A Convolutional Mixer-Based Deep Learning Network for Alzheimer’s Disease Classification from Structural Magnetic Resonance Imaging
Abstract
1. Introduction
- Optimizing neural network parameters: This includes selecting appropriate weight initialization methods. It reduces convergence and establishes stable learning biases within the network.
- Reducing trainable parameters: This approach aims to minimize the number of trainable parameters and computational complexity (reduces the number of floating-point operations).
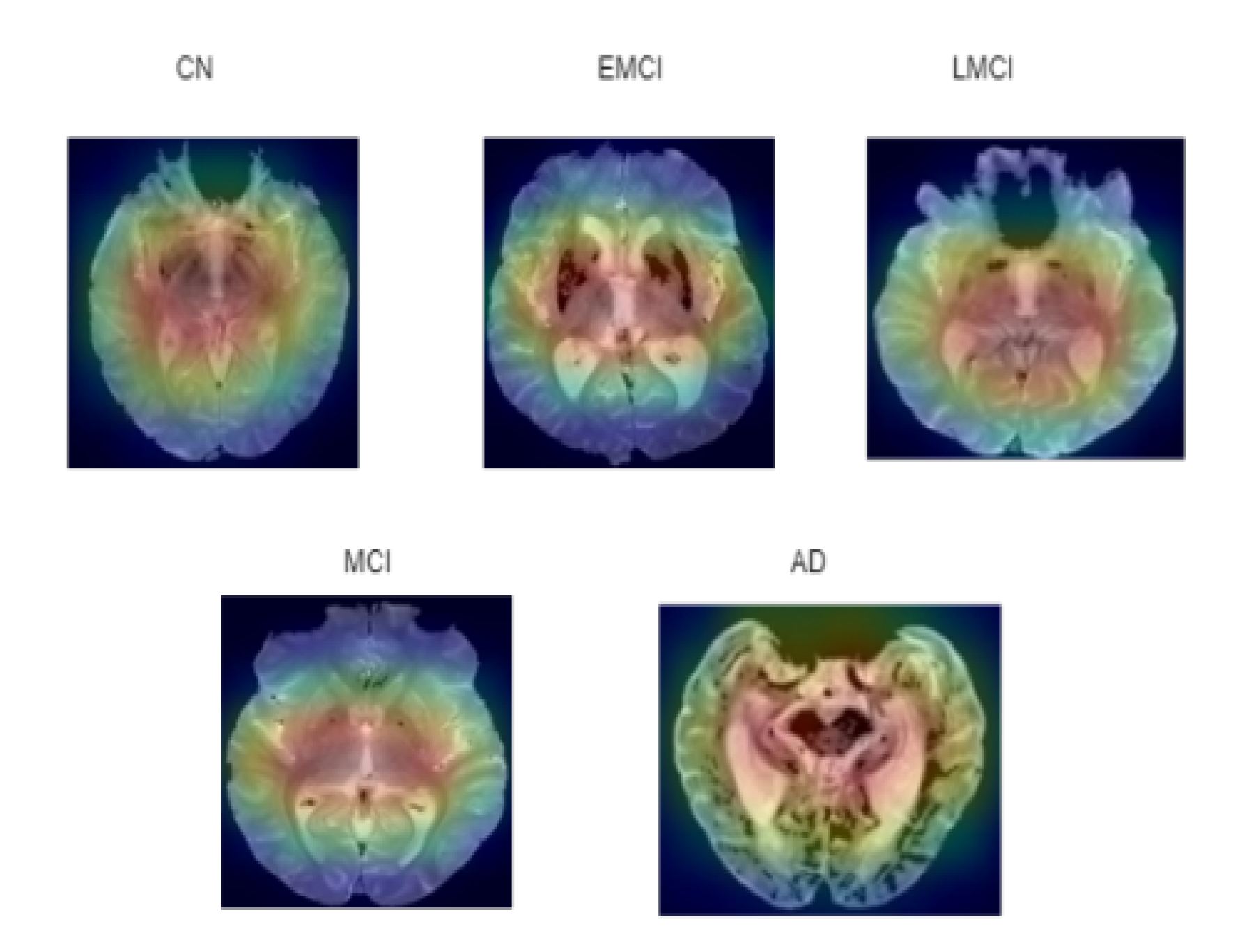
- Anatomical feature detection: Potential changes in anatomical features across different classes are detected using ASOP pixel attribution method This method enhances understanding of how neural networks interpret and distinguish features in an input image.
- Handling class imbalance: The class imbalance within the dataset can lead to biased learning outcomes if not balanced through a balancing technique. The synthetic minority