PAX Family, Master Regulator in Cancer
Abstract
1. PAX Structure, Subgroups and General Functions
- I (PAX1 and PAX9)
- II (PAX2, PAX5, PAX8)
- III (PAX3 and PAX7)
- IV (PAX4 and PAX6)
2. PAX5 in B-Cell Maturation and in Hematological Malignancies
2.1. PAX5 in Precursor B-Cell Neoplasm
2.1.1. PAX5 Deletions
2.1.2. PAX5 Translocation
2.1.3. PAX5 Point Mutation
2.1.4. Germline Mutation
2.2. PAX5 in Mature Cell Lymphoid Neoplasm
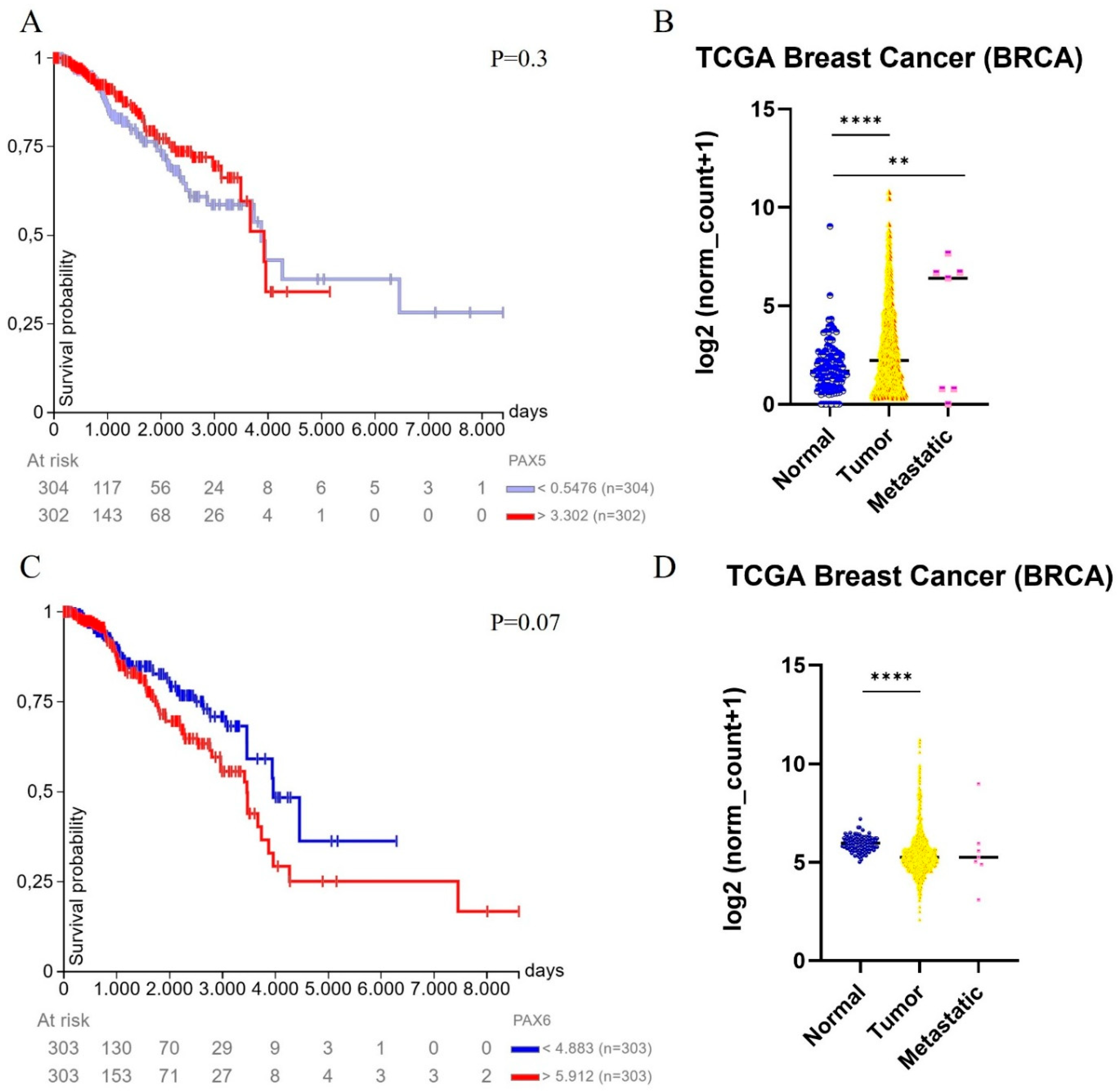
3. PAX5 and PAX6 in Breast Carcinomas
3.1. PAX5 as Tumor Suppressor in Breast Cancer
3.2. PAX6: A Double-Faced Actor in Breast Cancer
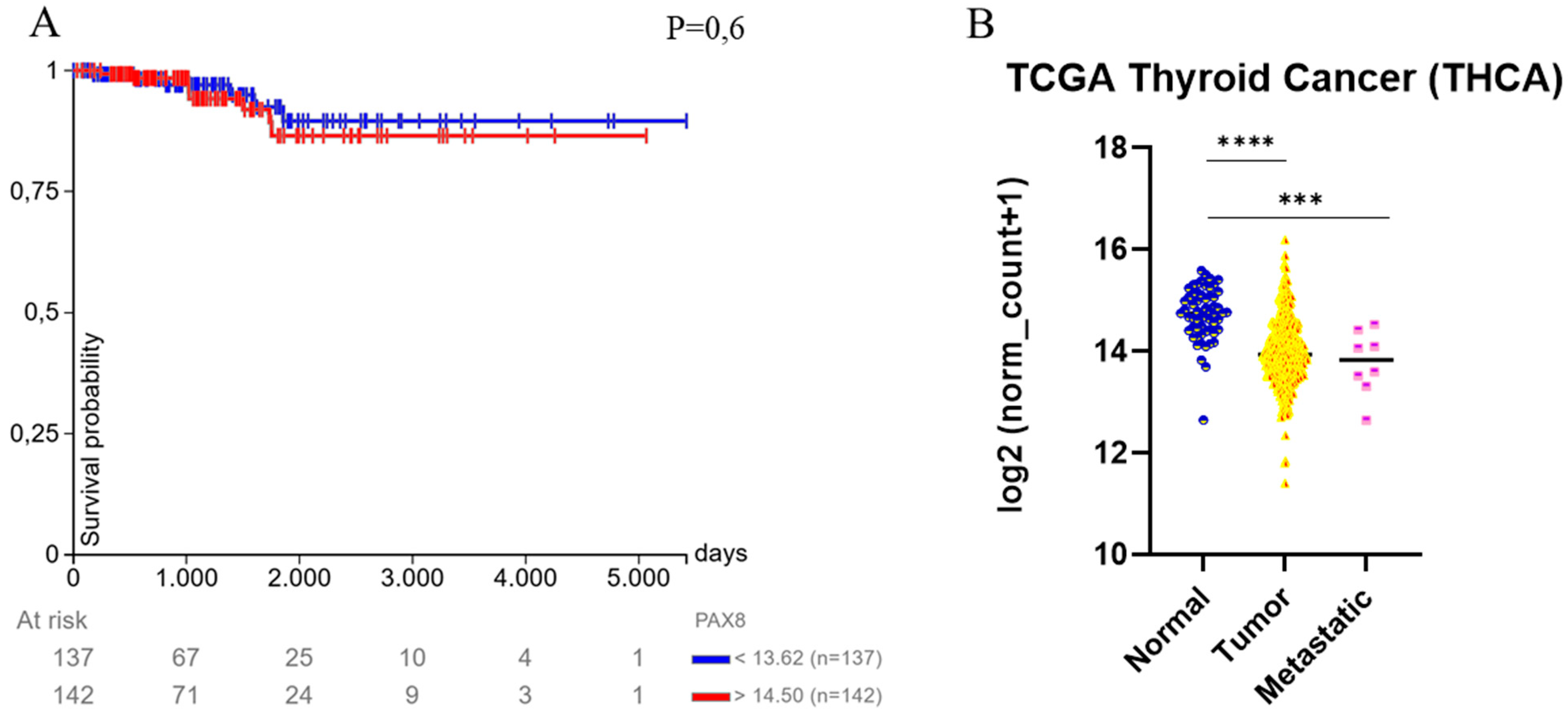
4. PAX8 and Thyroid Carcinoma
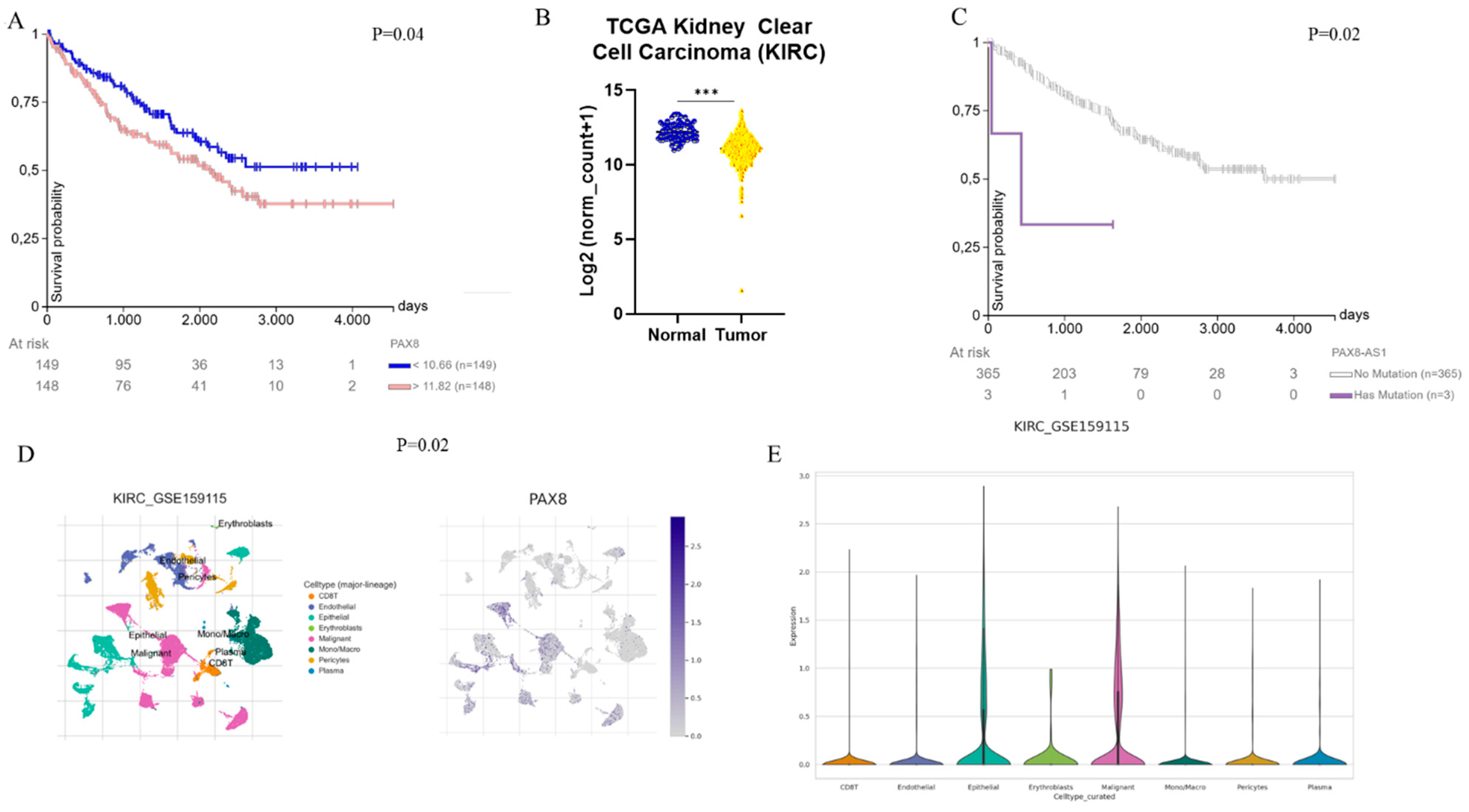
5. PAX2 and PAX8 in Tumors of the Urogenital Tract
Renal Cell Carcinoma
6. PAX8 and PAX9 in the Tumors of the Lung
7. PAX2 and PAX8 in Ovarian Cancer
8. Other PAX’s
9. Diagnostic Relevance and Clinical Applications of PAX Family Proteins
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Balczarek, K.A.; Lai, Z.C.; Kumar, S. Evolution of functional diversification of the paired box (Pax) DNA-binding domains. Mol. Biol. Evol. 1997, 14, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Rould, M.A.; Jun, S.; Desplan, C.; Pabo, C.O. Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell 1995, 80, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Bopp, D.; Burri, M.; Baumgartner, S.; Frigerio, G.; Noll, M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell 1986, 47, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.; Davidson, E.A.; Liu, W.; Nebert, D.W.; Bruford, E.A.; Zhao, H.; Dermitzakis, E.T.; Thompson, D.C.; Vasiliou, V. Overview of PAX gene family: Analysis of human tissue-specific variant expression and involvement in human disease. Hum. Genet. 2021, 140, 381–400. [Google Scholar] [CrossRef]
- Lang, D.; Powell, S.K.; Plummer, R.S.; Young, K.P.; Ruggeri, B.A. PAX genes: Roles in development, pathophysiology, and cancer. Biochem. Pharmacol. 2007, 73, 1–14. [Google Scholar] [CrossRef]
- Deutsch, U.; Dressler, G.R.; Gruss, P. Pax 1, a member of a paired box homologous murine gene family, is expressed in segmented structures during development. Cell 1988, 53, 617–625. [Google Scholar] [CrossRef]
- Neubüser, A.; Koseki, H.; Balling, R. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev. Biol. 1995, 170, 701–716. [Google Scholar] [CrossRef]
- Kan, Y.Y.; Liou, Y.L.; Wang, H.J.; Chen, C.Y.; Sung, L.C.; Chang, C.F.; Liao, C.I. PAX1 methylation as a potential biomarker for cervical cancer screening. Int. J. Gynecol. Cancer 2014, 24, 928–934. [Google Scholar] [CrossRef]
- Huang, T.H.; Lai, H.C.; Liu, H.W.; Lin, C.J.; Wang, K.H.; Ding, D.C.; Chu, T.Y. Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. Int. J. Gynecol. Cancer 2010, 20, 513–519. [Google Scholar] [CrossRef]
- Huang, J.; Tan, Z.R.; Yu, J.; Li, H.; Lv, Q.L.; Shao, Y.Y.; Zhou, H.H. DNA hypermethylated status and gene expression of PAX1/SOX1 in patients with colorectal carcinoma. OncoTargets Ther. 2017, 10, 4739–4751. [Google Scholar] [CrossRef]
- Peters, H.; Schuster, G.; Neubüser, A.; Richter, T.; Höfler, H.; Balling, R. Isolation of the Pax9 cDNA from adult human esophagus. Mamm. Genome 1997, 8, 62–64. [Google Scholar] [CrossRef] [PubMed]
- Gerber, J.K.; Richter, T.; Kremmer, E.; Adamski, J.; Höfler, H.; Balling, R.; Peters, H. Progressive loss of PAX9 expression correlates with increasing malignancy of dysplastic and cancerous epithelium of the human oesophagus. J. Pathol. 2002, 197, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Gómez-Pardo, E.; Dressler, G.R.; Gruss, P. Pax-2 controls multiple steps of urogenital development. Development 1995, 121, 4057–4065. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Bolado, G.; Schwarz, M.; Gruss, P. Pax-2 in the chiasm. Cell Tissue Res. 1997, 290, 197–200. [Google Scholar] [CrossRef]
- GETX Database. Available online: https://www.gtexportal.org/home/ (accessed on 28 March 2025).
- Gnarra, J.R.; Dressler, G.R. Expression of Pax-2 in human renal cell carcinoma and growth inhibition by antisense oligonucleotides. Cancer Res. 1995, 55, 4092–4098. [Google Scholar]
- Nutt, S.L.; Heavey, B.; Rolink, A.G.; Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 1999, 401, 556–562. [Google Scholar] [CrossRef]
- Adams, B.; Dörfler, P.; Aguzzi, A.; Kozmik, Z.; Urbanek, P.; Maurer-Fogy, I.; Busslinger, M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992, 6, 1589–1607. [Google Scholar] [CrossRef]
- Kaneko, H.; Ariyasu, T.; Inoue, R.; Fukao, T.; Kasahara, K.; Teramoto, T.; Matsui, E.; Hayakawa, S.; Kondo, N. Expression of Pax5 gene in human haematopoietic cells and tissues: Comparison with immunodeficient donors. Clin. Exp. Immunol. 1998, 111, 339–344. [Google Scholar] [CrossRef]
- Torlakovic, E.; Slipicevic, A.; Robinson, C.; DeCoteau, J.F.; Alfsen, G.C.; Vyberg, M.; Chibbar, R.; Flørenes, V.A. Pax-5 expression in nonhematopoietic tissues. Am. J. Clin. Pathol. 2006, 126, 798–804. [Google Scholar] [CrossRef]
- Krenacs, L.; Himmelmann, A.W.; Quintanilla-Martinez, L.; Fest, T.; Riva, A.; Wellmann, A.; Bagdi, E.; Kehrl, J.H.; Jaffe, E.S.; Raffeld, M. Transcription factor B-cell-specific activator protein (BSAP) is differentially expressed in B cells and in subsets of B-cell lymphomas. Blood 1998, 92, 1308–1316. [Google Scholar] [CrossRef]
- O’Brien, P.; Morin, P., Jr.; Ouellette, R.J.; Robichaud, G.A. The Pax-5 gene: A pluripotent regulator of B-cell differentiation and cancer disease. Cancer Res. 2011, 71, 7345–7350. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, M.; Souabni, A.; Mandler, M.; Neubüser, A.; Busslinger, M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002, 16, 2958–2970. [Google Scholar] [CrossRef] [PubMed]
