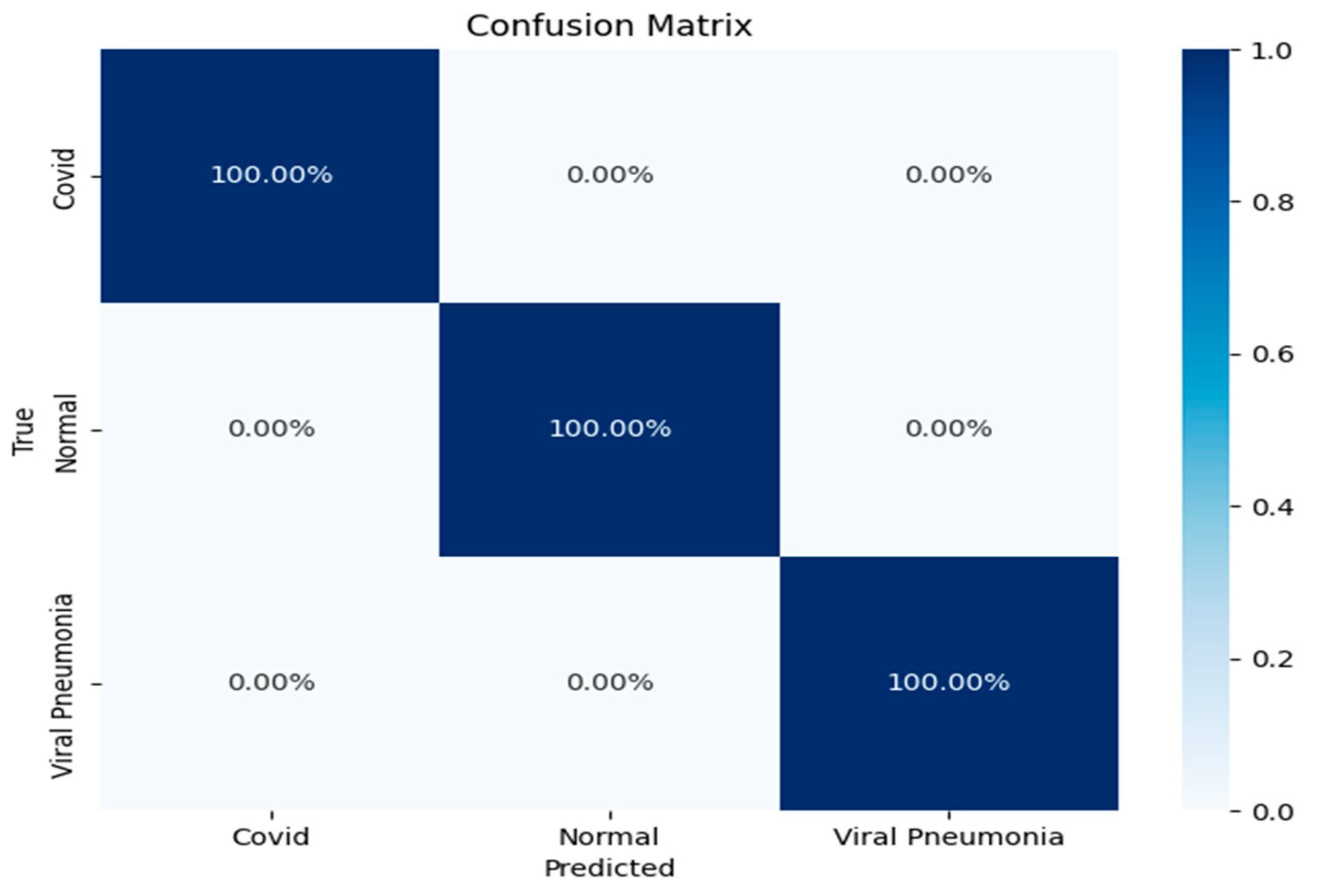
The confusion matrix further reinforced the model’s robustness, showing a perfect classification rate for all three categories (
Figure 8): COVID-19, viral pneumonia, and normal lung X-rays. Each of the classes was predicted with 100% accuracy, indicating that the model was able to distinguish between the subtle differences in the lung conditions without any misclassifications. This result underscores the CNN model’s strong generalization ability, especially in a domain where inter-class similarities might challenge less sophisticated models.
Overall, these results highlight the effectiveness of the CNN architecture in accurately classifying lung conditions from X-ray images. The model’s ability to maintain high accuracy across different conditions, coupled with the precise class predictions in the confusion matrix, suggests its potential utility in real-world clinical settings for rapid and reliable diagnosis. These findings demonstrate that the model could significantly enhance diagnostic workflows by providing accurate and automated image-based evaluations, contributing to more efficient disease detection and management.
To understand the source of the model’s high classification performance, we conducted an analysis comparing results with and without the proposed segmentation. The CNN architecture was kept identical in both scenarios. When trained on unsegmented chest X-ray images, the model reached an average accuracy of 89%, indicating moderate classification capability. However, when trained on images pre-processed through our optimized multi-threshold segmentation, the same model achieved accuracy values exceeding 99%. This significant improvement can be attributed to the enhanced clarity and separation of anatomical structures provided by the segmentation. By removing irrelevant background and enhancing the contrast of pathological regions, the segmentation step simplifies the learning task for the CNN. This finding suggests that the strength of our framework lies not in model complexity but in the quality of the image preparation pipeline, which allows even a relatively simple CNN to achieve high precision, recall, and overall accuracy.
The choice of a simple, custom-designed CNN in this study was guided by the goal of evaluating the contribution of the proposed segmentation strategy in a controlled setting. More complex architectures such as ResNet or vision transformers can marginally improve accuracy, but they also introduce considerable computational cost, increased inference time, and complexity, which can obscure the source of performance improvements. By using a lightweight CNN with approximately 11 million parameters, we ensured transparency in the results and demonstrated that high accuracy can be achieved when segmentation quality is maximized. Furthermore, the chosen architecture is more suitable for real-time diagnostic support in clinical environments with limited hardware resources, making it a practical and effective solution for deployment in diverse healthcare settings.
Although our proposed model demonstrated strong classification performance across the dataset, it does not currently distinguish between COVID-19 variants or recognize ARDS specific features, as the dataset lacked such annotations. Different COVID-19 variants tend to affect distinct anatomical regions within the lungs, and ARDS is associated with characteristic radiographic findings that significantly impact clinical outcomes. Future work should incorporate annotated clinical metadata, including variant and ARDS status, to enhance the model’s specificity and clinical utility. Additionally, integrating explainable AI techniques could provide valuable visual insights into the model’s decision-making process by highlighting which lung regions influenced predictions. This would not only improve interpretability but also assist clinicians in verifying that the model aligns with known pathological patterns.