oversampling technique (SMOTE) combines with edited nearest neighbors (ENN) to balance the distribution of classes in datasets, which overcomes the under-fitting predicament.
2. Related Works
- The existing CNN models are often trained with a large number of trainable parameters. In this research, the application of convolutional mixer architecture reduces the number of trainable parameters, thereby reducing computational demands without compromising performance.
- Existing convolutional mixers use multiple depthwise and pointwise convolution with residual connections. Our model simplifies this by using depthwise separable convolutions only in deeper layers, reducing trainable parameters and FLOPs.
- Many existing models are not trained from scratch and depend on pretrained resource-demanding architecture. These pretrained models are trained on datasets that differ from the AD dataset, which can introduce biases in the features extraction of grayscale medical images. In our research, deep neural networks are trained from scratch, enabling control over the training process and eliminating biases linked with pretrained weights.
- While existing AD classification models use oversampling techniques like SMOTE and SMOTE-Tomek to address class imbalance among four classes, this work employs the SMOTEENN (hybrid undersampling and oversampling) technique to handle class imbalance across five stages of AD: MCI, EMCI, LMCI, AD, and CN. SMOTEENN also addresses data overlapping issues and eliminates noise during data reconstruction.
- Most existing models do not emphasize skull stripping, a crucial step in medical image preprocessing. This works implements a skull stripping algorithm to ensure cleaner input data by removing unwanted non-brain features.
- Evaluation metrics such as accuracy, precision, F1 score, recall, and number of trainable parameters are compared with existing state-of-the-art (SOTA) deep learning models. To further assess the suggested model, the area under curve (AUC) value is calculated for each class. Existing works are summarized in Table 1.