- Pasca di Magliano, M.; Di Lauro, R.; Zannini, M. Pax8 has a key role in thyroid cell differentiation. Proc. Natl. Acad. Sci. USA 2000, 97, 13144–13149. [Google Scholar] [CrossRef]
- Castro, P.; Rebocho, A.P.; Soares, R.J.; Magalhaes, J.; Roque, L.; Trovisco, V.; Vieira de Castro, I.; Cardoso-de-Oliveira, M.; Fonseca, E.; Soares, P.; et al. PAX8-PPAR γ gamma rearrangement is frequently detected in the follicular variant of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2006, 91, 213–220. [Google Scholar] [CrossRef]
- Scouten, W.T.; Patel, A.; Terrell, R.; Burch, H.B.; Bernet, V.J.; Tuttle, R.M.; Francis, G.L. Cytoplasmic localization of the paired box gene, Pax-8, is found in pediatric thyroid cancer and may be associated with a greater risk of recurrence. Thyroid 2004, 14, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Hornyak, T.J.; Hayes, D.J.; Chiu, L.Y.; Ziff, E.B. Transcription factors in melanocyte development: Distinct roles for Pax-3 and Mitf. Mech. Dev. 2001, 101, 47–59. [Google Scholar] [CrossRef]
- Tsukamoto, K.; Nakamura, Y.; Niikawa, N. Isolation of two isoforms of the PAX3 gene transcripts and their tissue-specific alternative expression in human adult tissues. Hum. Genet. 1994, 93, 270–274. [Google Scholar] [CrossRef]
- Hoth, C.F.; Milunsky, A.; Lipsky, N.; Sheffer, R.; Clarren, S.K.; Baldwin, C.T. Mutations in the paired domain of the human PAX3 gene cause Klein-Waardenburg syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I). Am. J. Hum. Genet. 1993, 52, 455–462. [Google Scholar]
- Tiffin, N.; Williams, R.D.; Shipley, J.; Pritchard-Jones, K. PAX7 expression in embryonal rhabdomyosarcoma suggests an origin in muscle satellite cells. Br. J. Cancer 2003, 89, 327–332. [Google Scholar] [CrossRef]
- Cariati, I.; Scimeca, M.; Bonanni, R.; Triolo, R.; Naldi, V.; Toro, G.; Marini, M.; Tancredi, V.; Iundusi, R.; Gasbarra, E.; et al. Role of Myostatin in Muscle Degeneration by Random Positioning Machine Exposure: An in vitro Study for the Treatment of Sarcopenia. Front. Physiol. 2022, 13, 782000. [Google Scholar] [CrossRef]
- Scimeca, M.; Piccirilli, E.; Mastrangeli, F.; Rao, C.; Feola, M.; Orlandi, A.; Gasbarra, E.; Bonanno, E.; Tarantino, U. Bone Morphogenetic Proteins and myostatin pathways: Key mediator of human sarcopenia. J. Transl. Med. 2017, 15, 34. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Pineda, B. The gene Pax4 is an essential regulator of pancreatic beta-cell development. Mol. Cells 2004, 18, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Beucher, A.; Gjernes, E.; Collin, C.; Courtney, M.; Meunier, A.; Collombat, P.; Gradwohl, G. The homeodomain-containing transcription factors Arx and Pax4 control enteroendocrine subtype specification in mice. PLoS ONE 2012, 7, e36449. [Google Scholar] [CrossRef]
- Ashery-Padan, R.; Zhou, X.; Marquardt, T.; Herrera, P.; Toube, L.; Berry, A.; Gruss, P. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev. Biol. 2004, 269, 479–488. [Google Scholar] [CrossRef]
- Lau, H.H.; Krentz, N.A.; Abaitua, F.; Perez-Alcantara, M.; Chan, J.W.; Ajeian, J.; Ghosh, S.; Lee, Y.; Yang, J.; Thaman, S.; et al. PAX4 loss of function increases diabetes risk by altering human pancreatic endocrine cell development. Nat. Commun. 2023, 14, 6119. [Google Scholar] [CrossRef]
- St-Onge, L.; Sosa-Pineda, B.; Chowdhury, K.; Mansouri, A.; Gruss, P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 1997, 387, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.C.; Higgins, J.P.; Montgomery, K.; Kaygusuz, G.; van de Rijn, M.; Natkunam, Y. The utility of PAX5 immunohistochemistry in the diagnosis of undifferentiated malignant neoplasms. Mod. Pathol. 2007, 20, 871–877. [Google Scholar] [CrossRef]
- Cobaleda, C.; Schebesta, A.; Delogu, A.; Busslinger, M. Pax5: The guardian of B cell identity and function. Nat. Immunol. 2007, 8, 463–470. [Google Scholar] [CrossRef]
- Nasri Nasrabadi, P.; Martin, D.; Gharib, E.; Robichaud, G.A. The Pleiotropy of PAX5 Gene Products and Function. Int. J. Mol. Sci. 2022, 23, 10095. [Google Scholar] [CrossRef]
- Schatz, D.G.; Ji, Y. Recombination centres and the orchestration of V (D) J recombination. Nat. Rev. Immunol. 2011, 11, 251–263. [Google Scholar] [CrossRef]
- Fuxa, M.; Skok, J.; Souabni, A.; Salvagiotto, G.; Roldan, E.; Busslinger, M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004, 18, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Fedl, A.S.; Tagoh, H.; Gruenbacher, S.; Sun, Q.; Schenk, R.L.; Froussios, K.; Jaritz, M.; Busslinger, M.; Schwickert, T.A. Transcriptional function of E2A, Ebf1, Pax5, Ikaros and Aiolos analyzed by in vivo acute protein degradation in early B cell development. Nat. Immunol. 2024, 25, 1663–1677. [Google Scholar] [CrossRef] [PubMed]
- Malard, F.; Mohty, M. Acute lymphoblastic leukaemia. Lancet 2020, 395, 1146–1162. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, K.; Strid, T.; Kuruvilla, J.; Somasundaram, R.; Cristobal, S.; Smith, E.; Prasad, M.; Fioretos, T.; Lilljebjörn, H.; Soneji, S.; et al. PAX5 is part of a functional transcription factor network targeted in lymphoid leukemia. PLoS Genet. 2019, 15, e1008280. [Google Scholar] [CrossRef]
- Jia, Z.; Gu, Z. PAX5 alterations in B-cell acute lymphoblastic leukemia. Front. Oncol. 2022, 12, 1023606. [Google Scholar] [CrossRef]
- Kim, M.; Choi, J.E.; She, C.J.; Hwang, S.M.; Shin, H.Y.; Ahn, H.S.; Yoon, S.S.; Kim, B.K.; Park, M.H.; Lee, D.S. PAX5 deletion is common and concurrently occurs with CDKN2A deletion in B-lineage acute lymphoblastic leukemia. Blood Cells Mol. Dis. 2011, 47, 62–66. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Goorha, S.; Radtke, I.; Miller, C.B.; Coustan-Smith, E.; Dalton, J.D.; Girtman, K.; Mathew, S.; Ma, J.; Pounds, S.B.; et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007, 446, 758–764. [Google Scholar] [CrossRef]
- Familiades, J.; Bousquet, M.; Lafage-Pochitaloff, M.; Bene, M.C.; Beldjord, K.; De Vos, J.; Dastugue, N.; Coyaud, E.; Struski, S.; Quelen, C.; et al. PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploinsufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: A GRAALL study. Leukemia 2009, 23, 1989–1998. [Google Scholar] [CrossRef]
- Iacobucci, I.; Lonetti, A.; Paoloni, F.; Papayannidis, C.; Ferrari, A.; Storlazzi, C.T.; Vignetti, M.; Cilloni, D.; Messa, F.; Guadagnuolo, V.; et al. The PAX5 gene is frequently rearranged in BCR-ABL1-positive acute lymphoblastic leukemia but is not associated with outcome. A report on behalf of the GIMEMA Acute Leukemia Working Party. Haematologica 2010, 95, 1683–1690. [Google Scholar] [CrossRef]
- Urbánek, P.; Wang, Z.Q.; Fetka, I.; Wagner, E.F.; Busslinger, M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 1994, 79, 901–912. [Google Scholar] [CrossRef]
- Fouad, F.M.; Eid, J.I. PAX5 fusion genes in acute lymphoblastic leukemia: A literature review. Medicine 2023, 102, e33836. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, N.; Pennella, M.A.; Woo, J.L.; Berk, A.J.; Koeffler, H.P. Dominant-negative mechanism of leukemogenic PAX5 fusions. Oncogene 2012, 31, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Cazzaniga, G.; Daniotti, M.; Tosi, S.; Giudici, G.; Aloisi, A.; Pogliani, E.; Kearney, L.; Biondi, A. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001, 61, 4666–4670. [Google Scholar] [PubMed]
- Jones, C.L.; Kirkpatrick, G.; Fleenor, C.; Seth, W.; Noetzli, L.J.; Fosmire, S.; Baturin, D.; Liang, X.; Hernandez, G.; Pietras, E.M.; et al. ETV6 regulates Pax5 expression in early B cell development. Blood 2016, 128, 2655. [Google Scholar] [CrossRef]
- Fazio, G.; Palmi, C.; Rolink, A.; Biondi, A.; Cazzaniga, G. PAX5/TEL acts as a transcriptional repressor causing down-modulation of CD19, enhances migration to CXCL12, and confers survival advantage in pre-BI cells. Cancer Res. 2008, 68, 181–189. [Google Scholar] [CrossRef]
- Cazzaniga, V.; Bugarin, C.; Bardini, M.; Giordan, M.; te Kronnie, G.; Basso, G.; Biondi, A.; Fazio, G.; Cazzaniga, G. LCK over-expression drives STAT5 oncogenic signaling in PAX5 translocated BCP-ALL patients. Oncotarget 2015, 6, 1569–1581. [Google Scholar] [CrossRef]
- Fazio, G.; Bresolin, S.; Silvestri, D.; Quadri, M.; Saitta, C.; Vendramini, E.; Buldini, B.; Palmi, C.; Bardini, M.; Grioni, A.; et al. PAX5 fusion genes are frequent in poor risk childhood acute lymphoblastic leukaemia and can be targeted with BIBF1120. eBioMedicine 2022, 83, 104224. [Google Scholar] [CrossRef]
- Smeenk, L.; Fischer, M.; Jurado, S.; Jaritz, M.; Azaryan, A.; Werner, B.; Roth, M.; Zuber, J.; Stanulla, M.; den Boer, M.L.; et al. Molecular role of the PAX5-ETV6 oncoprotein in promoting B-cell acute lymphoblastic leukemia. EMBO J. 2017, 36, 718–735. [Google Scholar] [CrossRef]
- Denk, D.; Nebral, K.; Bradtke, J.; Pass, G.; Möricke, A.; Attarbaschi, A.; Strehl, S. PAX5-AUTS2: A recurrent fusion gene in childhood B-cell precursor acute lymphoblastic leukemia. Leuk. Res. 2012, 36, e178–e181. [Google Scholar] [CrossRef]
- Fazio, G.; Daniele, G.; Cazzaniga, V.; Impera, L.; Severgnini, M.; Iacobucci, I.; Galbiati, M.; Leszl, A.; Cifola, I.; De Bellis, G.; et al. Three novel fusion transcripts of the paired box 5 gene in B-cell precursor acute lymphoblastic leukemia. Haematologica 2015, 100, e14–e17. [Google Scholar] [CrossRef]
- Denk, D.; Bradtke, J.; König, M.; Strehl, S. PAX5 fusion genes in t (7;9) (q11.2;p13) leukemia: A case report and review of the literature. Mol. Cytogenet. 2014, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, M.; Broccardo, C.; Quelen, C.; Meggetto, F.; Kuhlein, E.; Delsol, G.; Dastugue, N.; Brousset, P. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood 2007, 109, 3417–3423. [Google Scholar] [CrossRef] [PubMed]
- Jamrog, L.; Chemin, G.; Fregona, V.; Coster, L.; Pasquet, M.; Oudinet, C.; Rouquié, N.; Prade, N.; Lagarde, S.; Cresson, C.; et al. PAX5-ELN oncoprotein promotes multistep B-cell acute lymphoblastic leukemia in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 10357–10362. [Google Scholar] [CrossRef] [PubMed]
- Nebral, K.; Denk, D.; Attarbaschi, A.; König, M.; Mann, G.; Haas, O.A.; Strehl, S. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia 2009, 23, 134–143. [Google Scholar] [CrossRef]
- Roberts, K.G.; Morin, R.D.; Zhang, J.; Hirst, M.; Zhao, Y.; Su, X.; Chen, S.C.; Payne-Turner, D.; Churchman, M.L.; Harvey, R.C.; et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 2012, 22, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Duffield, A.S.; Mullighan, C.G.; Borowitz, M.J. International Consensus Classification of acute lymphoblastic leukemia/lymphoma. Virchows Arch. 2023, 482, 11–26. [Google Scholar] [CrossRef]
- Jean-Loup Huret: T(9;9)(p13;p24) PAX5/JAK2 del(9)(p13p24) PAX5/JAK2 inv(9)(p13p24) PAX5/JAK2, Atlas Genet Cytogenet Oncol Haematol. 1 March 2014. Available online: http://atlasgeneticsoncology.org/haematological/1559/inv(9)(p13p24) (accessed on 28 March 2025).