3. Proposed Method
3.1. Dataset Collection
3.2. Preprocessing of Dataset
3.2.1. Data Cleaning
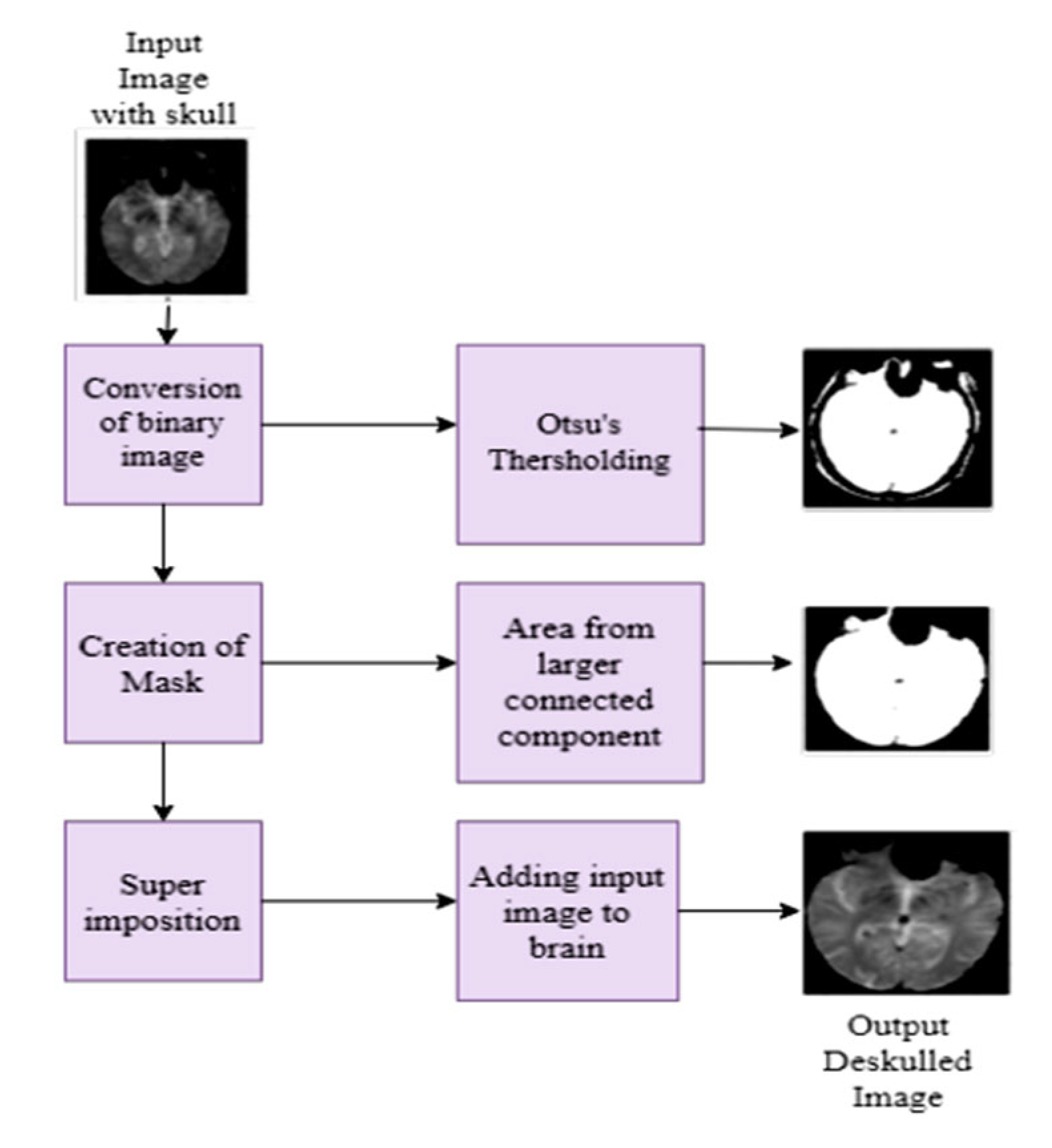
3.2.2. Skull Stripping
3.2.3. Balancing Data in All Classes
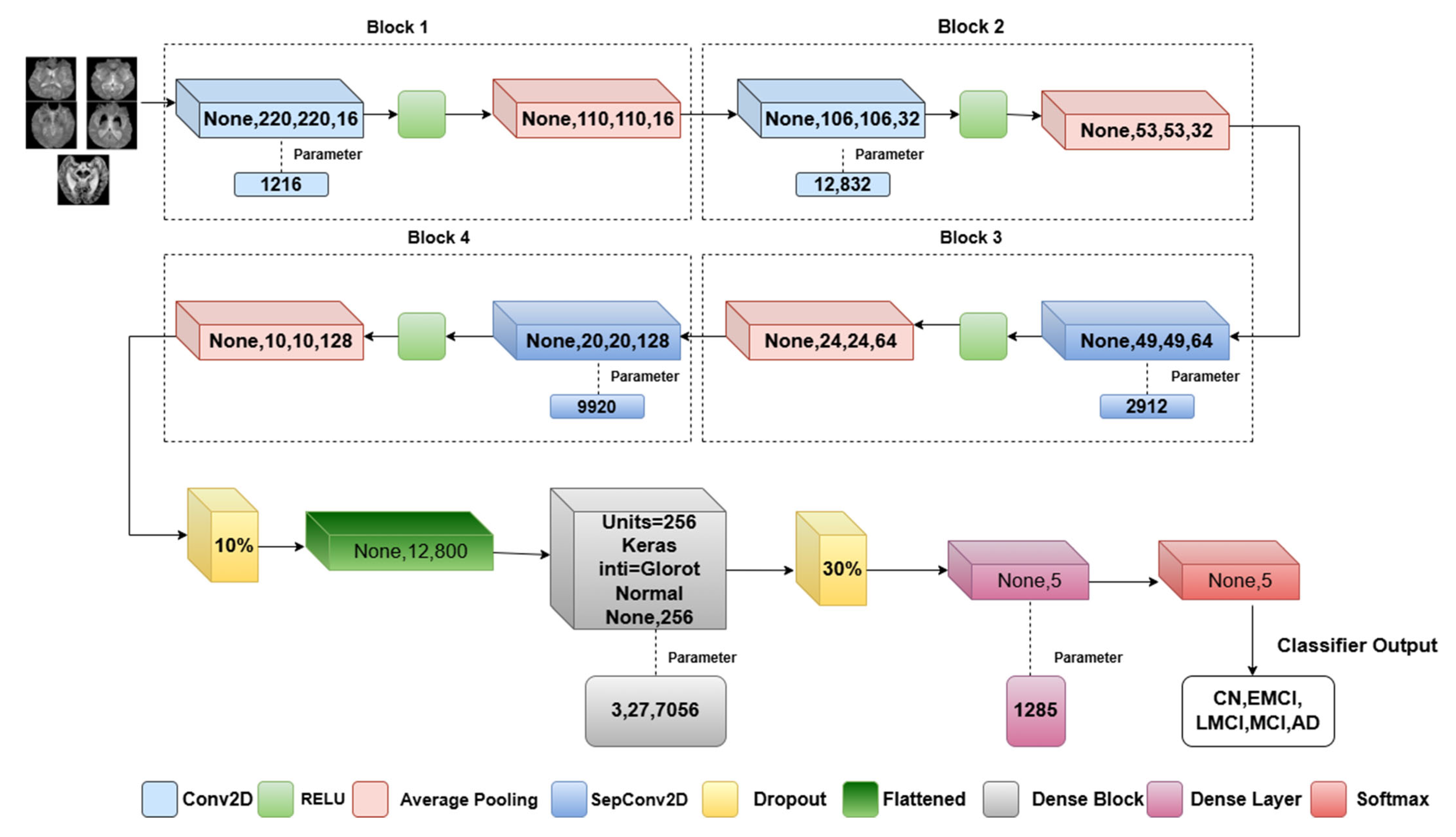
3.3. Proposed Model Architecture
| Algorithm 1. describes the proposed methodology |
| Input: X224 × 224 × 1(Grayscale brain Images) Step 1: Data preprocessing Normalize pixel value [0, 1] Data augmentation-SMOTEENN Train_Validation_Test split Step 2: Implementation of Convolutional mixer for each block in [1, 2]: Conv2D(I, filters = 16*block, kernel_size = 5× 5, activation = ReLU) AveragePooling2D(x, pool_size = 2 × 2) for each depth_block in [1, 2]: if depth_block == 1: DepthwiseSeparableConv(x, kernel = 5 × 5, filters = 64, activation = ReLU) else: DepthwiseSeparableConv(x, kernel = 5× 5, filters = 128, activation = ReLU) AveragePooling2D(x, pool_size = 2 × 2) Step3: Classification Layer # Apply Dropout and Flatten Dropout(x, rate = 0.5) Flatten(x) # Fully Connected Layers for i = 1 to 2: If i == 1: Dense(x, units = 256, activation = ReLU) Dense(x, units = 5) # Output Layer (Softmax activation) Softmax(x) Step4: Training Process #Model compile Optimizer-Adam,Epochs-50,Learning rate-0.01,Batch size-8. Step 5: Evaluation Metrics Plotting of Training,Validation-Accuracy,Recall,Loss,Precision,F1 Score,ROC plot,AUC. |
3.3.1. Convolutional Layer
3.3.2. Separable Convolutional Layer
3.3.3. Activation and Pooling Layer
3.3.4. Dropout and FCN
3.3.5. Loss Function
4. Ablation Study
4.1. Altering the Weight Initializer
4.1.1. Performance of Normal Initialization
4.1.2. Performance of Uniform Initialization
4.2. Altering Pooling Operation
4.3. Altering the Optimizers
4.4. Parameter Selection Based on Ablation Study
5. Performance Evaluation Metrics
6. Results and Discussion
6.1. Experimental Setup
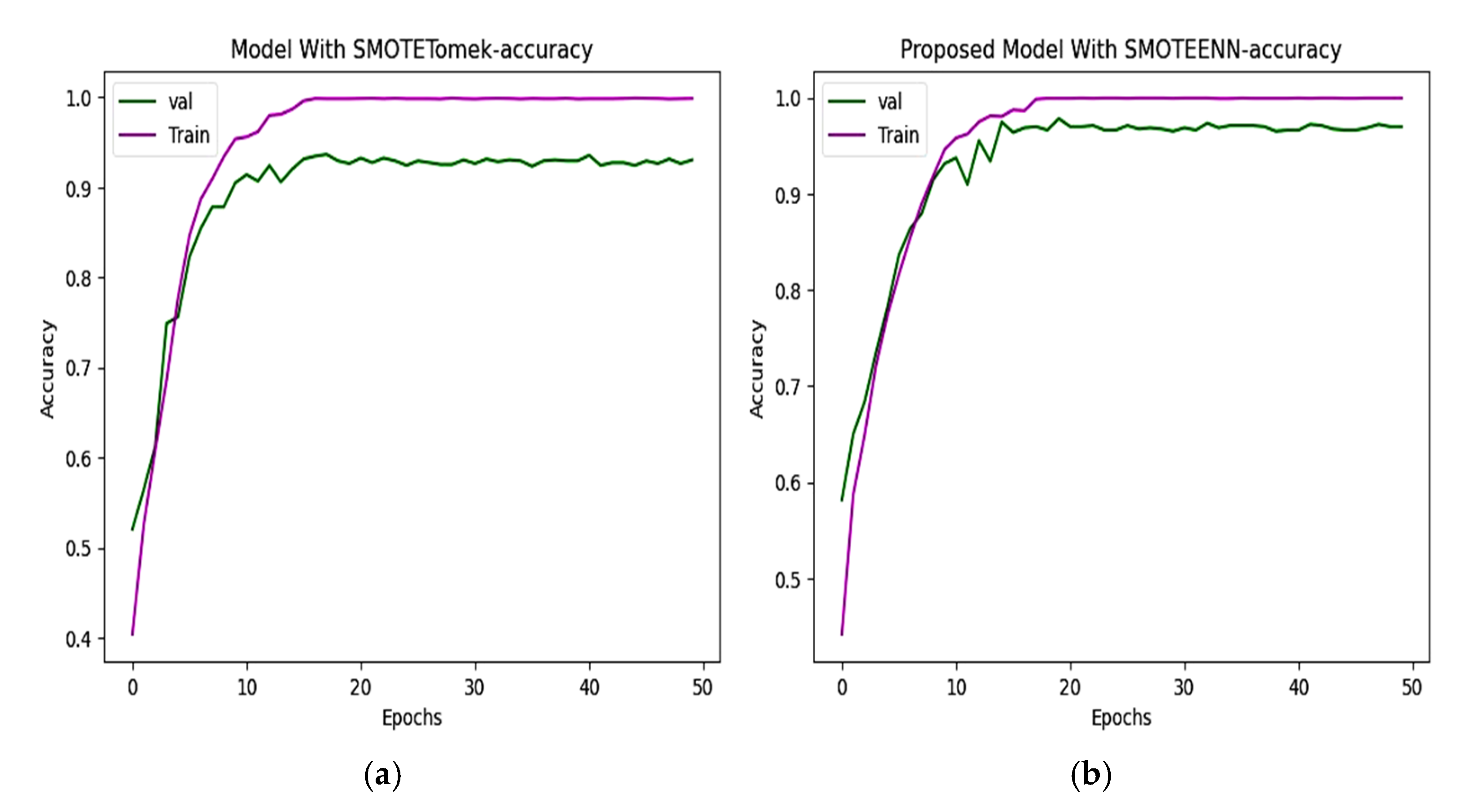