- Jurado, S.; Fedl, A.S.; Jaritz, M.; Kostanova-Poliakova, D.; Malin, S.G.; Mullighan, C.G.; Strehl, S.; Fischer, M.; Busslinger, M. The PAX5-JAK2 translocation acts as dual-hit mutation that promotes aggressive B-cell leukemia via nuclear STAT5 activation. EMBO J. 2022, 41, e108397. [Google Scholar] [CrossRef]
- Schinnerl, D.; Fortschegger, K.; Kauer, M.; Marchante, J.R.; Kofler, R.; Den Boer, M.L.; Strehl, S. The role of the Janus-faced transcription factor PAX5-JAK2 in acute lymphoblastic leukemia. Blood 2015, 125, 1282–1291. [Google Scholar] [CrossRef]
- Nebral, K.; König, M.; Harder, L.; Siebert, R.; Haas, O.A.; Strehl, S. Identification of PML as novel PAX5 fusion partner in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2007, 139, 269–274. [Google Scholar] [CrossRef]
- Gu, Z.; Churchman, M.L.; Roberts, K.G.; Moore, I.; Zhou, X.; Nakitandwe, J.; Hagiwara, K.; Pelletier, S.; Gingras, S.; Berns, H.; et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019, 51, 296–307. [Google Scholar] [CrossRef]
- Jung, M.; Schieck, M.; Hofmann, W.; Tauscher, M.; Lentes, J.; Bergmann, A.; Stelter, M.; Möricke, A.; Alten, J.; Schlegelberger, B.; et al. Frequency and prognostic impact of PAX5 p.P80R in pediatric acute lymphoblastic leukemia patients treated on an AIEOP-BFM acute lymphoblastic leukemia protocol. Genes Chromosomes Cancer 2020, 59, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Hu, Z.; Damirchi, B.; Han, T.T.; Gu, Z. Characterization of PAX5 Mutations in B Progenitor Acute Lymphoblastic Leukemia. Blood 2022, 140 (Suppl. S1), 1001–1002. [Google Scholar] [CrossRef]
- Strullu, M.; Caye-Eude, A.; Fenneteau, O.; Arfeuille, C.; Cuccuini, W.; Cavé, H.; Baruchel, A.; Lainey, E. A PAX5 P80R pediatric B acute lymphoblastic leukemia with monocytic lineage switch at diagnosis: Deciphering classification ambiguity. Pediatr. Blood Cancer 2024, 71, e30842. [Google Scholar] [CrossRef]
- Črepinšek, K.; Klobučar, N.; Tesovnik, T.; Šket, R.; Jenko Bizjan, B.; Kovač, J.; Kavčič, M.; Prelog, T.; Kitanovski, L.; Jazbec, J.; et al. PAX5 Alterations in a Consecutive Childhood B-Cell Acute Lymphoblastic Leukemia Cohort Treated Using the ALL IC-BFM 2009 Protocol. Cancers 2024, 16, 1164. [Google Scholar] [CrossRef]
- Passet, M.; Boissel, N.; Sigaux, F.; Saillard, C.; Bargetzi, M.; Ba, I.; Thomas, X.; Graux, C.; Chalandon, Y.; Leguay, T.; et al. PAX5 P80R mutation identifies a novel subtype of B-cell precursor acute lymphoblastic leukemia with favorable outcome. Blood 2019, 133, 280–284, Erratum in Blood 2020, 135, 2011. [Google Scholar] [CrossRef]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef]
- Ramamoorthy, S.; Kometani, K.; Herman, J.S.; Bayer, M.; Boller, S.; Edwards-Hicks, J.; Ramachandran, H.; Li, R.; Klein-Geltink, R.; Pearce, E.L.; et al. EBF1 and Pax5 safeguard leukemic transformation by limiting IL-7 signaling, Myc expression, and folate metabolism. Genes Dev. 2020, 34, 1503–1519. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, R.; Jensen, C.T.; Tingvall-Gustafsson, J.; Åhsberg, J.; Okuyama, K.; Prasad, M.; Hagman, J.R.; Wang, X.; Soneji, S.; Strid, T.; et al. EBF1 and PAX5 control pro-B cell expansion via opposing regulation of the Myc gene. Blood 2021, 137, 3037–3049. [Google Scholar] [CrossRef]
- Kaiser, F.M.; Janowska, I.; Menafra, R.; de Gier, M.; Korzhenevich, J.; Pico-Knijnenburg, I.; Khatri, I.; Schulz, A.; Kuijpers, T.W.; Lankester, A.C.; et al. IL-7 receptor signaling drives human B-cell progenitor differentiation and expansion. Blood 2023, 142, 1113–1130. [Google Scholar] [CrossRef]
- Hirokawa, S.; Sato, H.; Kato, I.; Kudo, A. EBF-regulating Pax5 transcription is enhanced by STAT5 in the early stage of B cells. Eur. J. Immunol. 2003, 33, 1824–1829. [Google Scholar] [CrossRef]
- Shah, S.; Schrader, K.A.; Waanders, E.; Timms, A.E.; Vijai, J.; Miething, C.; Wechsler, J.; Yang, J.; Hayes, J.; Klein, R.J.; et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 1226–1231. [Google Scholar] [CrossRef]
- Auer, F.; Rüschendorf, F.; Gombert, M.; Husemann, P.; Ginzel, S.; Izraeli, S.; Harit, M.; Weintraub, M.; Weinstein, O.Y.; Lerer, I.; et al. Inherited susceptibility to pre B-ALL caused by germline transmission of PAX5 c.547G>A. Leukemia 2014, 28, 1136–1138. [Google Scholar] [CrossRef] [PubMed]
- Yazdanparast, S.; Khatami, S.R.; Galehdari, H.; Jaseb, K. One missense mutation in exon 2 of the PAX5 gene in Iran. Genet. Mol. Res. 2015, 14, 17768–17775. [Google Scholar] [CrossRef] [PubMed]
- Van Engelen, N.; Roest, M.; van Dijk, F.; Sonneveld, E.; Bladergroen, R.; van Reijmersdal, S.V.; van der Velden, V.H.J.; Hoogeveen, P.G.; Kors, W.A.; Waanders, E.; et al. A novel germline PAX5 single exon deletion in a pediatric patient with precursor B-cell leukemia. Leukemia 2023, 37, 1908–1911. [Google Scholar] [CrossRef] [PubMed]
- Escudero, A.; Takagi, M.; Auer, F.; Friedrich, U.A.; Miyamoto, S.; Ogawa, A.; Imai, K.; Pascual, B.; Vela, M.; Stepensky, P.; et al. Clinical and immunophenotypic characteristics of familial leukemia predisposition caused by PAX5 germline variants. Leukemia 2022, 36, 2338–2342. [Google Scholar] [CrossRef]
- Duployez, N.; Jamrog, L.A.; Fregona, V.; Hamelle, C.; Fenwarth, L.; Lejeune, S.; Helevaut, N.; Geffroy, S.; Caillault, A.; Marceau-Renaut, A.; et al. Germline PAX5 mutation predisposes to familial B-cell precursor acute lymphoblastic leukemia. Blood 2021, 137, 1424–1428. [Google Scholar] [CrossRef]
- Bettini, L.R.; Fazio, G.; Saitta, C.; Piazza, R.; Palamini, S.; Buracchi, C.; Rebellato, S.; Santoro, N.; Simone, C.; Biondi, A.; et al. Diverse mechanisms of leukemogenesis associated with PAX5 germline mutation. Leukemia 2024, 38, 2479–2482. [Google Scholar] [CrossRef]
- Alfaifi, A.; Bahashwan, S.; Alsaadi, M.; Ageel, A.H.; Ahmed, H.H.; Fatima, K.; Malhan, H.; Qadri, I.; Almehdar, H. Advancements in B-Cell Non-Hodgkin’s Lymphoma: From Signaling Pathways to Targeted Therapies. Adv. Hematol. 2024, 2024, 5948170. [Google Scholar] [CrossRef]
- Miao, Y.; Medeiros, L.J.; Li, Y.; Li, J.; Young, K.H. Genetic alterations and their clinical implications in DLBCL. Nat. Rev. Clin. Oncol. 2019, 16, 634–652. [Google Scholar] [CrossRef]
- Ohno, H.; Nakagawa, M.; Kishimori, C.; Fukutsuka, K.; Maekawa, F.; Takeoka, K.; Hayashida, M.; Sakamoto, S.; Akasaka, T.; Honjo, G. Diffuse large B-cell lymphoma carrying t(9;14)(p13;q32)/PAX5-immunoglobulin heavy chain gene is characterized by nuclear positivity of MUM1 and PAX5 by immunohistochemistry. Hematol. Oncol. 2020, 38, 171–180. [Google Scholar] [CrossRef]
- Iida, S.; Rao, P.H.; Nallasivam, P.; Hibshoosh, H.; Butler, M.; Louie, D.C.; Dyomin, V.; Ohno, H.; Chaganti, R.S.; Dalla-Favera, R. The t(9;14)(p13;q32) chromosomal translocation associated with lymphoplasmacytoid lymphoma involves the PAX-5 gene. Blood 1996, 88, 4110–4117. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.M.; Jäger, U.; Chott, A.; Schebesta, M.; Haas, O.A.; Busslinger, M. Deregulated PAX-5 transcription from a translocated IgH promoter in marginal zone lymphoma. Blood 1998, 92, 3865–3878. [Google Scholar] [CrossRef]
- Balasenthil, S.; Gururaj, A.E.; Talukder, A.H.; Bagheri-Yarmand, R.; Arrington, T.; Haas, B.J.; Braisted, J.C.; Kim, I.; Lee, N.H.; Kumar, R. Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Res. 2007, 67, 7132–7138, Erratum in Cancer Res. 2015, 75, 2580–2581. [Google Scholar] [CrossRef]
- Wang, T.; Chen, Z.; Li, C.; Zhang, W.; Huang, W.; Xue, J.; Wang, J.; Li, S. PAX5 and circ1857 affected DLBCL progression and B-cell proliferation through regulating GINS1. Cancer Sci. 2023, 114, 3203–3215. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, T.; Li, C.; Zhang, W.; Huang, W.; Xue, J.; Wang, J.; Li, S. FOXP1-GINS1 axis promotes DLBCL proliferation and directs doxorubicin resistance. J. Cancer 2023, 14, 2289–2300. [Google Scholar] [CrossRef]
- Mohanty, A.; Sandoval, N.; Das, M.; Pillai, R.; Chen, L.; Chen, R.W.; Amin, H.M.; Wang, M.; Marcucci, G.; Weisenburger, D.D.; et al. CCND1 mutations increase protein stability and promote ibrutinib resistance in mantle cell lymphoma. Oncotarget 2016, 7, 73558–73572. [Google Scholar] [CrossRef]
- Mozos, A.; Royo, C.; Hartmann, E.; De Jong, D.; Baró, C.; Valera, A.; Fu, K.; Weisenburger, D.D.; Delabie, J.; Chuang, S.S.; et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica 2009, 94, 1555–1562. [Google Scholar] [CrossRef] [PubMed]
- Desouki, M.M.; Post, G.R.; Cherry, D.; Lazarchick, J. PAX-5: A valuable immunohistochemical marker in the differential diagnosis of lymphoid neoplasms. Clin. Med. Res. 2010, 8, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, Y.; Nie, Y.; Jiang, Y.; Sui, X.; Ge, X.; Liu, F.; Zhang, Y.; Wang, X. PAX5 aberrant expression incorporated in MIPI-SP risk scoring system exhibits additive value in mantle cell lymphoma. J. Mol. Med. 2023, 101, 595–606. [Google Scholar] [CrossRef]
- Vegliante, M.C.; Palomero, J.; Pérez-Galán, P.; Roué, G.; Castellano, G.; Navarro, A.; Clot, G.; Moros, A.; Suárez-Cisneros, H.; Bea, S.; et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood 2013, 121, 2175–2185. [Google Scholar] [CrossRef]
- Jing, C.; Zheng, Y.; Feng, Y.; Cao, X.; Xu, C. Prognostic significance of p53, Sox11, and Pax5 co-expression in mantle cell lymphoma. Sci. Rep. 2021, 11, 11896. [Google Scholar] [CrossRef]
- Zhang, X.R.; Chien, P.N.; Nam, S.Y.; Heo, C.Y. Anaplastic Large Cell Lymphoma: Molecular Pathogenesis and Treatment. Cancers 2022, 14, 1650. [Google Scholar] [CrossRef]
- Feldman, A.L.; Law, M.E.; Inwards, D.J.; Dogan, A.; McClure, R.F.; Macon, W.R. PAX5-positive T-cell anaplastic large cell lymphomas associated with extra copies of the PAX5 gene locus. Mod. Pathol. 2010, 23, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Fratoni, S.; Niscola, P.; Zhao, X.F.; Larocca, L.M.; Capalbo, A.; Fabbretti, M.; Bernardini, L.; Abruzzese, E. ALK-negative anaplastic large cell lymphoma with “Hodgkin-like” cytomorphology and nuclear expression of PAX5. Pathol. Res. Pract. 2020, 216, 152724. [Google Scholar] [CrossRef]
- Kawakami, K.; Yazaki, A.; Ito, R.; Tono, Y.; Murata, T.; Baba, Y.; Uchiyama, T.; Imai, H.; Nakamura, S. Refractory case of ALK-negative anaplastic large-cell lymphoma with PAX-5 expression and T-cell receptor-γ gene rearrangement. J. Clin. Exp. Hematop. 2013, 53, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Ong, D.M.; Cummins, K.D.; Pham, A.; Grigoriadis, G. PAX5-expressing ALK-negative anaplastic large cell lymphoma with extensive extranodal and nodal involvement. BMJ Case Rep. 2015, 2015, bcr2015211159. [Google Scholar] [CrossRef] [PubMed]
- Salyana, M.A.; Khan, S.; Zhang, X. ALK-negative anaplastic large cell lymphoma, null type with aberrant expression of PAX5 and CD138: A diagnostic pitfall. Diagn. Cytopathol. 2021, 49, E395–E399. [Google Scholar] [CrossRef]
- Bonfiglio, R.; Di Pietro, M.L. The impact of oral contraceptive use on breast cancer risk: State of the art and future perspectives in the era of 4P medicine. Semin. Cancer Biol. 2021, 72, 11–18. [Google Scholar] [CrossRef]
- Kim, J.; Harper, A.; McCormack, V.; Sung, H.; Houssami, N.; Morgan, E.; Mutebi, M.; Garvey, G.; Soerjomataram, I.; Fidler-Benaoudia, M.M. Global patterns and trends in breast cancer incidence and mortality across 185 countries. Nat. Med. 2025, 31, 1154–1162. [Google Scholar] [CrossRef]
- Yang, X.; Smirnov, A.; Buonomo, O.C.; Mauriello, A.; Shi, Y.; Bischof, J.; Woodsmith, J.; TOR CENTRE; Melino, G.; Candi, E.; et al. A primary luminal/HER2 negative breast cancer patient with mismatch repair deficiency. Cell Death Discov. 2023, 9, 365. [Google Scholar] [CrossRef]
- Vanni, G.; Costanzo, G.; Pellicciaro, M.; Materazzo, M.; Buonomo, C.; Federico, T.; Giacobbi, E.; Servadei, F.; Anemona, L.; Noce, A.; et al. Awake Breast Surgery: A Systematic Review. In Vivo 2023, 37, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Arciero, I.; Buonvino, S.; Palumbo, V.; Scimeca, M.; Melino, S. A 3D-Printable Cell Array for In Vitro Breast Cancer Modeling. Int. J. Mol. Sci. 2024, 25, 13068. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Rovella, V.; Smirnov, A.; Buonomo, O.C.; Mauriello, A.; Perretta, T.; Shi, Y.; Woodmsith, J.; Bischof, J.; TOR CENTRE; et al. A BRCA2 germline mutation and high expression of immune checkpoints in a TNBC patient. Cell Death Discov. 2023, 9, 370. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, N.; Harquail, J.; Crapoulet, N.; Ouellette, R.J.; Robichaud, G.A. Pax-5 Inhibits Breast Cancer Proliferation Through MiR-215 Up-regulation. Anticancer Res. 2018, 38, 5013–5026. [Google Scholar] [CrossRef]
- Scimeca, M.; Bonfiglio, R.; Varone, F.; Ciuffa, S.; Mauriello, A.; Bonanno, E. Calcifications in prostate cancer: An active phenomenon mediated by epithelial cells with osteoblast-phenotype. Microsc. Res. Tech. 2018, 81, 745–748. [Google Scholar] [CrossRef]
- Giacobbi, E.; Bonfiglio, R.; Rotondaro, G.; Servadei, F.; Smirnov, A.; Palumbo, V.; Scioli, M.P.; Bonanno, E.; Buonomo, C.O.; Vanni, G.; et al. Implications of Mineralization Biomarkers in Breast Cancer Outcomes Beyond Calcifications. Int. J. Mol. Sci. 2025, 26, 645. [Google Scholar] [CrossRef] [PubMed]
- Bonfiglio, R.; Sisto, R.; Casciardi, S.; Palumbo, V.; Scioli, M.P.; Giacobbi, E.; Servadei, F.; Melino, G.; Mauriello, A.; Scimeca, M. Aluminium bioaccumulation in colon cancer, impinging on epithelial-mesenchymal-transition and cell death. Sci. Total Environ. 2024, 908, 168335. [Google Scholar] [CrossRef]
- Scimeca, M.; Trivigno, D.; Bonfiglio, R.; Ciuffa, S.; Urbano, N.; Schillaci, O.; Bonanno, E. Breast cancer metastasis to bone: From epithelial to mesenchymal transition to breast osteoblast-like cells. Semin. Cancer Biol. 2021, 72, 155–164. [Google Scholar] [CrossRef]
- Bonfiglio, R.; Granaglia, A.; Giocondo, R.; Scimeca, M.; Bonanno, E. Molecular Aspects and Prognostic Significance of Microcalcifications in Human Pathology: A Narrative Review. Int. J. Mol. Sci. 2020, 22, 120. [Google Scholar] [CrossRef]
- Benzina, S.; Beauregard, A.P.; Guerrette, R.; Jean, S.; Faye, M.D.; Laflamme, M.; Maïcas, E.; Crapoulet, N.; Ouellette, R.J.; Robichaud, G.A. Pax-5 is a potent regulator of E-cadherin and breast cancer malignant processes. Oncotarget 2017, 8, 12052–12066. [Google Scholar] [CrossRef]
- Palmisano, W.A.; Crume, K.P.; Grimes, M.J.; Winters, S.A.; Toyota, M.; Esteller, M.; Joste, N.; Baylin, S.B.; Belinsky, S.A. Aberrant promoter methylation of the transcription factor genes PAX5 alpha and beta in human cancers. Cancer Res. 2003, 63, 4620–4625. [Google Scholar] [PubMed]
- Moelans, C.B.; Verschuur-Maes, A.H.; Van Diest, P.J. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J. Pathol. 2011, 225, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Chen, Y.B.; Yue, H.R.; Zhou, X.J.; Ma, H.Y.; Wang, X.; Cao, X.C.; Yu, Y. PAX5-miR-142 feedback loop promotes breast cancer proliferation by regulating DNMT1 and ZEB1. Mol. Med. 2023, 29, 89. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, J.; Luo, X.; Yang, D.; Yin, X.; Peng, W.; Bi, C.; Ren, G.; Xiang, T. Paired box 5 is a novel marker of breast cancers that is frequently downregulated by methylation. Int. J. Biol. Sci. 2018, 14, 1686–1695. [Google Scholar] [CrossRef]
- Luo, M.; Guan, J.L. Focal adhesion kinase: A prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett. 2010, 289, 127–139. [Google Scholar] [CrossRef]
- Crapoulet, N.; O’Brien, P.; Ouellette, R.J.; Robichaud, G.A. Coordinated expression of Pax-5 and FAK1 in metastasis. Anticancer Agents Med. Chem. 2011, 11, 643–649. [Google Scholar] [CrossRef]
- Van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef]
- Vidal, L.J.; Perry, J.K.; Vouyovitch, C.M.; Pandey, V.; Brunet-Dunand, S.E.; Mertani, H.C.; Liu, D.X.; Lobie, P.E. PAX5alpha enhances the epithelial behavior of human mammary carcinoma cells. Mol. Cancer Res. 2010, 8, 444–456. [Google Scholar] [CrossRef]
- Harquail, J.; LeBlanc, N.; Landry, C.; Crapoulet, N.; Robichaud, G.A. Pax-5 Inhibits NF-κB Activity in Breast Cancer Cells Through IKKε and miRNA-155 Effectors. J. Mammary Gland Biol. Neoplasia 2018, 23, 177–187. [Google Scholar] [CrossRef]
- Razaviyan, J.; Sirati-Sabet, M.; Hadavi, R.; Karima, S.; Rajabibazl, M.; Mohammadi-Yeganeh, S. Exosomal Delivery of miR-155 Inhibitor can Suppress Migration, Invasion, and Angiogenesis Via PTEN and DUSP14 in Triple-negative Breast Cancer. Curr. Med. Chem. 2024. [Google Scholar] [CrossRef]
- Chen, X.; Wu, W.; Jeong, J.H.; Rokavec, M.; Wei, R.; Feng, S.; Schroth, W.; Brauch, H.; Zhong, S.; Luo, J.L. Cytokines-activated nuclear IKKα-FAT10 pathway induces breast cancer tamoxifen-resistance. Sci. China Life Sci. 2024, 67, 1413–1426. [Google Scholar] [CrossRef] [PubMed]
- Georgala, P.A.; Carr, C.B.; Price, D.J. The role of Pax6 in forebrain development. Dev. Neurobiol. 2011, 71, 690–709. [Google Scholar] [CrossRef]
- Thakurela, S.; Tiwari, N.; Schick, S.; Garding, A.; Ivanek, R.; Berninger, B.; Tiwari, V.K. Mapping gene regulatory circuitry of Pax6 during neurogenesis. Cell Discov. 2016, 2, 15045. [Google Scholar] [CrossRef]
- Jones, L.; López-Bendito, G.; Gruss, P.; Stoykova, A.; Molnár, Z. Pax6 is required for the normal development of the forebrain axonal connections. Development 2002, 129, 5041–5052. [Google Scholar] [CrossRef]