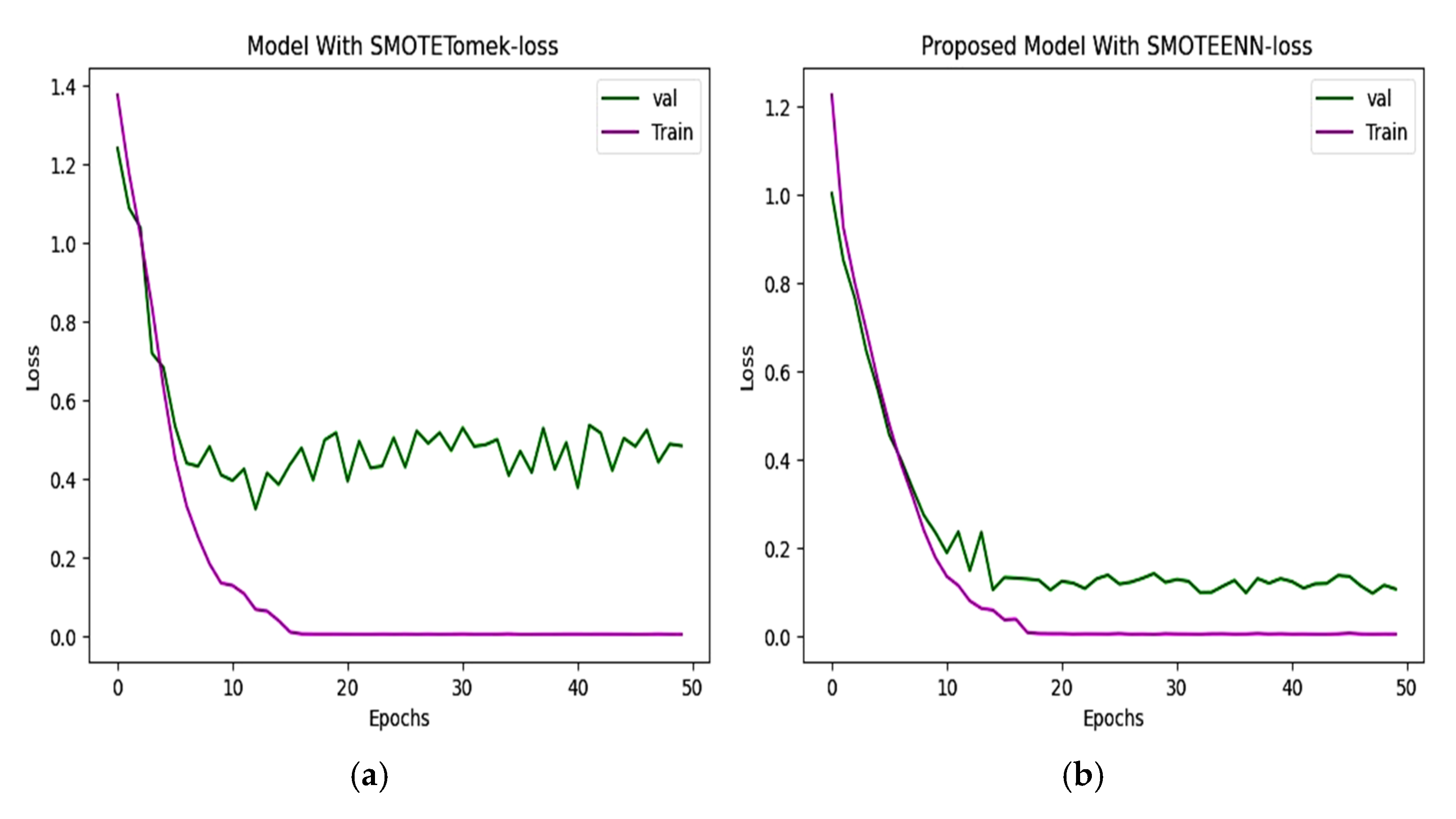
6.2. Accuracy and Loss
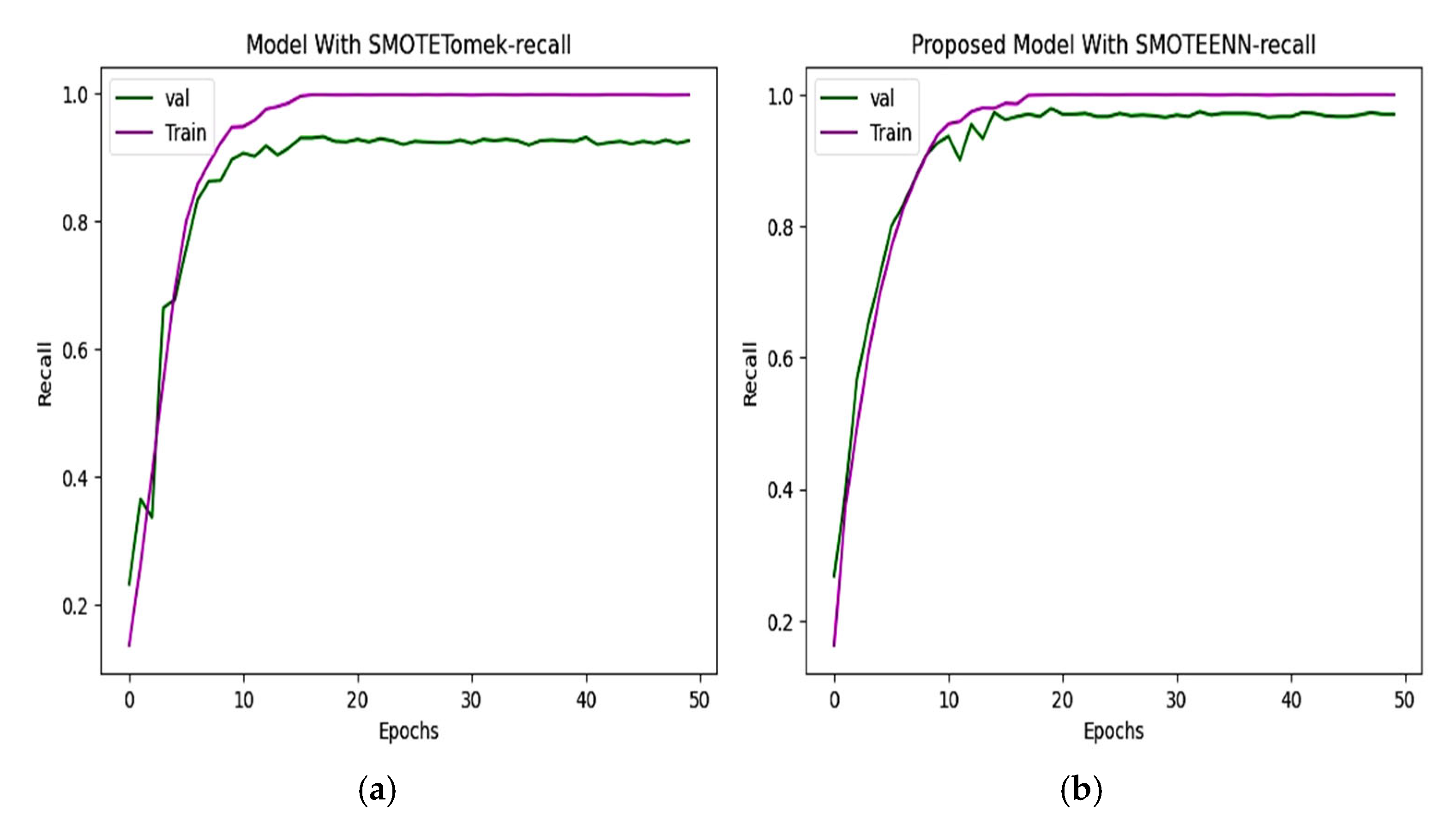
6.3. Precision and Recall
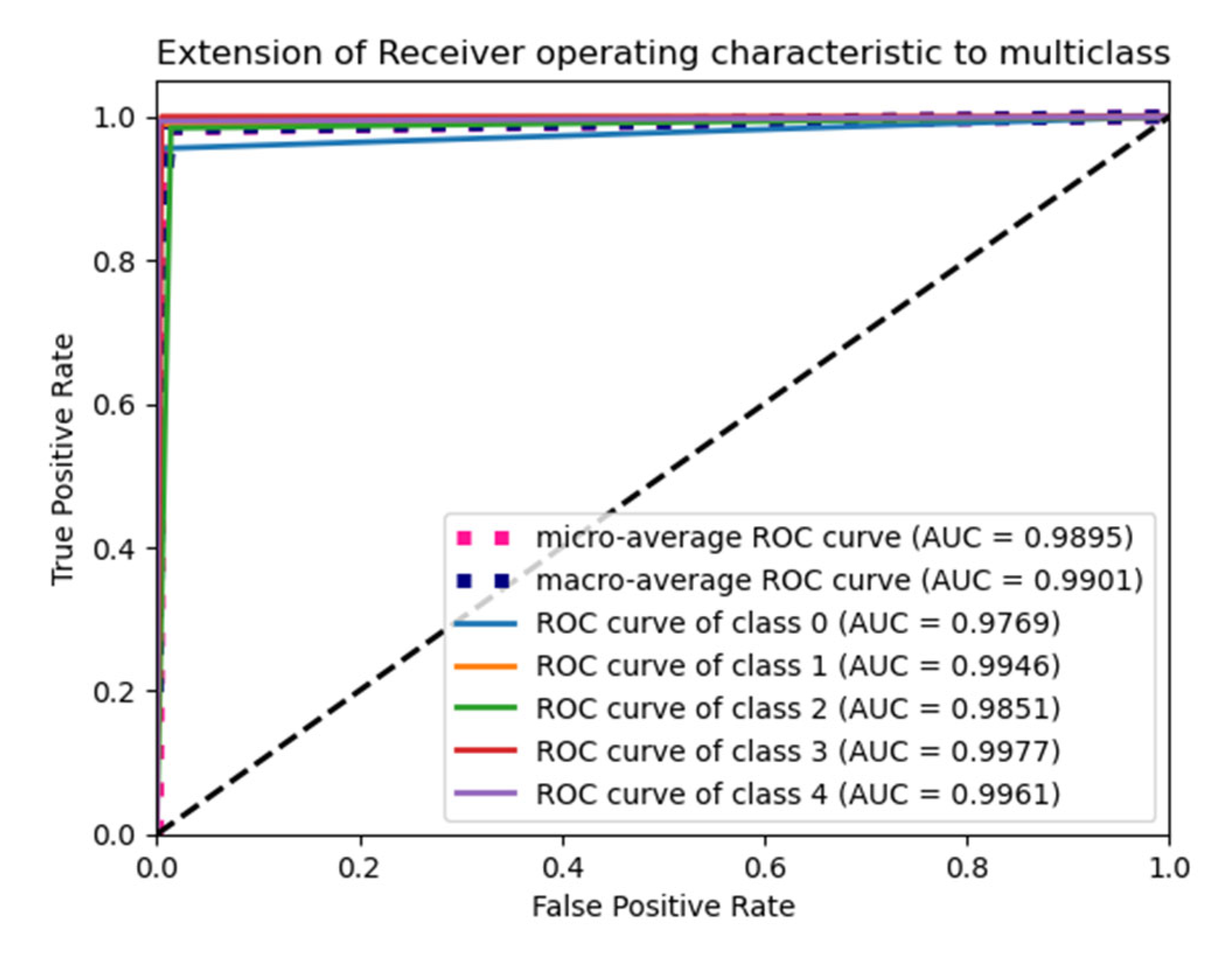
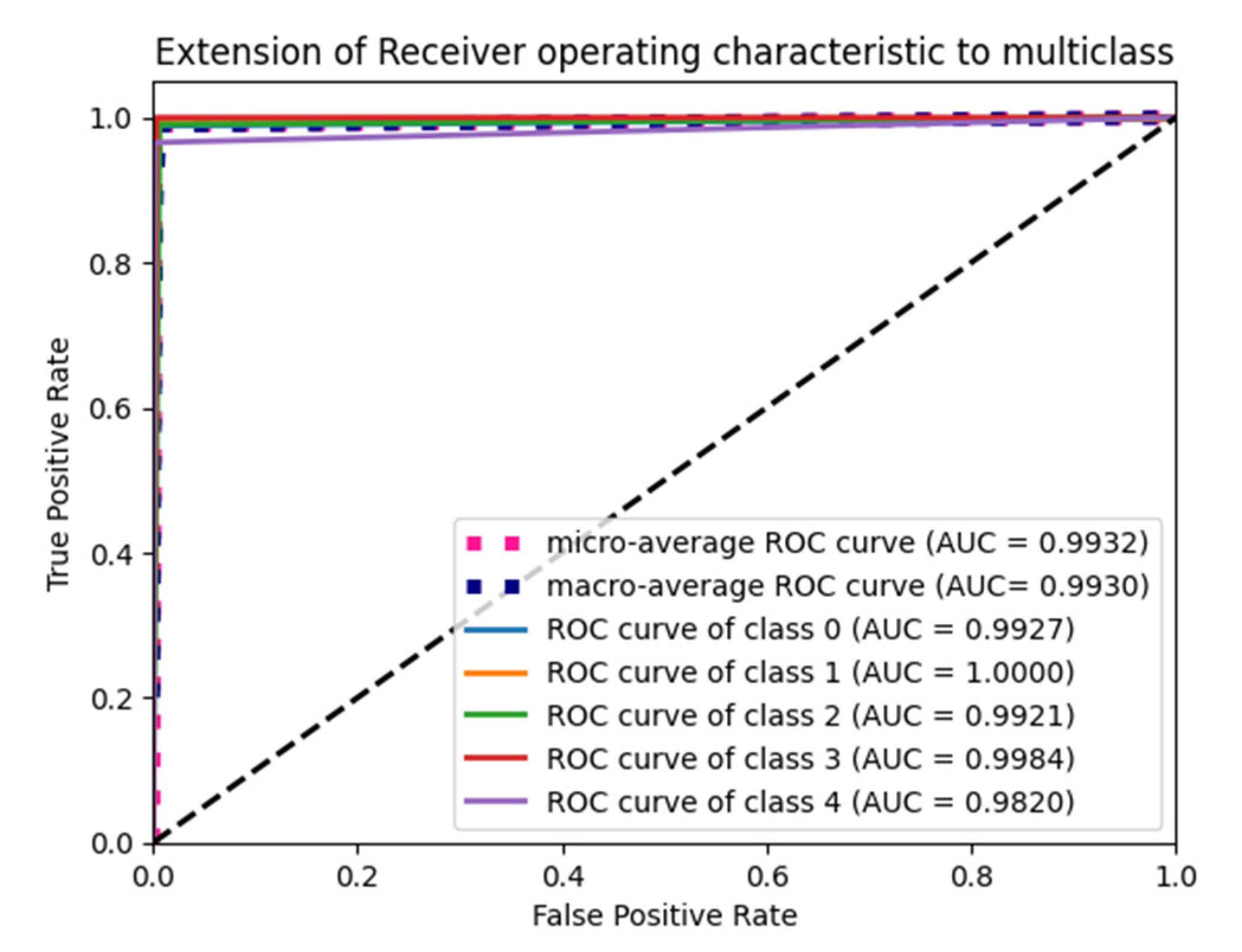
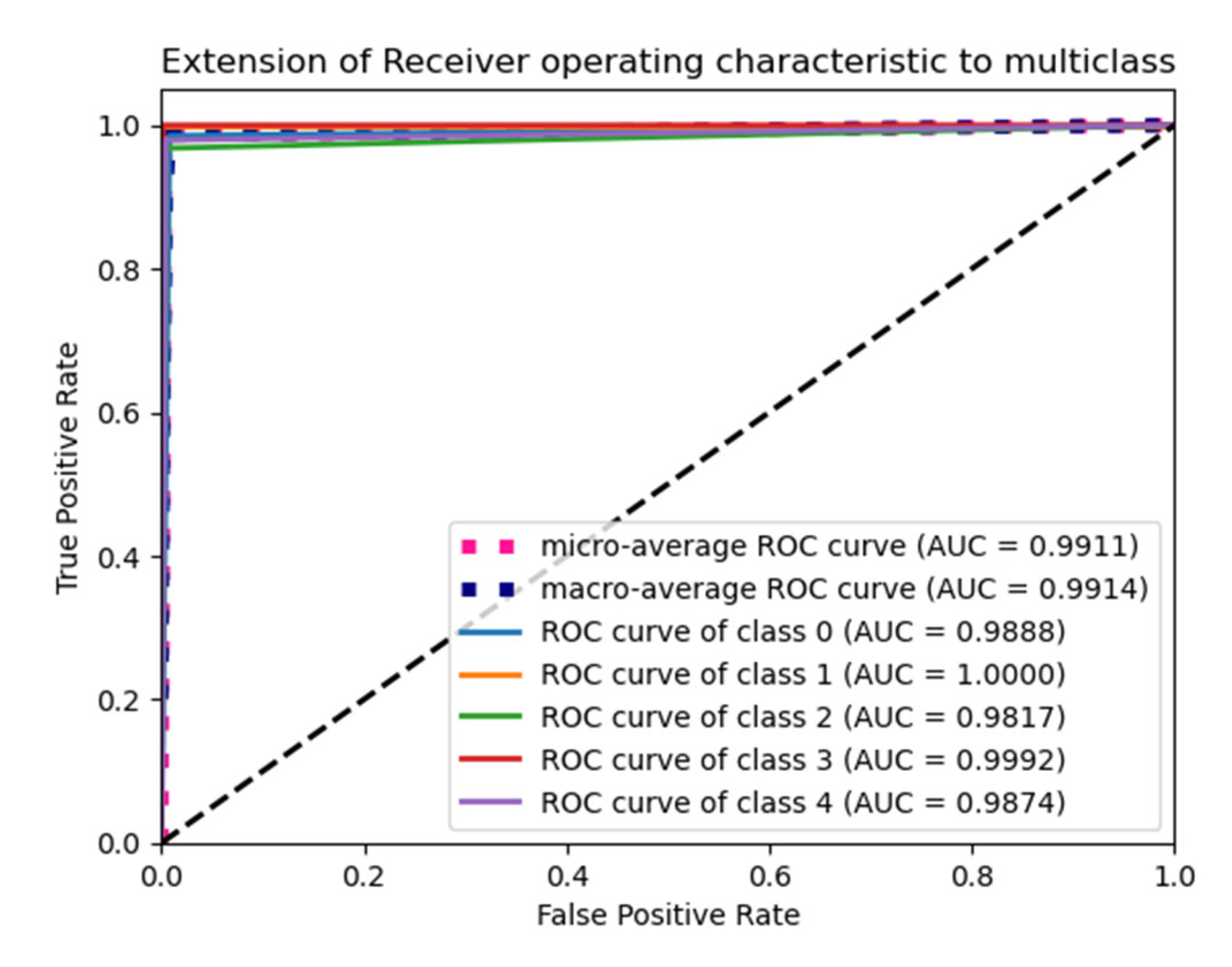
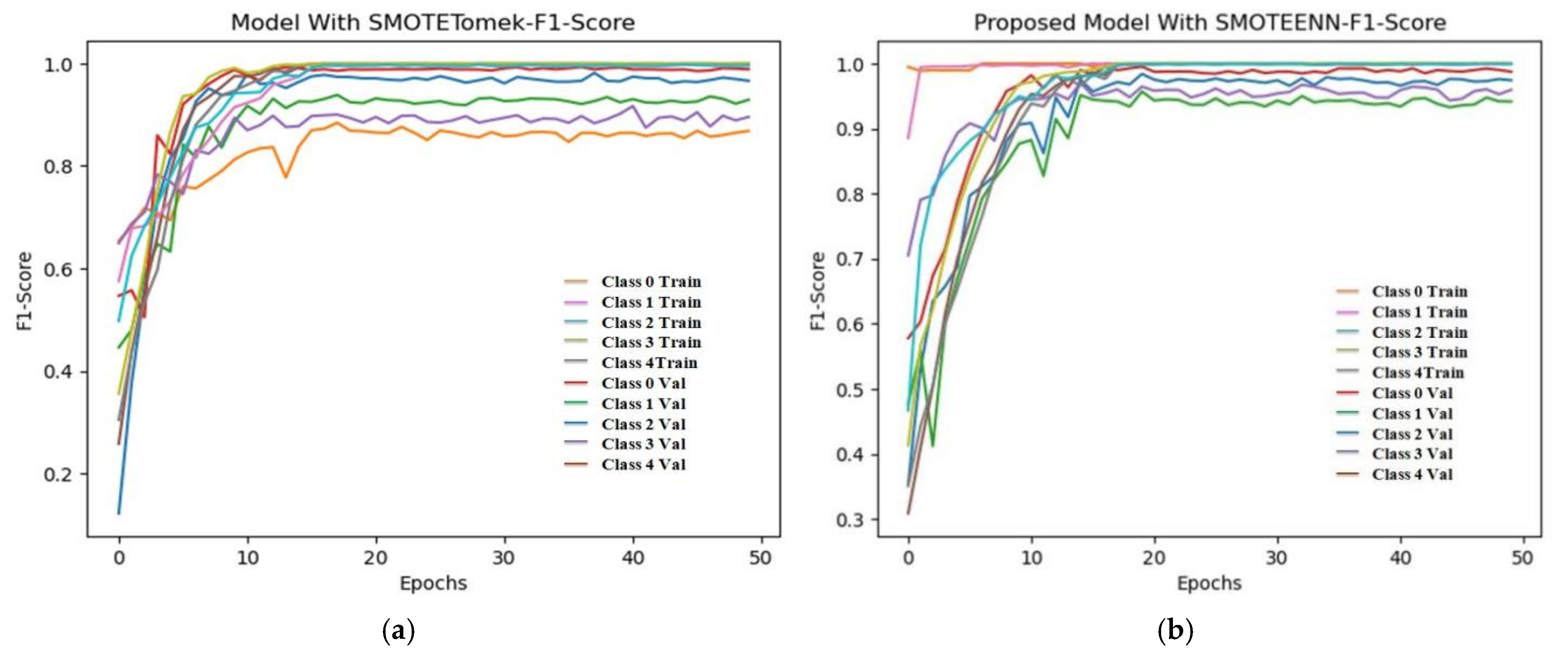
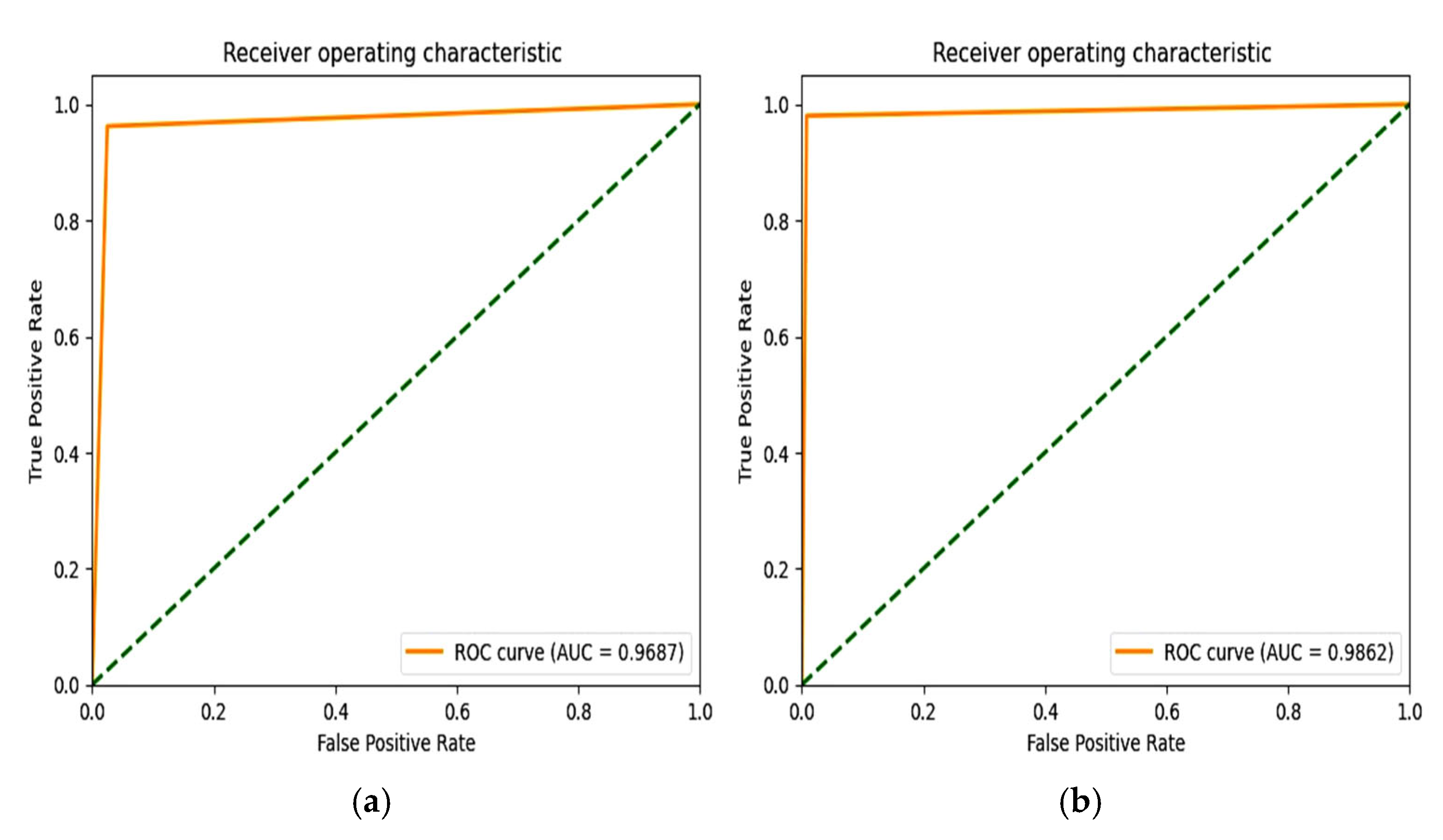
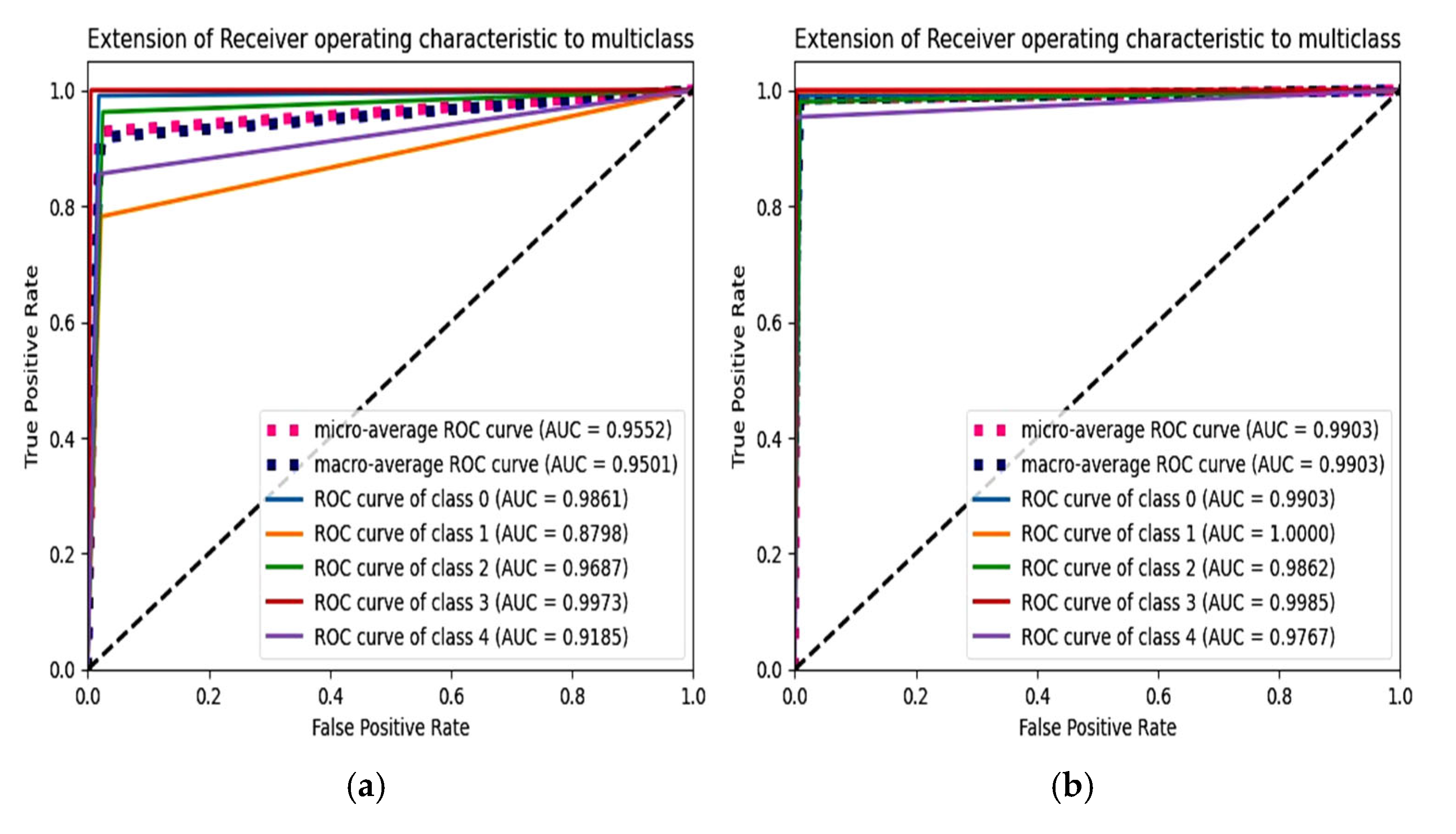
6.4. F1 Score and ROC Plot
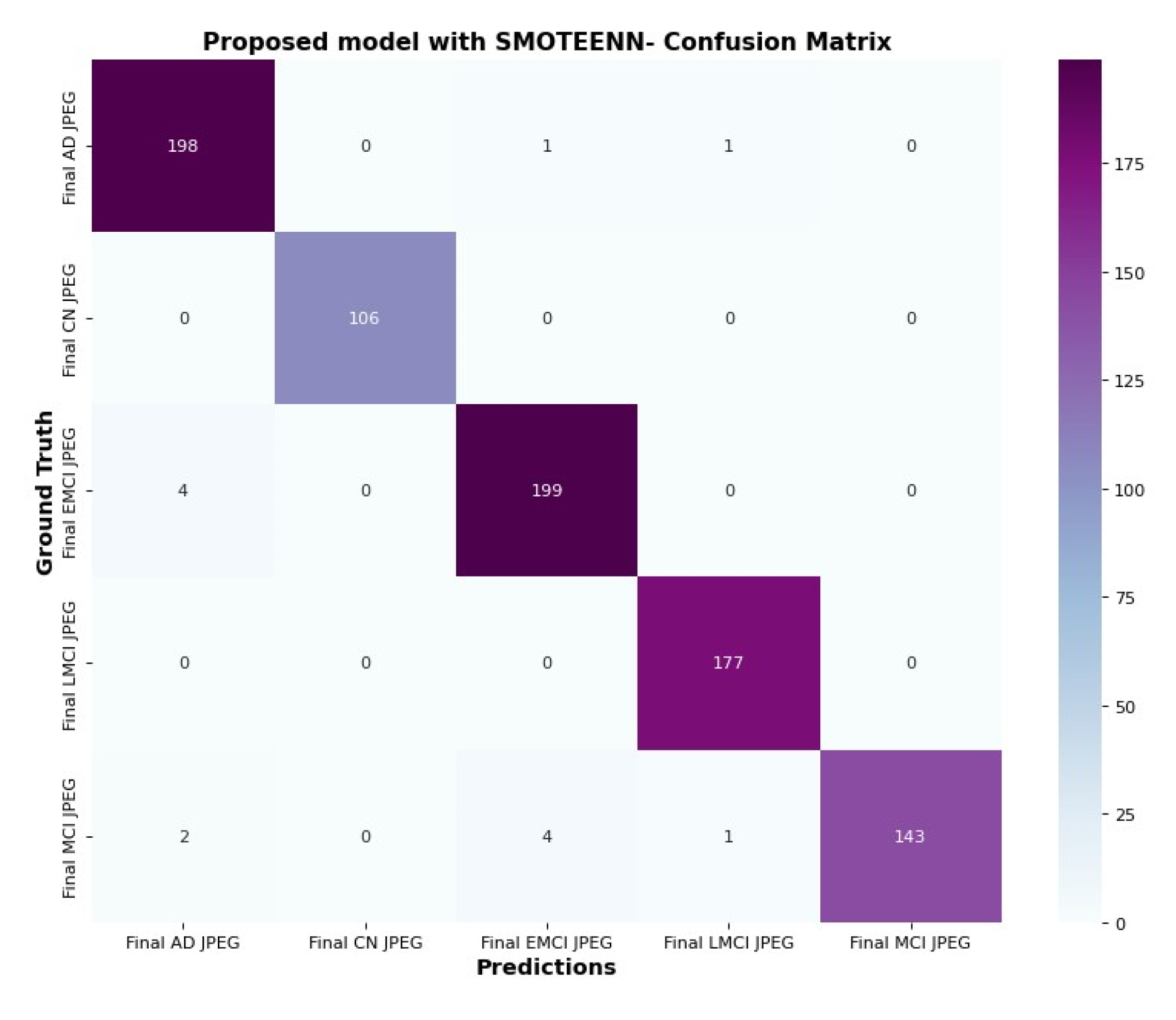
6.5. Confusion Matrix and Trainable Parameter
6.6. Visualization of ASOP
7. Discussion
7.1. Interpretation
7.2. Implication
7.3. Strength
7.4. Limitations
8. Conclusions
9. Future Work
Author Contributions
Funding
Institutional Review Board
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Fact Sheets Details on Dementia. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 15 March 2023).
- Alzheimer’s Association. Available online: https://www.alz.org/alzheimers-dementia/what-is-alzheimers/younger-early-onset (accessed on 12 August 2023).