- Manuel, M.; Tan, K.B.; Kozic, Z.; Molinek, M.; Marcos, T.S.; Razak, M.F.A.; Dobolyi, D.; Dobie, R.; Henderson, B.E.; Henderson, N.C.; et al. Pax6 limits the competence of developing cerebral cortical cells to respond to inductive intercellular signals. PLoS Biol. 2022, 20, e3001563. [Google Scholar] [CrossRef]
- Hart, A.W.; Mella, S.; Mendrychowski, J.; van Heyningen, V.; Kleinjan, D.A. The developmental regulator Pax6 is essential for maintenance of islet cell function in the adult mouse pancreas. PLoS ONE 2013, 8, e54173. [Google Scholar] [CrossRef] [PubMed]
- Lima Cunha, D.; Arno, G.; Corton, M.; Moosajee, M. The Spectrum of PAX6 Mutations and Genotype-Phenotype Correlations in the Eye. Genes 2019, 10, 1050. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.K.; Bobilev, A.M.; Branch, A.; Lauderdale, J.D. Structural and functional consequences of PAX6 mutations in the brain: Implications for aniridia. Brain Res. 2021, 1756, 147283. [Google Scholar] [CrossRef]
- Tian, N.M.L.; Price, D.J. Why cavefish are blind. Bioessays 2005, 27, 235–238. [Google Scholar] [CrossRef]
- Ninkovic, J.; Pinto, L.; Petricca, S.; Lepier, A.; Sun, J.; Rieger, M.A.; Schroeder, T.; Cvekl, A.; Favor, J.; Götz, M. The transcription factor Pax6 regulates survival of dopaminergic olfactory bulb neurons via crystallin αA. Neuron 2010, 68, 682–694. [Google Scholar] [CrossRef]
- Meng, B.; Wang, Y.; Li, B. Suppression of PAX6 promotes cell proliferation and inhibits apoptosis in human retinoblastoma cells. Int. J. Mol. Med. 2014, 34, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.B.; Young, K.P.; Littlejohn, E.L.; Yoo, B.K.; Salgia, R.; Lang, D. PAX6 is expressed in pancreatic cancer and actively participates in cancer progression through activation of the MET tyrosine kinase receptor gene. J. Biol. Chem. 2009, 284, 27524–27532. [Google Scholar] [CrossRef] [PubMed]
- Ooki, A.; Dinalankara, W.; Marchionni, L.; Tsay, J.C.J.; Goparaju, C.; Maleki, Z.; Rom, W.N.; Pass, H.I.; Hoque, M.O. Epigenetically regulated PAX6 drives cancer cells toward a stem-like state via GLI-SOX2 signaling axis in lung adenocarcinoma. Oncogene 2018, 37, 5967–5981. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.M.; Zhang, T.; Liu, Y.B.; Deng, S.H.; Han, R.; Liu, T.; Li, J.; Xu, Y. The PAX6-ZEB2 axis promotes metastasis and cisplatin resistance in non-small cell lung cancer through PI3K/AKT signaling. Cell Death Dis. 2019, 10, 349. [Google Scholar] [CrossRef]
- Jing, N.; Du, X.; Liang, Y.; Tao, Z.; Bao, S.; Xiao, H.; Dong, B.; Gao, W.Q.; Fang, Y.X. PAX6 promotes neuroendocrine phenotypes of prostate cancer via enhancing MET/STAT5A-mediated chromatin accessibility. J. Exp. Clin. Cancer Res. 2024, 43, 144, Erratum in J. Exp. Clin. Cancer Res. 2024, 43, 167. [Google Scholar] [CrossRef]
- Rønneberg, J.A.; Fleischer, T.; Solvang, H.K.; Nordgard, S.H.; Edvardsen, H.; Potapenko, I.; Nebdal, D.; Daviaud, C.; Gut, I.; Bukholm, I.; et al. Methylation profiling with a panel of cancer related genes: Association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol. Oncol. 2011, 5, 61–76. [Google Scholar] [CrossRef]
- Wang, D.; Yang, P.N.; Chen, J.; Zhou, X.Y.; Liu, Q.J.; Li, H.J.; Li, C.L. Promoter hypermethylation may be an important mechanism of the transcriptional inactivation of ARRDC3, GATA5, and ELP3 in invasive ductal breast carcinoma. Mol. Cell Biochem. 2014, 396, 67–77. [Google Scholar] [CrossRef]
- Zong, X.; Yang, H.; Yu, Y.; Zou, D.; Ling, Z.; He, X.; Meng, X. Possible role of Pax-6 in promoting breast cancer cell proliferation and tumorigenesis. BMB Rep. 2011, 44, 595–600. [Google Scholar] [CrossRef]
- Xia, X.; Yin, W.; Zhang, X.; Yu, X.; Wang, C.; Xu, S.; Feng, W.; Yang, H. PAX6 overexpression is associated with the poor prognosis of invasive ductal breast cancer. Oncol. Lett. 2015, 10, 1501–1506. [Google Scholar] [CrossRef]
- Urrutia, G.; Laurito, S.; Marzese, D.M.; Gago, F.; Orozco, J.; Tello, O.; Branham, T.; Campoy, E.M.; Roqué, M. Epigenetic variations in breast cancer progression to lymph node metastasis. Clin. Exp. Metastasis 2015, 32, 99–110. [Google Scholar] [CrossRef]
- Urrutia, G.; Laurito, S.; Campoy, E.; Nasif, D.; Branham, M.T.; Roqué, M. PAX6 Promoter Methylation Correlates with MDA-MB-231 Cell Migration, and Expression of MMP2 and MMP9. Asian Pac. J. Cancer Prev. 2018, 19, 2859–2866. [Google Scholar] [PubMed]
- Liu, X.; Zhou, Y.; Qin, C.; Zhu, X. TNFRSF9 Suppressed the Progression of Breast Cancer via the p38MAPK/PAX6 Signaling Pathway. J. Oncol. 2022, 2022, 8549781. [Google Scholar] [CrossRef]
- Zhang, X.H.; Li, B.F.; Ding, J.; Shi, L.; Ren, H.M.; Liu, K.; Huang, C.C.; Ma, F.X.; Wu, X.Y. LncRNA DANCR-miR-758-3p-PAX6 Molecular Network Regulates Apoptosis and Autophagy of Breast Cancer Cells. Cancer Manag. Res. 2020, 12, 4073–4084. [Google Scholar] [CrossRef] [PubMed]
- Zannini, M.; Francis-Lang, H.; Plachov, D.; Lauro, R.D. Pax-8, a paired domain-containing protein, binds to a sequence overlapping the recognition site of a homeodomain and activates transcription from two thyroid-specific promoters. Mol. Cell Biol. 1992, 12, 4230–4241. [Google Scholar]
- Santisteban, P.; Bernal, J. Thyroid development and effect on the nervous system. Rev. Endocr. Metab. Disord. 2005, 6, 217–228. [Google Scholar] [CrossRef]
- Di Palma, T.; Filippone, M.G.; Pierantoni, G.M.; Fusco, A.; Soddu, S.; Zannini, M. Pax8 has a critical role in epithelial cell survival and proliferation. Cell Death Dis. 2013, 4, e729. [Google Scholar] [CrossRef] [PubMed]
- Krol, T.G.; Sarraf, P.; Pecciarini, L.; Chen, C.J.; Mueller, E.; Spiegelman, B.M.; Fletcher, J.A. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma. Science 2000, 289, 1357–1360. [Google Scholar] [CrossRef]
- Gregory Powell, J.; Wang, X.; Allard, B.L.; Sahin, M.; Wang, X.L.; Hay, I.D.; Hiddinga, H.J.; Deshpande, S.S.; Kroll, T.G.; Grebe, S.K.; et al. The PAX8/PPARgamma fusion oncoprotein transforms immortalized human thyrocytes through a mechanism probably involving wild-type PPARgamma inhibition. Oncogene 2004, 23, 3634–3641. [Google Scholar] [CrossRef]
- Fan, T.; Zhu, W.; Kong, M.; Yang, X.; Wang, C.; Wang, M.; Wang, Z. The Significance of PAX8-PPARγ Expression in Thyroid Cancer and the Application of a PAX8-PPARγ-Targeted Ultrasound Contrast Agent in the Early Diagnosis of Thyroid Cancer. Contrast Media Mol. Imaging 2022, 2022, 3265342. [Google Scholar] [CrossRef]
- Zhou, P.; Xu, T.; Hu, H.; Hua, F. Overexpression of PAX8-AS1 Inhibits Malignant Phenotypes of Papillary Thyroid Carcinoma Cells via miR-96-5p/PKN2 Axis. Int. J. Endocrinol. 2021, 2021, 5499963. [Google Scholar] [CrossRef]
- Ráduly, G.; Pap, Z.; Dénes, L.; Szántó, A.; Sipos, T.C.; Pávai, Z. The immunoexpression of aquaporin 1, PAX2, PAX8, connexin 36, connexin 43 in human fetal kidney. Rom. J. Morphol. Embryol. 2019, 60, 437–444. [Google Scholar] [PubMed]
- Eccles, M.R.; Yun, K.; Reeve, A.E.; Fidler, A.E. Comparative in situ hybridization analysis of PAX2, PAX8, and WT1 gene transcription in human fetal kidney and Wilms’ tumors. Am. J. Pathol. 1995, 146, 40–45. [Google Scholar]
- Rothenpieler, U.W.; Dressler, G.R. Pax-2 is required for mesenchyme-to-epithelium conversion during kidney development. Development 1993, 119, 711–720. [Google Scholar] [CrossRef]
- Tong, G.X.; Woojin, M.Y.; Beaubier, N.T.; Weeden, E.M.; Hamele-Bena, D.; Mansukhani, M.M.; O’toole, K.M. Expression of PAX8 in normal and neoplastic renal tissues: An immunohistochemical study. Mod. Pathol. 2009, 22, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Bahadoram, S.; Davoodi, M.; Hassanzadeh, S.; Bahadoram, M.; Barahman, M.; Mafakher, L. Renal cell carcinoma: An overview of the epidemiology, diagnosis, and treatment. G. Ital. Nefrol. 2022, 39, 2022. [Google Scholar] [PubMed]
- Luu, V.D.; Boysen, G.; Struckmann, K.; Casagrande, S.; von Teichman, A.; Wild, P.J.; Sulser, T.; Schraml, P.; Moch, H. Loss of VHL and hypoxia provokes PAX2 up-regulation in clear cell renal cell carcinoma. Clin. Cancer Res. 2009, 15, 3297–3304. [Google Scholar] [CrossRef]
- Patel, S.A.; Hirosue, S.; Rodrigues, P.; Vojtasova, E.; Richardson, E.K.; Ge, J.; Syafruddin, S.E.; Speed, A.; Papachristou, E.K.; Baker, D.; et al. The renal lineage factor PAX8 controls oncogenic signalling in kidney cancer. Nature 2022, 606, 999–1006. [Google Scholar] [CrossRef]
- Bleu, M.; Gaulis, S.; Lopes, R.; Sprouffske, K.; Apfel, V.; Holwerda, S.; Pregnolato, M.; Yildiz, U.; Cordoʹ, V.; Dost, A.F.; et al. PAX8 activates metabolic genes via enhancer elements in Renal Cell Carcinoma. Nat. Commun. 2019, 10, 3739. [Google Scholar] [CrossRef]
- Doberstein, K.; Pfeilschifter, J.; Gutwein, P. The transcription factor PAX2 regulates ADAM10 expression in renal cell carcinoma. Carcinogenesis 2011, 32, 1713–1723. [Google Scholar] [CrossRef]
- Kaur, G.; Li, C.G.; Chantry, A.; Stayner, C.; Horsfield, J.; Eccles, M.R. SMAD proteins directly suppress PAX2 transcription downstream of transforming growth factor-beta 1 (TGF-β1) signalling in renal cell carcinoma. Oncotarget 2018, 9, 26852–26867. [Google Scholar] [CrossRef]
- Priyadarshini, K.; Ali, S.A.; Sivanandam, K.; Alagarsamy, M. Human lung cancer classification and comprehensive analysis using different machine learning techniques. Microsc. Res. Tech. 2024, 88, 234–250. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Hameed, O.; Findeis-Hosey, J.J.; Fan, L.; Li, F.; McMahon, L.A.; Yang, Q.; Wang, H.L.; Xu, H. Diagnostic utility of PAX8, TTF-1 and napsin A for discriminating metastatic carcinoma from primary adenocarcinoma of the lung. Biotech. Histochem. 2012, 87, 30–34. [Google Scholar] [CrossRef]
- Baston, C.; Parosanu, A.I.; Mihai, M.; Moldoveanu, O.; Stanciu, I.M.; Nitipir, C. Tumor-to-Tumor Metastasis of Lung Cancer to Kidney Cancer: A Review of the Literature and Our Experience. Diagnostics 2024, 14, 553. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Oba, T.; Chino, T.; Soma, A.; Ono, M.; Ito, T.; Kanai, T.; Maeno, K.; Sato, Y.; Uehara, T.; et al. Papillary thyroid microcarcinoma with lung metastases: A case report and review of the literature. Thyroid Res. 2021, 14, 15. [Google Scholar] [CrossRef]
- Asirvatham, J.R.; Esposito, M.J.; Bhuiya, T.A. Role of PAX-8, CD5, and CD117 in distinguishing thymic carcinoma from poorly differentiated lung carcinoma. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 372–376. [Google Scholar] [CrossRef]
- Weissferdt, A.; Tang, X.; Wistuba, I.I.; Moran, C.A. Comparative immunohistochemical analysis of pulmonary and thymic neuroendocrine carcinomas using PAX8 and TTF-1. Mod. Pathol. 2013, 26, 1554–1560. [Google Scholar] [CrossRef]
- Sharifai, N.; Abro, B.; Chen, J.F.; Zhao, M.; He, H.; Cao, D. Napsin A is a highly sensitive marker for nephrogenic adenoma: An immunohistochemical study with a specificity test in genitourinary tumors. Hum. Pathol. 2020, 102, 23–32. [Google Scholar] [CrossRef]
- Peters, H.; Neubüser, A.; Balling, R. Pax genes and organogenesis: Pax9 meets tooth development. Eur. J. Oral Sci. 1998, 106 (Suppl. S1), 38–43. [Google Scholar] [CrossRef]
- Sweat, Y.Y.; Sweat, M.; Mansaray, M.; Cao, H.; Eliason, S.; Adeyemo, W.L.; Gowans, L.J.; Eshete, M.A.; Anand, D.; Chalkley, C.; et al. Six2 regulates Pax9 expression, palatogenesis and craniofacial bone formation. Dev. Biol. 2020, 458, 246–256. [Google Scholar] [CrossRef]
- Chen, J.; Lan, Y.; Baek, J.A.; Gao, Y.; Jiang, R. Wnt/beta-catenin signaling plays an essential role in activation of odontogenic mesenchyme during early tooth development. Dev. Biol. 2009, 334, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Peters, H.; Neubüser, A.; Kratochwil, K.; Balling, R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998, 12, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Hetzer-Egger, C.; Schorpp, M.; Haas-Assenbaum, A.; Balling, R.; Peters, H.; Boehm, T. Thymopoiesis requires Pax9 function in thymic epithelial cells. Eur. J. Immunol. 2002, 32, 1175–1181. [Google Scholar] [CrossRef]
- Zhang, D.; Du, Y.; Zhang, W.; Ren, X.; Huang, C.; Qiu, F.; Zhou, J.; Zhang, X. PAX9 Overexpression in Lung Adenocarcinoma and Promotes Cancer Progression. In Proceedings of the 2024 16th International Conference on Bioinformatics and Biomedical Technology (ICBBT ′24). Association for Computing Machinery, New York, NY, USA, 24–26 May 2024; pp. 53–57. [Google Scholar]
- Hayashi, T.; Kishi, M.; Takamochi, K.; Hosoya, M.; Kohsaka, S.; Kishikawa, S.; Ura, A.; Sano, K.; Sasahara, N.; Suehara, Y.; et al. Expression of paired box 9 defines an aggressive subset of lung adenocarcinoma preferentially occurring in smokers. Histopathology 2023, 82, 672–683. [Google Scholar] [CrossRef]
- Zhao, Z.; Szczepanski, A.P.; Tsuboyama, N.; Abdala-Valencia, H.; Goo, Y.A.; Singer, B.D.; Bartom, E.T.; Yue, F.; Wang, L. PAX9 Determines Epigenetic State Transition and Cell Fate in Cancer. Cancer Res. 2021, 81, 4696–4708. [Google Scholar] [CrossRef]
- Kendall, J.; Liu, Q.; Bakleh, A.; Krasnitz, A.; Nguyen, K.C.; Lakshmi, B.; Gerald, W.L.; Powers, S.; Mu, D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 16663–16668. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.S.; Acharya, C.R.; Balakumaran, B.S.; Riedel, R.F.; Kim, M.K.; Stevenson, M.; Tuchman, S.; Mukherjee, S.; Barry, W.; Dressman, H.K.; et al. Characterizing the developmental pathways TTF-1, NKX2-8, and PAX9 in lung cancer. Proc. Natl. Acad. Sci. USA 2009, 106, 5312–5317. [Google Scholar] [CrossRef]
- Vainio, S.; Heikkilä, M.; Kispert, A.; Chin, N.; McMahon, A.P. Female development in mammals is regulated by Wnt-4 signalling. Nature 1999, 397, 405–409. [Google Scholar] [CrossRef]
- Zheng, L.; Cui, C.; Shi, O.; Lu, X.; Li, Y.K.; Wang, W.; Li, Y.; Wang, Q. Incidence and mortality of ovarian cancer at the global, regional, and national levels, 1990–2017. Gynecol. Oncol. 2020, 159, 239–247. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Laury, A.R.; Perets, R.; Piao, H.; Krane, J.F.; Barletta, J.A.; French, C.; Chirieac, L.R.; Lis, R.; Loda, M.; Hornick, J.L.; et al. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am. J. Surg. Pathol. 2011, 35, 816–826. [Google Scholar] [CrossRef]
- Chaves-Moreira, D.; Mitchell, M.A.; Arruza, C.; Rawat, P.; Sidoli, S.; Nameki, R.; Reddy, J.; Corona, R.I.; Afeyan, L.K.; Klein, I.A.; et al. The transcription factor PAX8 promotes angiogenesis in ovarian cancer through interaction with SOX17. Sci. Signal 2022, 15, eabm2496. [Google Scholar] [CrossRef] [PubMed]
- Ghannam-Shahbari, D.; Jacob, E.; Kakun, R.R.; Wasserman, T.; Korsensky, L.; Sternfeld, O.; Kagan, J.; Bublik, D.R.; Aviel-Ronen, S.; Levanon, K.; et al. PAX8 activates a p53-p21-dependent pro-proliferative effect in high grade serous ovarian carcinoma. Oncogene 2018, 37, 2213–2224. [Google Scholar] [CrossRef]
- Kakun, R.R.; Melamed, Z.; Perets, R. PAX8 in the Junction between Development and Tumorigenesis. Int. J. Mol. Sci. 2022, 23, 7410. [Google Scholar] [CrossRef] [PubMed]
- Li, C.G.; Nyman, J.E.; Braithwaite, A.W.; Eccles, M.R. PAX8 promotes tumor cell growth by transcriptionally regulating E2F1 and stabilizing RB protein. Oncogene 2011, 30, 4824–4834. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Tang, Y.; Mao, Y.; Liu, Y.; Yao, D.; Yang, L.; Garson, K.; Vanderhyden, B.C.; Wang, Q. PAX2 promotes epithelial ovarian cancer progression involving fatty acid metabolic reprogramming. Int. J. Oncol. 2020, 56, 697–708. [Google Scholar] [CrossRef]
- Tung, C.S.; Mok, S.C.; Tsang, Y.; Zu, Z.; Song, H.; Liu, J.; Deavers, M.T.; Malpica, A.; Wolf, J.K.; Lu, K.H.; et al. PAX2 expression in low malignant potential ovarian tumors and low-grade ovarian serous carcinomas. Mod. Pathol. 2009, 22, 1243–1250. [Google Scholar] [CrossRef]
- Manderfield, L.J.; Engleka, K.A.; Aghajanian, H.; Gupta, M.; Yang, S.; Li, L.; Baggs, J.E.; Hogenesch, J.B.; Olson, E.N.; Epstein, J.A. Pax3 and Hippo Signaling Coordinate Melanocyte Gene Expression in Neural Crest. Cell Rep. 2015, 10, 841. [Google Scholar] [CrossRef]
- Panatta, E.; Zampieri, C.; Melino, G.; Amelio, I. Understanding p53 tumour suppressor network. Biol. Direct. 2021, 16, 14. [Google Scholar] [CrossRef]
- Liang, X.; Dong, Z.; Bin, W.; Dekang, N.; Xuhang, Z.; Shuyuan, Z.; Liwen, L.; Kai, J.; Caixing, S. PAX3 Promotes Proliferation of Human Glioma Cells by WNT/β-Catenin Signaling Pathways. J. Mol. Neurosci. 2019, 68, 66–77. [Google Scholar] [CrossRef]
- Shaw, T.; Barr, F.G.; Üren, A. The PAX Genes: Roles in Development, Cancer, and Other Diseases. Cancers 2024, 16, 1022. [Google Scholar] [CrossRef] [PubMed]
- Stuart, E.T.; Haffner, R.; Oren, M.; Gruss, P. Loss of p53 function through PAX-mediated transcriptional repression. EMBO J. 1995, 14, 5638–5645. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, M.; Remppis, A.; Fredericks, W.J.; Rauscher, F.J., III; Schäfer, B.W. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 13164–13169. [Google Scholar] [CrossRef]
- Zhang, S.L.; Guo, J.; Moini, B.; Ingelfinger, J.R. Angiotensin II stimulates Pax-2 in rat kidney proximal tubular cells: Impact on proliferation and apoptosis. Kidney Int. 2004, 66, 2181–2192. [Google Scholar] [CrossRef]
- Marquardt, T.; Ashery-Padan, R.; Andrejewski, N.; Scardigli, R.; Guillemot, F.; Gruss, P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell 2001, 105, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Du, Y.; Zhao, C.; Wu, Y. PAX2 may induce ADAM10 expression in renal tubular epithelial cells and contribute to epithelial-to-mesenchymal transition. Int. Urol. Nephrol. 2018, 50, 1729–1741. [Google Scholar] [CrossRef]