- Jeste, D.V.; Mausbach, B.; Lee, E.E. Caring for caregivers/care partners of persons with dementia. Int. Psychogeriatr. 2021, 33, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Jitsuishi, T.; Yamaguchi, A. Searching for optimal machine learning model to classify mild cognitive impairment (mci) subtypes using multimodal mri data. Sci. Rep. 2022, 12, 4284. [Google Scholar] [CrossRef] [PubMed]
- Tufail, A.B.; Ma, Y.-K.; Zhang, Q.-N. Binary classification of alzheimer’s disease using smri imaging modality and deep learning. J. Digit. Imaging 2020, 33, 1073–1090. [Google Scholar] [CrossRef]
- Tiwari, S.; Atluri, V.; Kaushik, A.; Yndart, A.; Nair, M. Alzheimer’s disease: Pathogenesis, diagnostics, and therapeutics. Int. J. Nano Med. 2019, 14, 5541–5554. [Google Scholar] [CrossRef]
- Alsubaie, M.G.; Luo, S.; Shaukat, K. Alzheimer’s disease detection using deep learning on neuroimaging: A systematic review. Mach. Learn. Knowl. Extr. 2024, 6, 464–505. [Google Scholar] [CrossRef]
- Suh, C.; Shim, W.; Kim, S.; Roh, J.; Lee, J.-H.; Kim, M.-J.; Park, S.; Jung, W.; Sung, J.; Jahng, G.-H.; et al. Development and validation of a deep learning–based automatic brain segmentation and classification algorithm for alzheimer disease using 3d t1-weighted volumetric images. Am. J. Neuroradiol. 2020, 41, 2227–2234. [Google Scholar] [CrossRef]
- Khojaste-Sarakhsi, M.; Haghighi, S.S.; Ghomi, S.M.F.; Marchiori, E. Deep learning for Alzheimer′s disease diagnosis: A survey. Artif. Intell. Med. 2020, 130, 102332. [Google Scholar] [CrossRef]
- Peng, A.Y.; Sing Koh, Y.; Riddle, P.; Pfahringer, B. Using supervised pretraining to improvegeneralization of neural networks on binary classification problems. In Machine Learning and Knowledge Discovery in Databases, Proceedings of the European Conference, ECML PKDD 2018, Dublin, Ireland, 10–14 September 2018; Springer: Berlin/Heidelberg, Germany, 2019; Part I 18, pp. 410–425. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Gülmez, B. A novel deep neural network model based Xception and genetic algorithm for detection of COVID-19 from X-ray images. Ann. Oper. Res. 2023, 328, 617–641. [Google Scholar] [CrossRef]
- Jonathan, C.; Araujo, D.A.; Saha, S.; Kabir, M.F. CS-Mixer: A Cross-Scale Vision Multi-Layer Perceptron with Spatial–Channel Mixing. IEEE Trans. Artif. Intell. 2024, 5, 4915–4927. [Google Scholar]
- Üzen, H.; Fırat, H. A hybrid approach based on multipath Swin transformer and ConvMixer for white blood cells classification. Health Inf. Sci. Syst. 2024, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Begüm, Ş.; Acici, K.; Sümer, E. Categorization of Alzheimer’s disease stages using deep learning approaches with McNemar’s test. PeerJ Comput. Sci. 2024, 10, e1877. [Google Scholar]
- Gayathri, P.; Geetha, N.; Sridhar, M.; Kuchipudi, R.; Babu, K.S.; Maguluri, L.P.; Bala, B.K. Deep Learning Augmented with SMOTE for Timely Alzheimer’s Disease Detection in MRI Images. Int. J. Adv. Comput. Sci. Appl. 2024, 15. [Google Scholar] [CrossRef]
- AbdulAzeem, Y.; Bahgat, W.M.; Badawy, M. A CNN based framework for classification of Alzheimer’s disease. Neural Comput. Appl. 2021, 33, 10415–10428. [Google Scholar] [CrossRef]
- Shamrat, F.J.M.; Akter, S.; Azam, S.; Karim, A.; Ghosh, P.; Tasnim, Z.; Hasib, K.M.; De Boer, F.; Ahmed, K. Alzheimer net: An effective deep learning based proposition for alzheimer’s disease stages classification from functional brain changes in magnetic resonance images. IEEE Access 2023, 11, 16376–16395. [Google Scholar] [CrossRef]
- Hajamohideen, F.; Shaffi, N.; Mahmud, M.; Subramanian, K.; Al Sariri, A.; Vimbi, V.; Abdesselam, A. Four-way classification of alzheimer’s disease using deep siamese convolutional neural network with triplet-loss function. Brain Inform. 2023, 10, 5. [Google Scholar] [CrossRef]
- Srividhya, L.; Sowmya, V.; Ravi, V.; Gopalakrishnan, E.A.; Soman, K.P. Deep learning-based approach for multi-stage diagnosis of alzheimer’s disease. Multimed. Tools Appl. 2023, 83, 16799–16822. [Google Scholar]
- Dar, G.; Bhagat, A.; Ansarullah, S.I.; Othman, M.T.B.; Hamid, Y.; Alkahtani, H.K.; Ullah, I.; Hamam, H. A novel framework for classification of different alzheimer’s disease stages using cnn model. Electronics 2023, 12, 469. [Google Scholar] [CrossRef]
- Chabib, C.; Hadjileontiadis, L.J.; Al Shehhi, A. Deepcurvmri: Deep convolutional curvelet transform-based mri approach for early detection of alzheimer’s disease. IEEE Access 2023, 11, 44650–44659. [Google Scholar] [CrossRef]
- Murugan, S.; Venkatesan, C.; Sumithra, M.; Gao, X.-Z.; Elakkiya, B.; Akila, M.; Manoharan, S. DEMNET: A Deep Learning Model for Early Diagnosis of Alzheimer Diseases and Dementia from MR Images. IEEE Access 2021, 9, 90319–90329. [Google Scholar] [CrossRef]
- Fareed, M.S.; Zikria, S.; Ahmed, G.; Mui-zzud-din; Mahmood, S.; Aslam, M.; Jilani, S.F.; Moustafa, A.; Asad, M. Add-net: An effective deep learning model for early detection of alzheimer disease in mri scans. IEEE Access 2022, 10, 96930–96951. [Google Scholar] [CrossRef]
- Helaly, H.A.; Badawy, M.; Haikal, A.Y. Deep learning approach for early detection of alzheimer’s disease. Cogn. Comput. 2022, 14, 1711–1727. [Google Scholar] [CrossRef] [PubMed]
- Heising, L.; Angelopoulos, S. Operationalising fairness in medical ai adoption: Detection of early alzheimer’s disease with 2D CNN. BMJ Health Care Inform. 2022, 29, e100485. [Google Scholar] [CrossRef]
- Hazarika, R.A.; Kandar, D.; Maji, A.K. A deep convolutional neural networks based approach for alzheimer’s disease and mild cognitive impairment classification using brain images. IEEE Access 2022, 10, 99066–99076. [Google Scholar] [CrossRef]
- Ashtari-Majlan, M.; Seifi, A.; Dehshibi, M.M. A multi-stream convolutional neural network for classification of progressive mci in alzheimer’s disease using structural mri images. IEEE J. Biomed. Health Inform. 2022, 26, 3918–3926. [Google Scholar] [CrossRef]
- Nawaz, H.; Maqsood, M.; Afzal, S.; Aadil, F.; Mehmood, I.; Rho, S. A deep feature-based real-time system for alzheimer disease stage detection. Multimed. Tools Appl. 2021, 80, 35789–35807. [Google Scholar] [CrossRef]
- Basheera, S.; Ram, M.S.S. Deep learning based alzheimer’s disease early diagnosis using t2w segmented gray matter mri. Int. J. Imaging Syst. Technol. 2021, 31, 1692–1710. [Google Scholar] [CrossRef]
- Papanastasiou, G.; Dikaios, N.; Huang, J.; Wang, C.; Yang, G. Is attention all you need in medical image analysis? A review. IEEE J. Biomed. Health Inform. 2023, 28, 1398–1411. [Google Scholar] [CrossRef]
- Maurício, J.; Domingues, I.; Bernardino, J. Comparing vision transformers and convolutional neural networks for image classification: A literature review. Appl. Sci. 2023, 13, 5521. [Google Scholar] [CrossRef]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis. Intell. Med. 2023, 3, 59–78. [Google Scholar] [CrossRef]
- Alzheimers Disease 5 Class Dataset ADNI. Available online: https://www.kaggle.com/datasets/madhucharan (accessed on 7 July 2023).