- Tong, H.; Liu, X.; Li, T.; Qiu, W.; Peng, C.; Shen, B.; Zhu, Z. MACC1-AS1 promotes hepatocellular carcinoma cell invasion and proliferation by regulating PAX8. Aging 2020, 12, 70–79. [Google Scholar] [CrossRef]
- Searcy, M.B.; Larsen, R.K., IV; Stevens, B.T.; Zhang, Y.; Jin, H.; Drummond, C.J.; Langdon, C.G.; Gadek, K.E.; Vuong, K.; Reed, K.B.; et al. PAX3-FOXO1 dictates myogenic reprogramming and rhabdomyosarcoma identity in endothelial progenitors. Nat. Commun. 2023, 14, 7291. [Google Scholar] [CrossRef]
- Charytonowicz, E.; Matushansky, I.; Domingo-Doménech, J.; Castillo-Martín, M.; Ladanyi, M.; Cordon-Cardo, C.; Ziman, M. PAX7-FKHR fusion gene inhibits myogenic differentiation via NF-kappaB upregulation. Clin. Transl. Oncol. 2012, 14, 197–206. [Google Scholar] [CrossRef]
- Vu-Phan, D.; Grachtchouk, V.; Yu, J.; Colby, L.A.; Wicha, M.S.; Koenig, R.J. The thyroid cancer PAX8-PPARG fusion protein activates Wnt/TCF-responsive cells that have a transformed phenotype. Endocr. Relat. Cancer 2013, 20, 725–739. [Google Scholar] [CrossRef]
- Mentrikoski, M.J.; Wendroth, S.M.; Wick, M.R. Immunohistochemical distinction of renal cell carcinoma from other carcinomas with clear-cell histomorphology: Utility of CD10 and CA-125 in addition to PAX-2, PAX-8, RCCma, and adipophilin. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Sangoi, A.R.; Karamchandani, J.; Kim, J.; Pai, R.K.; McKenney, J.K. The use of immunohistochemistry in the diagnosis of metastatic clear cell renal cell carcinoma: A review of PAX-8, PAX-2, hKIM-1, RCCma, and CD10. Adv. Anat. Pathol. 2010, 17, 377–393. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, A.; De La Roza, G.; Ro, J.Y.; Shen, S.S.; Truong, L.D. PAX2 and PAX8 expression in primary and metastatic renal tumors: A comprehensive comparison. Arch. Pathol. Lab. Med. 2012, 136, 1541–1551. [Google Scholar] [CrossRef] [PubMed]
- Gorbokon, N.; Baltruschat, S.; Lennartz, M.; Luebke, A.M.; Höflmayer, D.; Kluth, M.; Hube-Magg, C.; Hinsch, A.; Fraune, C.; Lebok, P.; et al. PAX8 expression in cancerous and non-neoplastic tissue: A tissue microarray study on more than 17,000 tumors from 149 different tumor entities. Virchows Arch. 2024, 485, 491–507. [Google Scholar] [CrossRef]
- Scimeca, M.; Urbano, N.; Bonfiglio, R.; Schillaci, O.; Bonanno, E. Management of oncological patients in the digital era: Anatomic pathology and nuclear medicine teamwork. Future Oncol. 2018, 14, 1013–1015. [Google Scholar] [CrossRef]
- Melino, G.; Bischof, J.; Chen, W.L.; Jia, W.; Juhl, H.; Kopeina, G.S.; Mauriello, A.; Novelli, F.; Scimeca, M.; Shi, Y.; et al. New hope for the world cancer day. Biol. Direct. 2025, 20, 14. [Google Scholar] [CrossRef]
- Agostini, M.; Giacobbi, E.; Servadei, F.; Bishof, J.; Funke, L.; Sica, G.; Rovella, V.; Carilli, M.; Iacovelli, V.; Shi, Y.; et al. Unveiling the molecular profile of a prostate carcinoma: Implications for personalized medicine. Biol. Direct. 2024, 19, 146. [Google Scholar] [CrossRef]
- Amelio, I.; Bertolo, R.; Bove, P.; Candi, E.; Chiocchi, M.; Cipriani, C.; Di Daniele, N.; Ganini, C.; Juhl, H.; Mauriello, A.; et al. Cancer predictive studies. Biol. Direct. 2020, 15, 18. [Google Scholar] [CrossRef]
- Amelio, I.; Bertolo, R.; Bove, P.; Buonomo, O.C.; Candi, E.; Chiocchi, M.; Cipriani, C.; Di Daniele, N.; Ganini, C.; Juhl, H.; et al. Liquid biopsies and cancer omics. Cell Death Discov. 2020, 6, 131. [Google Scholar] [CrossRef]
| Group I | Expression During Development | Expression in Adults | Role in Tumorigenesis |
|---|---|---|---|
| PAX1 | Human skeleton, parathyroid glands and thymus [6,7] | Low levels in esophagus, skeletal muscle, kidneys, pituitary gland, skin and thyroid [8,9,10] | Tumor suppressor role in various cancers (e.g., cervical cancer, ovarian cancer, colorectal cancer). Highly methylated in these tumors [8,9,10] |
| PAX9 | Human skeleton, parathyroid glands and thymus [7] | PAX9 expression is restricted to the endocrine tissues, lymphatic system, cervix, bronchus, tongue, esophagus and salivary gland [11] | Genetic alterations contribute to carcinogenesis, common in lung adenocarcinoma and squamous cell carcinoma [12] |
| Group II | Expression during development | Expression in adults | Role in tumorigenesis |
| PAX2 | Central nervous system, optic vesicle, optic disk, optic nerve, ears, kidneys, pancreas, female reproductive tract (cervix, fallopian tubes, uterus) and adult testis [13,14] | Low expression in the brain, pancreas, pituitary gland, testis, uterus; moderate levels in cervix, fallopian tubes and kidneys [15] | Highly expressed in primary renal carcinomas. Inhibition induces rapid apoptosis in bladder carcinoma cell lines. Important for growth and survival of several urogenital cancers [16] |
| PAX5 | Involved in the differentiation of hematopoietic stem cells into mature B cells [17] | Expressed in adult human brain, spleen, colon and testis [18,19,20] | A potent oncogene in hematological cancers, particularly lymphoma and lymphocytic leukemia. Down-regulation blocks B-cell differentiation, giving cells the ability to proliferate, evade, and resist apoptosis [21,22] |
| PAX8 | Necessary for proper development of thyroid, testis, kidneys and fallopian tubes [23,24] | Some isoforms expressed in adult tissues [23,24] | Expressed in most thyroid cancers, correlating with higher risk of tumor recurrence and plays a role in progression of Follicular Thyroid Carcinoma [25,26] |
| Group III | Expression during development | Expression in adults | Role in tumorigenesis |
| PAX3 | Important regulator of neural tube development, neural crest formation, skeletal muscle development [27] | Expressed in adult human adipose tissue, arterial tissue, brain, breast, cervix, minor salivary gland, skeletal muscle, prostate, skin, and testis [28] | Mutations associated with Waardenburg syndrome (types I and III), embryonal and alveolar rhabdomyosarcoma [29,30] |
| PAX7 | Important regulator of neural tube development, neural crest formation, skeletal muscle development [27] | Satellite cells in skeletal muscle [31,32] | Mutations associated with Waardenburg syndrome (types I and III), embryonal and alveolar rhabdomyosarcoma [29,30] |
| Group IV | Expression during development | Expression in adults | Role in tumorigenesis |
| PAX4 | Plays a fundamental role in the development of pancreas and gastrointestinal tract cells [33] | Colon and small intestine [34] | Dysregulation linked to developmental disorders, diabetes, and tumors of the exocrine pancreas and intestine [35,36] |
| PAX6 | Important for the development of eyes, brain, and pituitary gland | Brain, kidneys, pituitary gland, pancreas, testis [37] | Dysregulation linked to developmental disorders, diabetes, and tumors of the exocrine pancreas and intestine [35,36] |
| Molecular Mechanism | Description | Examples |
|---|---|---|
| Regulation of Cell Survival and Proliferation | PAX proteins can inhibit p53 expression, influencing cell survival and proliferation |
|
| Regulation of Cell Fate and Differentiation | Many PAX proteins influence specific cell lineage differentiation and maturation. | |
| Regulation of Invasion and Metastasis | PAX proteins can promote invasion and metastasis by regulating cell adhesion and migration processes. |
|
| Oncogenic Fusion Proteins | PAX proteins can form fusion proteins due to chromosomal translocations, leading to aberrant activation of oncogenic pathways. |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giacobbi, E.; Scioli, M.P.; Servadei, F.; Palumbo, V.; Bonfiglio, R.; Bove, P.; Mauriello, A.; Scimeca, M. PAX Family, Master Regulator in Cancer. Diagnostics 2025, 15, 1420. https://doi.org/10.3390/diagnostics15111420
Giacobbi E, Scioli MP, Servadei F, Palumbo V, Bonfiglio R, Bove P, Mauriello A, Scimeca M. PAX Family, Master Regulator in Cancer. Diagnostics. 2025; 15(11):1420. https://doi.org/10.3390/diagnostics15111420
Chicago/Turabian StyleGiacobbi, Erica, Maria Paola Scioli, Francesca Servadei, Valeria Palumbo, Rita Bonfiglio, Pierluigi Bove, Alessandro Mauriello, and Manuel Scimeca. 2025. "PAX Family, Master Regulator in Cancer" Diagnostics 15, no. 11: 1420. https://doi.org/10.3390/diagnostics15111420
APA StyleGiacobbi, E., Scioli, M. P., Servadei, F., Palumbo, V., Bonfiglio, R., Bove, P., Mauriello, A., & Scimeca, M. (2025). PAX Family, Master Regulator in Cancer. Diagnostics, 15(11), 1420. https://doi.org/10.3390/diagnostics15111420