- Roy, S.; Maji, P. A simple skull stripping algorithm for brain mri. In Proceedings of the 2015 Eighth International Conference on Advances in Pattern Recognition (ICAPR), Kolkata, India, 4–7 January 2015; IEEE: New York, NY, USA, 2015; pp. 1–6. [Google Scholar]
- Swiebocka-Wiek, J. Skull stripping for mri images using morphological operators. In Computer Information Systems and Industrial Management, Proceedings of the 15th IFIP TC8 International Conference, CISIM 2016, Vilnius, Lithuania, 14–16 September 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 172–182. [Google Scholar]
- Wang, K.; Tian, J.; Zheng, C.; Yang, H.; Ren, J.; Li, C.; Han, Q.; Zhang, Y. Improving risk identification of adverse outcomes in chronic heart failure using SMOTE+ ENN and machine learning. Risk Manag. Healthc. Policy 2021, 14, 2453–2463. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shen, D.; Nie, T.; Kou, Y. A hybrid sampling algorithm combining M-SMOTE and ENN based on random forest for medical imbalanced data. J. Biomed. Inform. 2020, 107, 103465. [Google Scholar] [CrossRef]
- Gu, Q.; Zhu, L.; Cai, Z. Evaluation Measures of the Classification Performance of Imbalanced Data Sets. In Computational Intelligence and Intelligent Systems; ISICA 2009; Communications in Computer and Information Science; Cai, Z., Li, Z., Kang, Z., Liu, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 51. [Google Scholar] [CrossRef]
- Aminu, M.; Ahmad, N.A.; Noor, M.H.M. COVID-19 detection via deep neural network and occlusion sensitivity maps. Alex. Eng. J. 2021, 60, 4829–4855. [Google Scholar] [CrossRef]




| Year | Technique | Dataset | Classification | Model Accuracy % | Imbalance Handling |
|---|---|---|---|---|---|
| 2024 | EfficientNetB0 [11] | ADNI | 2-way | 98.94 | - |
| 2024 | CNN [12] | ADNI | 4-way | 97.5% | Data augmentation |
| 2024 | STCNN [13] | OASIS-kaggle | 4-way | 99.36% | SMOTETomek |
| 2023 | Modified Inception V3 [14] | ADNI | 6-way | 98.67 | Data augmentation |
| 2023 | SCNN [15] | ADNI | 4-way | 91.83 | - |
| OASIS | 93.85 | - | |||
| 2023 | Resnet50v2 [16] | ADNI | 5-way | 91.84 | - |
| 2023 | Mobile Net [17] | Private Dataset | 5-way | 96.6 | - |
| 2023 | DeepCurvelet convolutional [18] | ADNI-kaggle | 98.6 | - | |
| 2022 | Modified CNN-DEMNET [19] | ADNI | 4-way | 84.83 | SMOTE |
| 2022 | Modified CNNADD-NET [20] | Kaggle-OASIS | 4-way | 98.63 | SMOTETomek |
| 2022 | VGG19 [21] | ADNI | 4-way | 97 | - |
| 2022 | LeNet [22] | ADNI | 2-way | 83.7 | - |
| 2022 | DCNN [23] | ADNI | 2-way | 95.2 | - |
| 2022 | MCNN [24] | ADNI | AD and CN | 97.78 | - |
| pMCI and MCI | 79.90 | - | |||
| 2021 | CNN with inception [25] | ADNI | 3-way | 94.9 | - |
| 2021 | Alexnet [24] | ADNI | 4-way | 99.61 | - |
| Class | Dice Coefficient |
|---|---|
| AD | 96.1819 |
| CN | 94.2001 |
| LMCI | 96.8123 |
| EMCI | 95.612 |
| MCI | 93.457 |
| Overall | 95.257 |
| K-Value | Validation_Accuracy | Testing_Accuracy |
|---|---|---|
| 3 | 98.92 | 98.81 |
| 5 | 61.86 | 61.86 |
| 7 | 56.08 | 56.08 |
| Optimizer | Pooling | Validation Accuracy | Testing Accuracy | Validation Loss | Testing Loss | Validation F1Score | Testing F1Score |
|---|---|---|---|---|---|---|---|
| Adam | Max Pooling | 98.32 | 98.68 | 0.1045 | 0.0707 | 98.34 | 98.76 |
| Average Pooling | 98.92 | 98.81 | 0.0863 | 0.0724 | 98.93 | 98.86 | |
| Adagrad | Max Pooling | 59.74 | 65.39 | 0.9454 | 0.8834 | 63.52 | 68.69 |
| Average Pooling | 59.74 | 65.39 | 0.9454 | 0.8834 | 63.52 | 68.69 | |
| Adamax | Max Pooling | 98.32 | 98.21 | 0.1002 | 0.0956 | 98.27 | 98.36 |
| Average Pooling | 97.72 | 98.09 | 0.1018 | 0.0751 | 97.63 | 98.19 | |
| Nadam | Max Pooling | 98.32 | 97.85 | 0.0539 | 0.1211 | 98.43 | 98.03 |
| Average Pooling | 92.01 | 92.62 | 0.3819 | 0.3766 | 91.88 | 92.65 |
| Optimizer | Pooling | Validation Accuracy | Testing Accuracy | Validation Loss | Testing Loss | Validation F1Score | Testing F1Score |
|---|---|---|---|---|---|---|---|
| Adam | Max Pooling | 98.24 | 99.18 | 0.0747 | 0.0168 | 98.22 | 99.18 |
| Average Pooling | 97.84 | 98.44 | 0.1308 | 0.1544 | 98.02 | 98.59 | |
| Adagrad | Max Pooling | 93.99 | 71.60 | 0.7248 | 0.7086 | 75.13 | 74.02 |
| Average Pooling | 70.19 | 71.89 | 0.7373 | 0.7053 | 72.47 | 74.02 | |
| Adamax | Max Pooling | 98.20 | 97.96 | 0.0896 | 0,1047 | 98.25 | 98.10 |
| Average Pooling | 98.80 | 98.57 | 0.0811 | 0.0769 | 98.85 | 98.61 | |
| Nadam | Max Pooling | 97.96 | 98.80 | 0.1282 | 0.0378 | 98.04 | 98.91 |
| Average Pooling | 99.52 | 98.92 | 0.0356 | 0.0514 | 99.54 | 98.92 |
| Optimizer | Pooling | Validation | Testing | Validation | Testing | Validation | Testing |
|---|---|---|---|---|---|---|---|
| Accuracy | Accuracy | Loss | Loss | F1Score | F1Score | ||
| Adam | Max Pooling | 98.80 | 98.91 | 0.1011 | 0.0402 | 98.92 | 98.97 |
| Average Pooling | 98.44 | 97.84 | 0.0998 | 0.1045 | 98.39 | 97.98 | |
| Adagrad | Max pooling | 59.98 | 62.96 | 0.9730 | 0.9572 | 63.47 | 66.04 |
| Average Pooling | 64.90 | 66.94 | 0.8655 | 0.8492 | 69.09 | 70.05 | |
| Adamax | Max pooling | 98.56 | 98.69 | 0.0899 | 0.1029 | 98.55 | 98.71 |
| Average Pooling | 98.32 | 97.96 | 0.1253 | 0.1342 | 98.42 | 97.88 | |
| Nadam | Max pooling | 98.80 | 97.84 | 0.0715 | 0.1591 | 98.84 | 97.81 |
| Average Pooling | 97.38 | 98.33 | 0.1815 | 0.0860 | 97.56 | 98.45 |
| Optimizer | Pooling | Validation Accuracy | Testing Accuracy | Validation Loss | Testing Loss | Validation F1Score | Testing F1Score |
|---|---|---|---|---|---|---|---|
| Adam | Max Pooling | 98.80 | 98.09 | 0.0515 | 0.1581 | 98.77 | 98.09 |
| Average Pooling | 99.16 | 98.45 | 0.0516 | 0.1371 | 99.20 | 98.59 | |
| Adagrad | Max Pooling | 78.25 | 79.67 | 0.6275 | 0.5887 | 80.75 | 81.37 |
| Average Pooling | 70.19 | 72.23 | 0.7936 | 0.7878 | 73.43 | 74.96 | |
| Adamax | Max Pooling | 97.96 | 99.04 | 0.1031 | 0.07004 | 98.02 | 99.00 |
| Average Pooling | 98.20 | 98.44 | 0.0940 | 0.12104 | 98.21 | 98.52 | |
| Nadam | Max Pooling | 98.65 | 98.67 | 0.0715 | 0.04966 | 98.67 | 98.65 |
| Average Pooling | 97.36 | 98.19 | 0.1551 | 0.0719 | 97.42 | 98.16 |
| Parameter | Selection Value | Optimal Value |
|---|---|---|
| Learning rate | (0.01, 0.001, 0.00001) | 0.01 |
| Batch size | (8 or 16) | 8 |
| Epochs | (10–50) | 50 |
| Models | Accuracy % | Precision % | Recall % | F1 Score % | Trainable Parameter | FLOPS (M-Million and B-Billion) | Computation Time(s) |
|---|---|---|---|---|---|---|---|
| Mobile Net [14] | 96.0 | 96.0 | 96.0 | 96.0 | 25,958,917 | - | - |
| Existing model with SMOTETOMEK | 92.82 | 93.49 | 92.72 | 92.84 | 3,548,581 | 717.59 M | 75.28 |
| DenseNet201 | 91.0 | 90.0 | 89.0 | 89.0 | 18,926,389 | 4 B | 419.50 |
| VGG19 | 89.0 | 90.0 | 86.0 | 88.0 | 20,188,229 | 19.6 B | 2056.42 |
| Mobile Net | 88.0 | 87.0 | 84.0 | 85.0 | 3,556,349 | 314 M | 32.96 |
| ResNet152 | 91.0 | 90.0 | 89.0 | 89.0 | 59,026,309 | 11 B | 1153.94 |
| Inception V3 | 90.0 | 85.0 | 87.0 | 86.0 | 22,171,429 | 5.7 B | 597.40 |
| Xception | 85.0 | 87.0 | 82.0 | 84.0 | 21,516,845 | 8.4 B | 880.49 |
| EfficientNetV2S | 90.0 | 89.0 | 87.0 | 88.0 | 20,740,965 | 8.4 B | 888.89 |
| Proposed Model without SMOTEENN | 70.19 | 77.81 | 66.59 | 41.93 | 3,305,221 | 126.4 M | 13.28 |
| Proposed Model with SMOTEENN | 98.87 | 98.80 | 98.60 | 98.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krithika Alias Anbu Devi, M.; Suganthi, K. A Convolutional Mixer-Based Deep Learning Network for Alzheimer’s Disease Classification from Structural Magnetic Resonance Imaging. Diagnostics 2025, 15, 1318. https://doi.org/10.3390/diagnostics15111318
Krithika Alias Anbu Devi M, Suganthi K. A Convolutional Mixer-Based Deep Learning Network for Alzheimer’s Disease Classification from Structural Magnetic Resonance Imaging. Diagnostics. 2025; 15(11):1318. https://doi.org/10.3390/diagnostics15111318
Chicago/Turabian StyleKrithika Alias Anbu Devi, M., and K. Suganthi. 2025. "A Convolutional Mixer-Based Deep Learning Network for Alzheimer’s Disease Classification from Structural Magnetic Resonance Imaging" Diagnostics 15, no. 11: 1318. https://doi.org/10.3390/diagnostics15111318
APA StyleKrithika Alias Anbu Devi, M., & Suganthi, K. (2025). A Convolutional Mixer-Based Deep Learning Network for Alzheimer’s Disease Classification from Structural Magnetic Resonance Imaging. Diagnostics, 15(11), 1318. https://doi.org/10.3390/diagnostics15111318







